Abstract
Glucosinolates are natural plant products that function in the defense toward herbivores and pathogens. Plant defense is regulated by multiple signal transduction pathways in which salicylic acid (SA), jasmonic acid, and ethylene function as signaling molecules. Glucosinolate content was analyzed in Arabidopsis wild-type plants in response to single or combinatorial treatments with methyljasmonate (MeJA), 2,6-dichloro-isonicotinic acid, ethylene, and 2,4-dichloro-phenoxyacetic acid, or by wounding. In addition, several signal transduction mutants and the SA-depleted transgenic NahG line were analyzed. In parallel, expression of glucosinolate biosynthetic genes of the CYP79 gene family and the UDPG:thiohydroximate glucosyltransferase was monitored. After MeJA treatment, the amount of indole glucosinolates increased 3- to 4-fold, and the corresponding Trp-metabolizing genes CYP79B2 and CYP79B3 were both highly induced. Specifically, the indole glucosinolate N-methoxy-indol-3-ylmethylglucosinolate accumulated 10-fold in response to MeJA treatment, whereas 4-methoxy-indol-3-ylmethylglucosinolate accumulated 1.5-fold in response to 2,6-dichloro-isonicotinic acid. In general, few changes were seen for the levels of aliphatic glucosinolates, although increases in the levels of 8-methylthiooctyl glucosinolate and 8-methylsulfinyloctyl glucosinolate were observed, particularly after MeJA treatments. The findings were supported by the composition of glucosinolates in the coronatine-insensitive mutant coi1, the ctr1 mutant displaying constitutive triple response, and the SA-overproducing mpk4 and cpr1 mutants. The present data indicate that different indole glucosinolate methoxylating enzymes are induced by the jasmonate and the SA signal transduction pathways, whereas the aliphatic glucosinolates appear to be primarily genetically and not environmentally controlled. Thus, different defense pathways activate subsets of biosynthetic enzymes, leading to the accumulation of specific glucosinolates.
Glucosinolates are amino acid-derived natural plant products that function in the defense against herbivores and microorganisms. Upon tissue disruption, e.g. caused by insect feeding, glucosinolates are hydrolyzed by specific thioglucosidases called myrosinases to produce an array of biologically active compounds, typically isothiocyanates, nitriles, and thiocyanates (for review, see Halkier, 1999; Rask et al., 2000). These compounds function as inhibitors of microbial growth (Mari et al., 1993; Manici et al., 1997), as attractants for specialist insects, and as deterrents of generalist herbivores. For humans, glucosinolates are important as flavor compounds, as cancer-preventive agents, and as biopesticides in agriculture.
Glucosinolate biosynthesis is considered a three-step process: First, the amino acid may enter the chain elongation pathway, in which the condensing enzymes MAM1 and MAM-L have recently been identified (de Quiros et al., 2000; Kroymann et al., 2001). Second, the core glucosinolate structure is formed (see below); and third, secondary modifications of the core structure can take place, which include e.g. methoxylation of the indole structure of Trp-derived glucosinolates and oxidation of the Met-derived side chains. 2-Oxoglutarate-dependent dioxygenases catalyzing conversion of methylsulfinylalkyl glucosinolates to alkenyl or hydroxyalkenyl glucosinolates were recently identified (Kliebenstein et al., 2001). The first committed step in biosynthesis of the core structure of glucosinolates is the conversion of amino acids to the corresponding aldoximes. This step is catalyzed by cytochromes P450 from the CYP79 family, which in Arabidopsis includes seven members. CYP79B2 and CYP79B3 metabolize Trp (Hull et al., 2000; Mikkelsen et al., 2000), CYP79F1 and CYP79F2 metabolize chain-elongated Met derivatives with respectively 1-6 or 5-6 additional methylene groups in the side chain (Hansen et al., 2001b; Reintanz et al., 2001; S. Chen, E. Glawischnig, and B.A. Halkier, unpublished data), CYP79A2 metabolizes Phe (Wittstock and Halkier, 2000), and CYP79C1 and CYP79C2 have not yet been assigned a function. Because CYP79s catalyze a key step in the biosynthesis of glucosinolates, alteration of the expression of these genes often has dramatic effects on the profile of glucosinolates (for review, see Mikkelsen et al., 2002). The aldoxime-metabolizing enzymes are cytochromes P450 belonging to the CYP83 family (Bak et al., 2001; Hansen et al., 2001a). CYP83A1 metabolizes the aliphatic aldoximes, whereas CYP83B1 metabolizes the indole and aromatic aldoximes (Bak and Feyereisen, 2001; P. Naur and B.A. Halkier, unpublished data). In the remaining part of the core pathway, a candidate Arabidopsis UDP-Glc:S-thiohydroximic acid glucosyl transferase (S-GT) has been identified (Petersen et al., 2001) based on homology to a putative Brassica sp. S-GT (Marillia et al., 2001).
Several signaling molecules have been identified in plant defense responses. These include jasmonic acid (JA), salicylic acid (SA), and ethylene, which have been shown to operate independently and/or synergistically in different signal transduction pathways (for review, see Thomma et al., 2001). Jasmonates affect a wide range of processes in plants, including root growth, fruit ripening, senescence, pollen development, tuber formation, and defense against insects and pathogens (for review, see Reymond and Farmer, 1998; Xie et al., 1998). SA has mainly been associated with resistance to bacterial pathogens (Pieterse and van Loon, 1999; Feys and Parker, 2000), but evidence that SA is involved in responses to insects is emerging (Moran and Thompson, 2001). 2,6-Dichloro-isonicotinic acid (INA) is a functional homolog of SA and induces the same subset of genes (Uknes et al., 1992). Ethylene is a plant hormone involved in developmental processes such as e.g. hypocotyl and root elongation, but also in defense responses (Johnson and Ecker, 1998; Chang and Shockey, 1999). Many defense response genes, including PDF1.2, PR5, and basic chitinase, are activated synergistically by JA and ethylene (Xu et al., 1994; Penninckx et al., 1998; Norman-Setterblad et al., 2000). However, JA-dependent activation of vegetative storage protein (VSP) expression is suppressed by ethylene as evidenced by increased VSP expression in ethylene-insensitive mutants (Rojo et al., 1999; Norman-Setterblad et al., 2000). JA-dependent defense against certain insects is similarly suppressed by ethylene (Stotz et al., 2000). Auxins have been shown to antagonize JA-dependent signal transduction (Rojo et al., 1998) and formation of Trp-derived secondary metabolites (Goddijn et al., 1992).
Several mutants in the plant defense signal transduction pathways have been identified (Glazebrook, 2001). In the JA insensitive coi1 mutant, JA is synthesized at normal levels (Feys et al., 1994), but the induction of genes normally induced by JA such as PDF1.2 and THI2.1 is suppressed (Penninckx et al., 1996, 1998; Xie et al., 1998; Pieterse and van Loon, 1999). The ctr1 (constitutive triple response) mutant exerts a constitutive triple response, which includes a constitutive ethylene response, inhibition of hypocotyl and root elongation, and constitutive expression of PDF1.2. The mutants cpr1 (constitutive expression of PR proteins) and mpk4 (map kinase 4), accumulate increased levels of SA, exhibit constitutive expression of PR genes, and have enhanced resistance to certain virulent strains of Pseudomonas spp. (Bowling et al., 1994; Petersen et al., 2000). Furthermore, mpk4 is insensitive to JA (Petersen et al., 2000). Introduction of the bacterial salicylate hydroxylase encoding NahG gene under control of the cauliflower mosaic virus 35S promoter into Arabidopsis has generated a SA-deficient transgenic line unable to accumulate SA (Delaney et al., 1994).
Plant species belonging to the Brassicaceae family produce aliphatic, aromatic, and indole glucosinolates. In general, glucosinolates are believed to play a role in plant defense and are classified both as phytoanticipines and as phytoalexins. This poses the question of whether specific glucosinolates are constitutively present and others inducible, and in the latter case, which signaling pathways are activated. The available mutants are valuable tools for elucidating the roles of different signal transduction pathways in the induction of defense compounds such as e.g. glucosinolates.
In white mustard (Sinapis alba), methyljasmonate (MeJA) has been shown to increase the incorporation rate of 14C-Tyr into p-hydroxybenzylglucosinolate, (Du et al., 1995). White mustard and oilseed rape (Brassica napus) either treated with MeJA or wounded have been shown to accumulate increased levels of indol-3-ylmethyl glucosinolate (i-3ym; Bodnaryk, 1992; Bodnaryk, 1994). In other studies, oilseed rape and Arabidopsis treated with MeJA were shown to accumulate both i-3ym and N-methoxyindol-3-ylmethyl glucosinolate (Nmi-3ym; Doughty et al., 1995; Brader et al., 2001). In leaves of oilseed rape, SA was shown to increase the overall level of glucosinolates, of which the major Phe-derived 2-phenylethyl glucosinolate showed the highest accumulation (Kiddle et al., 1994).
In the present study, we have determined the glucosinolate composition and content in response to treatment with MeJA, ethylene, and INA in Arabidopsis wild-type plants and in several signaling mutants to elucidate the potential role of different defense pathways in induction or suppression of specific glucosinolates and biosynthetic genes.
RESULTS
Reverse Transcriptase (RT)-PCR Analysis of Elicitor-Treated Wild-Type Arabidopsis
Expression levels of CYP79 genes were monitored by semiquantitative RT-PCR because northern analysis was hampered by low abundance of the transcripts and possible cross-hybridization between closely related gene family members of the CYP79 family. For each primer set, the optimal number of cycles was determined based on 20 to 30 cycles for Actin1, PDF1.2, and PR1, and 32 to 38 cycles for the individual CYP79s (for details, see “Materials and Methods”). RT-PCR was performed on RNA isolated from Arabidopsis rosette leaves treated with MeJA, INA, 1-aminocyclopropane-1-carboxylate (ACC, an ethylene precursor), MeJA in combination with ACC, and mechanical wounding. The plants were not visibly affected by any of the treatments.
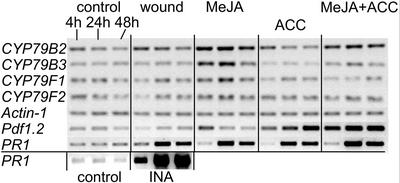
PDF1.2 was induced transiently up to 1.5 ± 0.2-fold by MeJA, 4.8 ± 0.6-fold by ACC, and 6.1 ± 0.5-fold by the combination of ACC and MeJA (Fig. 1). The induction by ACC and MeJA was both stronger and faster than for any of single hormone treatments. PR1 was used as positive control for the INA treatment (Uknes et al., 1992). PR1 was induced 16.8 ± 0.9-fold by INA treatment, and 3.3 ± 0.6-fold and 3.6 ± 0.2-fold by ACC and MeJA treatments, respectively. PR1 is generally referred to as a SA inducible gene, although it was previously shown to be induced by a combination of ethylene and MeJA (Xu et al., 1994), as was also observed in the present study. Lack of ethylene- and MeJA-dependent induction of PR1 in many studies might be attributable to limited sensitivity of northern analysis.
Figure 1.
Expression of CYP79 genes in Arabidopsis leaves after treatment with different signaling molecules. Semiquantitative RT-PCR analysis was performed on leaves of 6-week-old Arabidopsis plants 4, 24, and 48 h after treatment with INA, MeJA, ACC, MeJA, and ACC or after wounding. Actin1 was used as control of equal RNA loading and RT-reaction efficiency. PDF1.2 was used as a positive control for ACC and MeJA treatments, and PR1 was used as a positive control for INA treatment. The experiments were performed twice on three independent sets of plants.
CYP79B2 expression was induced up to 1.5 ± 0.2-fold by wounding, 2.8 ± 0.4-fold by MeJA in combination with ACC, and 4.6 ± 0.6-fold after MeJA treatment alone (Fig. 1). The induction after wounding peaked at 4 h. After both MeJA treatments, a strong induction of CYP79B2 was evident after 4 h and peaked at 24 h, after which the expression level remained constant. ACC appeared to suppress the induction of CYP79B2 caused by MeJA treatment because the combination of MeJA and ACC gave rise to a smaller induction than MeJA treatment alone. A slight decrease in expression of CYP79B2 during the course of the experiments was seen in control plants, which suggests a developmentally determined down-regulation.
CYP79B3 was induced 3.5 ± 0.4-fold by MeJA and up to 1.7 ± 0.4 by MeJA in combination with ACC, and the induction levels were lower than those observed for CYP79B2 (Fig. 1). MeJA treatment gave the highest induction, and this induction was suppressed when added in combination with ACC. The inductions after both MeJA treatments peaked at 24 h. Interestingly, the basal expression level of CYP79B3 was suppressed to 0.6 ± 0.1-fold that of control levels by ACC treatment.
The level of CYP79F1 mRNA was induced 2.0 ± 0.4 by MeJA. The induction peaked at 24 h. The combined treatment of MeJA and ACC suppressed the induction of CYP79F1 expression after MeJA treatment to control levels (1.1 ± 0.2). CYP79F2 was induced 1.7 ± 0.1-fold by the MeJA treatment and 1.4 ± 0.2-fold after the combined MeJA and ACC treatment. Expression of CYP79A2 and CYP79C1 was undetectable after 45 cycles with primer sets that readily amplify the expected fragments from gDNA (data not shown). No change in the expression pattern of any of the CYP79s after INA treatment was detected (data not shown).
Glucosinolate Analysis of Arabidopsis after Treatment with Signaling Molecules
Glucosinolates were extracted from freeze-dried rosette leaves subjected to the various treatments. HPLC analysis of the most dominant glucosinolates in rosette leaves of Arabidopsis included 3-methylsulfinylpropyl- (3-msp), 3-hydroxypropyl- (3-OHp), 4-methylthiobutyl- (4-mtb), 4-methylsulfinylbutyl- (4-msb), 4-benzoyloxybutyl- (4-bzb), 4-hydroxybutyl-, 5-methylsulfinylpentyl- (5-msp), 7-methylsulfinylheptyl- (7-msh), 7-methylthioheptyl- (7-mth), 8-methylthiooctyl- (8-mto), 8-methylsulfinyloctyl- (8-mso), i-3ym, Nmi-3ym, and 4-methoxyindol-3-ylmethyl- (4mi-3ym) glucosinolate, which were identified and quantified as described previously (Petersen et al., 2001, 2002).
For short-chain Met-derived glucosinolates (C3-C6 side chains), no significant increase was seen for 3-msp, 4-mtb, or 4-msb (data not shown). 5-msp was increased 2-fold from 0.27 ± 0.1 to 0.53 ± 0.1 nmol mg dry weight−1 after MeJA treatment. 5-msp was the only short-chain Met-derived glucosinolate that was affected significantly by the treatments (data not shown).
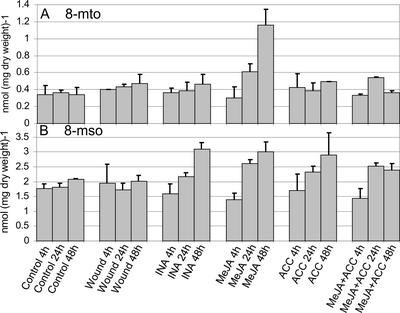
For long-chain Met-derived glucosinolates (C7 and C8 side chains), no significant increase was seen for 7-msh after any of the treatments (data not shown). After MeJA treatment, the concentration of 8-mto increased approximately 3-fold compared with control plants (Fig. 2A). This increase was apparent after 24 h and at its highest at 48 h. 8-mto was efficiently oxidized into 8-mso, that was present in concentrations of approximately 5-fold that of 8-mto (Fig. 2B). 8-mso was present at higher concentrations after INA treatment, MeJA treatment, and ACC treatment and after the combined treatment of MeJA and ACC. A slight 8-mso accumulation was observed after INA, MeJA, and ACC treatments. After the combined treatment with MeJA and ACC, the concentration of 8-mso was constant at 24 and 48 h and reached a concentration that was slightly below that seen for the other three treatments. The increase in 8-mso in response to several of the treatments without a simultaneous increase in 8-mto suggests that the enzyme activities responsible for synthesizing 8-mto and oxidizing 8-mto into 8-mso are induced by the same treatments. One exception, however, was observed following MeJA treatment, after which the 8-mto concentration increased approximately 4-fold from 0.3 ± 0.1 to 1.2 ± 0.2 nmol mg−1 dry weight. This shows that under these conditions, 8-mto accumulates faster than the oxidizing enzyme is able to convert 8-mto to 8-mso.
Figure 2.
Analysis of long-chain aliphatic glucosinolates in Arabidopsis leaves after treatment with signaling molecules or wounding. Six-week-old plants were treated with MeJA, ACC, INA, MeJA, and ACC or subjected to wounding. Glucosinolates were extracted from rosette leaves harvested at 4, 24, and 48 h after treatment and analyzed by HPLC. The analyses were done in triplicates on pools of eight to 12 plants grown independently. The error bars shown denote sd.
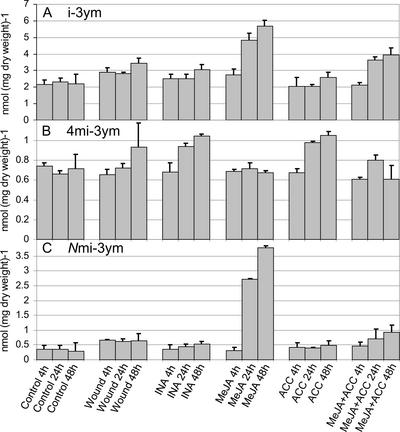
After MeJA treatment, the concentration of i-3ym in rosette leaves increased more than 2-fold compared with that of control plants (Fig. 3A). Upon treatment with both MeJA and ACC, the concentration of i-3ym increased to approximately two times that of control plants. In both cases, the accumulation was evident after 24 h, at which point the concentration reached approximately three-quarters of the level seen at 48 h. A slight increase in the i-3ym concentration was seen after wounding. i-3ym may be methoxylated at the 4′ and N′ position of the indole ring structure leading to the formation of 4mi-3ym and Nmi-3ym, respectively. The 4mi-3ym concentration was shown to increase moderately after INA treatment and after ACC treatment (Fig. 3B). Furthermore, the increased accumulation of 4 mi-3ym in response to ACC treatment was absent when MeJA was added in concert. This indicates that the enzyme activities responsible for methoxylation of i-3ym to 4mi-3ym are induced by INA and ACC and suppressed by MeJA. Nmi-3ym was shown to accumulate dramatically after MeJA treatment (Fig. 3C). The concentration of Nmi-3ym increased more than 10-fold from 0.3 ± 0.1 to 3.8 ± 0.1 nmol mg−1. At 24 h, the Nmi-3ym concentration reached three-quarters of that at 48 h. Interestingly, the Nmi-3ym concentration was only increased approximately 2-fold upon a combined MeJA and ACC treatment. This indicates that the enzyme activities responsible for the N-methoxylation of i-3ym are strongly induced by MeJA and that this induction is suppressed by ACC.
Figure 3.
Analysis of indole glucosinolates in Arabidopsis leaves after treatment with signaling molecules or wounding. Six-week-old Arabidopsis plants were treated with MeJA, ACC, INA, MeJA, and ACC or wounded. For details, see Figure 2 legend.
The total levels of indole and aliphatic glucosinolates were determined from the concentrations of the individual glucosinolates depicted in Figures 2 and 3. After MeJA treatment, indole glucosinolate concentration increased 3-fold from 3.3 ± 0.2 to 10.2 ± 0.1 nmol mg−1. Even though long-chain aliphatic glucosinolates increased from 2.4 ± 0.2 to 4.5 ± 0.2 nmol mg−1, the percentage of indole glucosinolates of the total glucosinolate content doubled from 18% ± 2% to approximately 34% ± 3%. The combined treatment of MeJA and ACC resulted in an increase in the total concentration of indole glucosinolates from 3.3 ± 0.2 to 5.5 ± 0.8 nmol mg−1. The concentration of long-chain aliphatic glucosinolates increased from 2.4 ± 0.2 to 3.3 ± 0.4 nmol mg−1.
Effects of Auxin on CYP79s and Glucosinolates
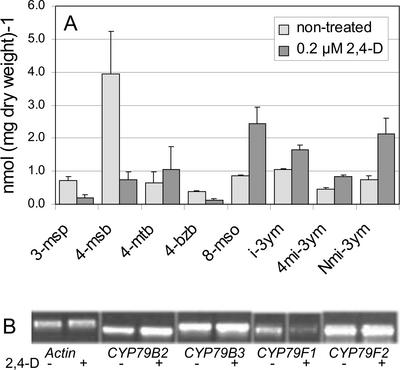
The effect of auxin treatment on the glucosinolate profile was analyzed in Arabidopsis seedlings grown on medium with or without 2,4-dichloro-phenoxyacetic acid (2,4-D; Fig. 4A). In untreated plants, the total concentration of short-chain aliphatic glucosinolates was 5.7 ± 1.1 nmol mg−1. This value was reduced significantly when seedlings where grown on 0.2 or 1 μm 2,4-D (2.1 ± 0.9 nmol mg−1 and 1.6 ± 1.0 nmol mg−1, respectively). The most dramatic decrease was observed for 4-msb, which was reduced 5-fold from 4.0 ± 1.3 to 0.7 ± 0.02 nmol mg−1 in seedlings grown on 0.2 μm 2,4-D. In contrast, the concentrations of long-chain aliphatic and indole glucosinolates were increased from 0.9 ± 0.04 and 2.2 ± 0.1 nmol mg−1 to 2.4 ± 0.5 and 4.6 ± 0.5 nmol mg−1, respectively.
Figure 4.
Effect of 2,4-D treatment on glucosinolate composition and CYP79 gene expression in 15-d-old seedlings. A, Concentration of the major glucosinolates in seedlings grown on Murashige and Skoog medium (light gray) or Murashige and Skoog medium supplemented with 0.2 μm 2,4-D (dark gray). The experiments were done in triplicates. Error bars denote sd. B, Relative gene expression of Actin1, CYP79B2, CYP79B3, CYP79F1, and CYP79F2, as determined by RT-PCR.
In parallel, the expression of CYP79B2, CYP79B3, CYP79F1, and CYP79F2 was monitored by RT-PCR (Fig. 4B). After treatment with 0.2 μm 2,4-D, expression of CYP79B2 and CYP79B3 was induced 1.6 ± 0.2- and 1.3 ± 0.2-fold, respectively, whereas expression of CYP79F1 was reduced to 0.4 ± 0.1-fold of the level found in control plants. This indicates that auxin-induced changes of the glucosinolate profile are mainly mediated on the level of CYP79 transcription.
Signal Transduction Mutants
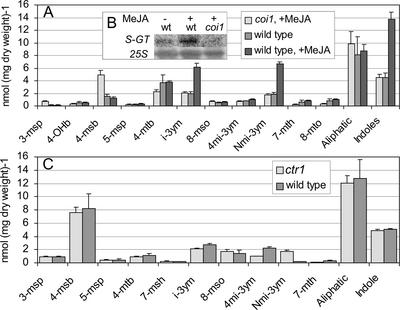
The possible role of different signaling pathways in induction or suppression of glucosinolate accumulation was studied using available well-characterized mutants and transgenic lines affecting the signaling of JA (coi1), ethylene (ein2 and ctr1), and SA (mpk4, cpr1, and NahG). As expected, the glucosinolate profile of the JA-insensitive coi1 mutant treated with MeJA (Fig. 5A) was similar to that of untreated coi1 and to that of untreated wild type (data not shown). Interestingly, coi1 accumulated 3-fold more 3-msp and 4-msb than wild type, whereas the level of 4-mtb was reduced by almost 50%. These glucosinolates were unaffected by all of the treatments examined in wild-type plants. This difference may reflect activation or suppression of pathways in coi1, which are not affected by the different hormonal treatments. In agreement with the induction of the indole glucosinolates in wild type treated with MeJA, the putative S-GT, which is believed to be specific for the glucosinolate biosynthetic pathway, was induced in response to the MeJA treatment (Fig. 5B). In coi1 treated with MeJA, S-GT mRNA was expressed at a level comparable with that of untreated wild-type plants.
Figure 5.
Analysis of glucosinolate profile in seedlings of the coi1 and ctr1 mutants. A, Concentration of major glucosinolates in 3-week-old plants of coi1 and wild type grown on Murashige and Skoog medium in the absence or presence of 50 μm MeJA. B, Northern analysis of S-GT expression in wild type and coi1 after MeJA treatment. C, Major glucosinolates in 3-week-old plants of ctr1 and wild type. The glucosinolates were analyzed by HPLC and the experiments were done in triplicates. Error bars denote sd.
The glucosinolate profile of the ethylene response mutant ctr1 was similar to wild type with respect to aliphatic glucosinolates (Fig. 5C). However, whereas the level of 4mi-3ym was reduced by 50% and the level of i-3ym was slightly decreased, the level of Nmi-3ym was significantly increased. The response differed significantly from that of ACC-treated wild-type plants and indicates that the response of ctr1 is more complex than a simple constitutive up-regulation of the ethylene signal transduction pathway.
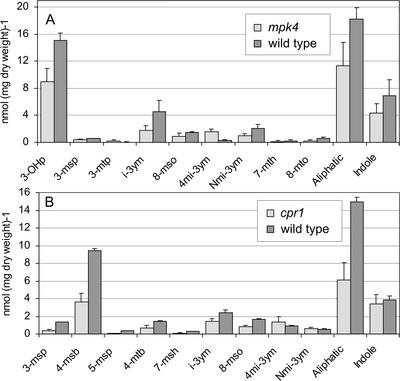
In the SA-overproducing mpk4 and cpr1 mutants, the total amount of glucosinolates was reduced approximately 50% compared with wild type (Fig. 6, A and B). For cpr1, this was primarily because of a reduction of aliphatic glucosinolates, whereas in mpk4, the level of both indole- and aliphatic glucosinolates was reduced. With the exception of 3-mtp in mpk4, the level of individual aliphatic glucosinolates in both mutants was approximately one-half that of wild type. In mpk4 and to a lesser extend in cpr1, 4mi-3ym accumulated to significantly higher levels than in wild type. The increase was not seen in mpk4NahG plants, which further confirms that induction of 4mi-3ym is SA dependent (Fig. 7). This is in agreement with the result that methoxylation at the 4′ position was induced by INA, which indicates that 4mi-3ym accumulated in mpk4 and cpr1 because of their elevated levels of endogenous SA. In cpr1, the concentration of i-3ym was slightly below that of wild type (Fig. 6B). Treatment of mpk4 with MeJA did not result in increases of i-3ym or Nmi-3ym, which supports the JA insensitivity of mpk4 (Fig. 7). In contrast to the mpk4 mutant, MeJA treatment of cpr1 resulted in elevated levels of i-3ym and Nmi-3ym. The increase of both i-3ym and Nmi-3ym was approximately 2-fold compared with uninduced levels, whereas in the wild-type i-3ym and Nmi-3ym were induced approximately 3- and 7-fold, respectively. This indicates that the increased SA level in the mutant suppresses MeJA induction.
Figure 6.
Analysis of glucosinolate profile in mpk4 and cpr1 mutants and wild type. Glucosinolates were extracted from 3-week-old plants and analyzed by HPLC. A, mpk4, ecotype Ler. B, cpr1, ecotype Col-0. The experiments were done in triplicates. Error bars denote sd.
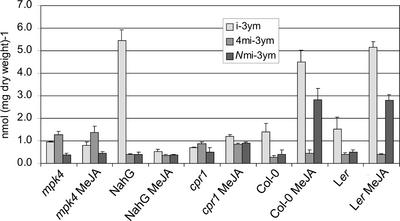
Figure 7.
The content of indole glucosinolates in signaling mutants after treatment with MeJA. Three-week-old mpk4, NahG, cpr1, mpk4NahG plants, and Col-0 were sprayed with 250 μm MeJA and harvested after 24 h. Glucosinolates were extracted and analyzed by HPLC. The experiments were done in triplicates. Error bars denote sd.
In NahG plants, the level of i-3ym was increased dramatically, whereas the other MeJA inducible indole glucosinolate, Nmi-3ym, was not. A similar increase of i-3ym was not observed in mpk4NahG plants.
DISCUSSION
Hormonal Treatments of Wild-Type Arabidopsis
In the present paper, we have studied the expression of major CYP79 genes and the accumulation of the corresponding glucosinolates in response to various hormonal treatments of Arabidopsis wild-type plants and mutants in signaling pathways. In general, the induction of CYP79s correlated with accumulation of the corresponding glucosinolates. CYP79B2 and CYP79B3 were highly induced by MeJA, and the concentration of indole glucosinolates increased in response to the treatment. The combination of MeJA and ACC induced CYP79B2 and to a lesser extent CYP79B3. This was reflected in the accumulation of a correspondingly smaller amount of indole glucosinolates. Accumulation of CYP79B2 and CYP79B3 mRNA was induced already after 4 h, whereas the accumulation of the corresponding glucosinolates was evident after 24 h showing an expected delay as gene expression precedes metabolite accumulation. In a recent study, northern analysis of Arabidopsis treated with a culture filtrate of Erwinia carotovora or MeJA showed strong induction of CYP79B3, whereas CYP79B2 was only weakly induced under these conditions (Brader et al., 2001). This observation may reflect the lower expression level of CYP79B2 combined with detection limitations of northern analysis.
In the present study, expression of CYP79B3 was induced by MeJA and suppressed by ACC. CYP79B3 is the first gene conclusively shown to be regulated in this way. The reduced induction of CYP79B3 after the combined MeJA and ACC treatment is likely attributable to suppression by ethylene. VSP shows similar regulation as CYP79B3 after combined MeJA and ACC treatments. It is, however, unclear if VSP is down-regulated by ethylene because the limited sensitivity of the northern analysis did not allow detection of VSP during uninduced conditions. The induction patterns of VSP and CYP79B3 differ in that CYP79B3 is not induced by wounding whereas VSP is heavily wound induced. The nature of the CYP79B3 expression pattern indicates that at least two signaling pathways interact to mediate CYP79B3 expression. CYP79B3 may accordingly function as a marker for genes that are induced by MeJA and suppressed by ethylene. CYP79B2 has been reported to be induced by treatment with Pseudomonas syringae pv maculicola (Hull et al., 2000) and wounding (Mikkelsen et al., 2000), as evidenced by northern analysis. The reported expression data may, however, not be CYP79B2 specific. In the present study, RT-PCR shows that both CYP79B genes are inducible and that CYP79B2 undergoes higher levels of induction than CYP79B3. The data suggest that both CYP79B2 and CYP79B3 are responsible for maintaining a basal level of indole glucosinolates and that both are induced following pathogen attack to increase accumulation of indole glucosinolates.
CYP79F1 and CYP79F2, which are involved in biosynthesis of aliphatic glucosinolates, were induced by MeJA. The concentration of long-chain aliphatic glucosinolates increased after MeJA treatment. This could be part of the response to make the plant more resistant to pathogens as the toxicity of the glucosinolate breakdown products in general increases with increasing side-chain length (Borek et al., 1998). MeJA has not previously been identified as an inducer of long-chain aliphatic glucosinolates. Interestingly, both 8-mto and 8-mso accumulated after MeJA treatment, whereas the only short-chain aliphatic glucosinolate to accumulate was 5-msp. The increase in 8-mto and 8-mso was approximately 10-fold higher than that of 5-msp. After MeJA treatment, CYP79F1 was induced more heavily than CYP79F2. It was expected that CYP79F2 and not CYP79F1 was induced by the treatment because CYP79F2 specifically metabolizes long-chain Met-derived amino acids, whereas CYP79F1 metabolizes all chain-elongated Met-derived amino acids. The data suggest that under induced conditions, the products of the chain elongation pathway and not the CYP79s determine the profile of aliphatic glucosinolates. If separate chain elongation pathways exist for short- and long-chain Met derivatives (Kroymann et al., 2001), the machinery for long-chain Met derivatives would thus be induced by MeJA. Further studies are needed to clarify this issue.
In our study, MeJA was shown to be the most potent elicitor of glucosinolate biosynthesis. Treatment of oilseed rape and Arabidopsis with MeJA has previously been shown to induce i-3ym and Nmi-3ym biosynthesis (Bodnaryk, 1992, 1994; Doughty et al., 1995; Brader et al., 2001). Our data supported these findings and showed that specifically Nmi-3ym biosynthesis was induced strongly in response to MeJA treatment and that this induction was suppressed by the addition of ACC. 4mi-3ym did not accumulate after this treatment but was induced by INA and ACC, and the ACC induction was suppressed by the addition of MeJA. This indicates that the enzyme activities responsible for converting i-3ym to respectively 4mi-3ym and Nmi-3ym are induced by different signaling pathways. Furthermore, this suggests that these compounds function in different defense pathways and that these pathways produce specific effector molecules.
Many processes in plants are regulated by auxin, and auxin biosynthesis has been proposed to be interconnected with the biosynthesis of indole glucosinolates (Hull et al., 2000; Mikkelsen et al., 2000; Bak et al., 2001). In addition, knockout mutants in the glucosinolate biosynthetic genes CYP83B1 and CYP79F1 showed “high” auxin phenotypes (Barlier et al., 2000; Bak et al., 2001; Reintanz et al., 2001; Tantikanjana et al., 2001). Seedlings grown on Murashige and Skoog medium supplemented with 2,4-D showed a dramatic reduction in the level of short-chain aliphatic glucosinolates and of gene expression of CYP79F1. In contrast, the level of indole glucosinolates and the expression of the CYP79B genes were induced. This observation was consistent with the finding that a CYP79F1 knockout mutant, had both higher levels of IAA and increased levels of indole glucosinolates (Reintanz et al., 2001). Because growth on 0.2 μm 2,4-D affected the morphology of the seedling by increasing the formation of roots, it is not clear to which extent auxin directly affects glucosinolate biosynthesis. The decreased level of short-chain aliphatic glucosinolates and the elevated level of Nmi-3ym could partly reflect the enhanced root formation. As an alternative, auxin could be an inducer of CYP79B2 expression and therefore of indole glucosinolate biosynthesis. The hypothesis is supported by the presence of a putative auxin responsive element (GAGACA, reverse orientation) located just 51 bp upstream of the start codon of CYP79B2. No auxin responsive elements were found in the promoter of CYP79B3.
Signal Transduction Mutants
Signal transduction mutants were analyzed for their glucosinolate content and the modulation of this in response to treatment with various hormones. The presence of a basal level of glucosinolates in coi1 shows that MeJA is not required for biosynthesis. MeJA induces glucosinolate biosynthesis by increasing the transcription of biosynthetic genes such as the CYP79 genes and S-GT (Figs. 1 and 5). These results were confirmed by lack of induction of S-GT (Fig. 5) and CYP79B3 (Brader et al., 2001) in the coi1 mutant.
In ctr1, the level of Nmi-3ym was increased compared with wild-type plants. This could be attributable to suppression of the N-methoxylating enzymes in wild-type plants with normal levels of ethylene. This is supported by the finding that ethylene perception negatively affects JA-induced gene expression in Arabidopsis, because higher concentrations of JA were needed to induce e.g. VSP in wild-type plants than in the ethylene-insensitive ein2, ein3, and etr1 mutants (Rojo et al., 1999).
In the SA-overproducing mpk4 and cpr1, the level of i-3ym and Nmi-3ym was reduced, whereas the level of 4mi-3ym was increased. This is in agreement with results from wild-type plants showing that 4mi-3ym was induced by INA. In support of a SA-mediated accumulation of 4mi-3ym, the level of 4mi-3ym was reduced in mpk4NahG plants. In a recent study, Brader et al. (2001) found that the level of 4mi-3ym was decreased slightly in NahG plants. However, they did not report increased accumulation of 4mi-3ym upon SA treatment. This may be because of a weaker induction caused by SA compared with INA due to metabolism of SA as shown previously (Uknes et al., 1992). NahG plants have higher PDF1.2 background than wild-type plants (Petersen et al., 2000). A basal level of SA could be required to inhibit JA production, in which case the JA level would be higher in NahG plants than in wild type. This suggests that absence of SA results in an induced JA pathway. However, the level of Nmi-3ym was not increased in NahG plants, which suggests that a repression of the JA pathway by SA is downstream of JA itself and does therefore only induce a subset of genes. SA could alternatively repress part of both the JA and the ethylene pathway. In accordance with data obtained from induction of wild type with MeJA and ACC, this would result in increased levels of i-3ym and unaltered levels of Nmi-3ym as was seen for NahG plants. This is supported by several reports including Norman-Setterblad et al. (2000) who showed that NahG plants treated with a culture filtrate of E. carotovora induced VSP to higher levels than similarly treated wild-type plants. Other reports have demonstrated a similar SA-dependent inhibition of the JA pathway (Peña-Cortés et al., 1993; Doares et al., 1995). i-3ym and Nmi-3ym biosynthesis was induced by MeJA in cpr1, which also has elevated levels of SA. The approximately 50% reduction of total glucosinolates in both mpk4 and cpr1, combined with induction of i-3ym and Nmi-3ym by MeJA in cpr1 indicates that elevated endogenous SA level suppresses overall glucosinolate biosynthesis and antagonizes JA-dependent induction of indole glucosinolates, although it does not abolish the JA response. The mpk4 mutant has previously been suggested to be insensitive to JA (Petersen et al., 2000). Our data support this, because induction of indole glucosinolates by JA is abolished in this mutant.
CONCLUSIONS
Glucosinolates constitute final products in the signaling cascades. There is basically no information available about the regulators that activate the glucosinolate biosynthetic genes. In a recent paper, Chen et al. (2002) took a genomics approach and monitored by microarray analysis the regulation of one-third of the transcription factors in Arabidopsis at 81 different experimental conditions, including treatment with SA, JA, and ethylene, in both wild-type and signaling mutants. The study identified a number of transcription factors that are induced under particular stress conditions, which might be candidate genes for activation of the glucosinolate pathway. Future research will focus on the regulation of glucosinolate biosynthesis in response to signaling molecules.
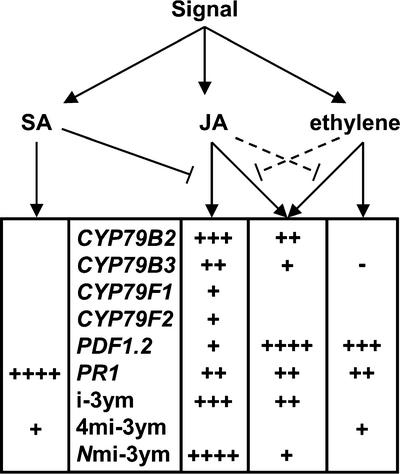
The present data show that different signal transduction pathways activate specific biosynthetic and secondary modifying enzymes, leading to altered levels of specific glucosinolates (Fig. 8). The induction of glucosinolates by several defense pathways strongly indicates that glucosinolates play a role in plant defense, although knowledge on the biological function of the individual glucosinolates in interaction between plant-herbivores or plant-microorganisms is limited.
Figure 8.
Signal transduction pathways and their effects on glucosinolate biosynthetic genes and products. The upper part of the figure represents an overview of the plant defense signal transduction pathways and their (synergistic and antagonistic) crosstalks. The lower table summarizes the major effects on glucosinolate profiles and the expression of glucosinolate biosynthetic genes in Arabidopsis treated with SA, MeJA, and ethylene. The level of indole glucosinolates and expression of CYP79B genes is highly affected, whereas expression of CYP79F genes is barely affected. PDF1.2 and PR1 are included as reference genes.
In recent years, major breakthroughs have been made with respect to identification of glucosinolate biosynthetic genes and glucosinolate degradation enzymes (Wittstock and Halkier, 2002). This has provided the tools necessary for generation of plants with specific glucosinolate profiles for study of the role of glucosinolates in plant-insect and plant-microorganism interactions. A basic understanding of the biological role of specific glucosinolates will enable the design of crop plants with improved pest resistance. The recent advances have made this goal a realistic possibility.
MATERIALS AND METHODS
Growth and Treatments of Arabidopsis
Arabidopsis ecotypes Columbia (Col-0) and Landsberg erecta (Ler) were grown in soil in a controlled-environment growth chamber (AR-60 I, Percival, Boone, IA) at a photosynthetic flux of 100 to 120 μmol photons m−2 s−1 at 12 h light, 20°C, and 70% relative humidity.
When rosettes were between 3 and 4 cm in diameter, the plants were sprayed with 250 μm MeJA, 12.5 mm INA, 2.5 mm ACC, combinations hereof (all in 0.25% [v/v] ethanol), 0.25% (v/v) ethanol (control), or mechanically wounded with a pair of ribbed forceps. Each tray of plants contained 30 to 40 plants, which were sprayed with approximately 10 mL of the appropriate solutions. Plants were harvested 4, 24, and 48 h after treatments. For glucosinolate analysis, the plants were lyophilized, and for RNA extractions, the plants were harvested directly into liquid nitrogen. In three independent experiments, eight to 12 plants were used for each measurement.
For analysis of the effect of auxin on the glucosinolate profile and the expression of CYP79 genes, Arabidopsis Col-0 was grown for 15 d at a 12-h light period on Murashige and Skoog medium or Murashige and Skoog supplemented with 0.2 or 1 μm 2,4-D. Approximately 50 seedlings, four- to eight-leaf stage, were pooled and subjected to glucosinolate analysis (triplicate) and RT-PCR (n = 6).
Seeds of cpr1-1 (Col-0; Dr. Xinnian Dong, Duke University, Durham, NC), ctr1-1 (Col-0; CS8057), mpk4-1 (Ler; CS5205), the NahG transgenic line (Col-0, Dr. John Ryals, Paradigm Genetics, Research Triangle Park, NC), mpk4NahG (segregating with respect to the parental Ler and Col-0 background; Petersen et al., 2000), and Col-0 and Ler wild types were sown in peat-based soil (Enhets K-jord, Weibulls, Sweden) supplemented with osmocote, vermiculate, and perlite. The plants were cultivated in 21°C at a 24-h light period at 100 to 120 μmol photons m−2 s−1 and analyzed in the seedling stage. MeJA was applied as described above and glucosinolates were extracted after 24 h. The coi1-1 (Col-0; Professor John G. Turner, University of East Anglia, Norwich, UK) mutant is male sterile, and therefore the glucosinolate study was made on seedlings that were selected on Murashige and Skoog plates with or without 50 μm MeJA. Furthermore, seeds of coi1 were sown on soil and grown as described for wild-type Col-0, and the glucosinolate profile of individual plants was determined. These plants did not exhibit significant changes in the glucosinolate profiles.
RT-PCR
RNA was extracted from Arabidopsis rosette leaves using the TRIzol reagent (Invitrogen, Carlsbad, CA). Five micrograms of total RNA was heated to 65°C for 3 min. cDNA was synthesized from the 5 μg of RNA in RT-buffer (New England Biolabs, Beverly, MA) containing 0.5 mm dNTPs, 200 ng of random hexamers (Amersham Biosciences AB, Uppsala), 10 units of RNAsin (Promega, Madison, WI), and 25 units of Moloney murine leukemia virus reverse transcriptase (New England Biolabs) in a total volume of 20 μL. The mixture was incubated at 37°C for 1 h and heated to 95°C for 5 min.
PCR reactions were done in a total volume of 50 μL in PCR buffer (Invitrogen) containing 200 μm dNTPs, 1.5 mm MgCl2, 50 pmol of the forward and reverse primers, and 2.5 units of Taq DNA polymerase (Invitrogen). The PCR programs were as follows: 2 min at 94°C, 23 cycles (Actin1; U39449), 27 cycles (PR1; AT2G14580), 28 cycles (PDF1.2; AT5G44420), 32 cycles (CYP79B3 [At2g22330] and CYP79F1 [At1g16410]), or 33 cycles (CYP79B2 [At4g39950] and CYP79F2 [At1g16400]) of: 94°C for 10 s, 57°C (Actin1) or 53°C (PR1, PDF1.2) or 54°C (all other primers) for 15 s and 72°C for 45 s. The following primers were used (all listed from 5′ to 3′): Actin1 forward, TGGAACTGGAATGGTTAAGGCTGG; Actin1 reverse, TCTCCAGAGTCGAGCACAATACCG; PDF1.2 forward, TCATGGCTAAGTTTGCTTCC; PDF1.2 reverse, AATACACACGATTTAGCACC; PR1 forward, GCCCACAAGATTATCTAAGGG; PR1 reverse, ACCTCCTGCATATGATGCTCCT; CYP79B2 forward, AACCCACCATTAAGGAGC; CYP79B2 reverse, TCATAAAATATATACGGCGTCG; CYP79B3 forward, AAACCAACCATTAAGGAACT; CYP79B3 reverse, TCCTCGCCGTACGTCACCG; CYP79F1 forward, TTTTTAGACACCATCTTGTTTTCTTCTTC; CYP79F1 reverse, AAAGCTCAATGGGTAGAAT; CYP79F2 forward, AAAGCTCAATGCGTCGAAT; and CYP79F2 reverse, GCGTCGAAACACATCACAGAG. Most primer sets were designed to be intron spanning, and with these primers, no PCR products from genomic DNA were detected. All primers successfully amplified a band of the correct size when cDNA clones were used as template. A 10× loading buffer was added to the PCR reactions, and 10 μL was analyzed by gel electrophoresis on a 1% (w/v) agarose gel. Bands were visualized by ethidium bromide staining and quantified on a Gel Doc 2000 Transilluminator (Bio-Rad, Hercules, CA). PCR with Actin1-specific primers was used to ensure that an equal amount of RNA was used for all samples and to ensure that RT reactions were equally effective. PDF1.2- and PR1-specific primers were used as positive controls for MeJA- and SA-dependent induction, respectively. RT-PCR was performed twice on each of three sets of plants.
Northern Analysis
Total RNA was extracted from 0.5 g fresh weight of 3-week-old rosette leaves by use of the Trizol Reagent (Invitrogen). Twenty-five micrograms of RNA was separated on 0.8% (v/v) formaldehyde agarose gels, blotted onto a Hybond-N (Amersham Biosciences AB) filter, and cross-linked at 0.6 J cm−2. Approximately 50 ng of the first exon of the S-GT (EMBL accession no. AC002396, Arabidopsis I BAC F316) and the 25S rRNA (EMBL accession no. X52320) gene, respectively, were labeled with [α-32P]dCTP using the Megaprime labeling kit (Amersham Biosciences AB). High-stringency washes were performed at 68°C with 0.1× SSPE (1× SSPE: 0.18 m NaCl, 10 mm sodium phosphate, pH 7.7, and 1 mm EDTA) and 0.1% (w/v) SDS in the final wash. The filter was first probed with the S-GT probe and stripped according to Sambrook et al. (1989) and re-probed with the 25S rRNA probe. Bands were visualized and quantified on a STORM 840 PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Glucosinolate Analysis by HPLC
Glucosinolates were extracted from approximately 20 mg of slightly homogenized freeze-dried rosette leaves by boiling 4 min in 4 mL of 70% (v/v) methanol. The supernatant was collected, and the plant material was washed with 2 mL of 70% (v/v) methanol. The extracts were combined and applied to 200-μL DEAE Sephadex CL-6B (Pharmacia AB, Uppsala) columns (Bio-Rad-Polyprep) equilibrated with 1 mL of 0.02 m KOAc, pH 5.0, and washed with 1 mL of water. The columns were washed with 2 mL of 70% (v/v) methanol, 2 mL of water, and 0.02 m KOAc, pH 5. After the addition of 100 μL of 2.5 mg mL−1 Helix pomatia sulfatase (Sigma-Aldrich, St. Louis), the columns were sealed and left overnight. The resulting desulfoglucosinolates were eluted with 2 × 1 mL of water. The eluate was lyophilized until dryness and resuspended in 200 μL of water. Aliquots of 100 μL were applied to a Spectachrom HPLC system (Shimadzu, Kyoto) equipped with a Supelco supelcosil LC-ABZ 59142 RP-amid C-16 (25 cm × 4.6 mm, 5 mm; Supelco; Holm and Halby, Brøndby, Denmark) and an SPD-M10AVP photodiode array detector (Shimadzu). The flow rate was 1 mL min−1. Desulphoglucosinolates were eluted with water for 2 min followed by a linear gradient from 0% to 60% (v/v) methanol in water (48 min), a linear gradient from 60% to 100% (v/v) methanol in water (3 min), and with 100% methanol (14 min). Detection was performed at 229 and 260 nm using a photodiode array. Desulphoglucosinolates were quantified based on response factors (Buchner, 1987; Haughn et al., 1991) and internal benzylglucosinolate- (Merck, Darmstadt, Germany) or p-hydroxybenzylglucosinolate (Bioraf, Åkirkeby, Denmark) standards as previously described (Petersen et al., 2001, 2002). The standard was added at the beginning of the extraction procedure and analyses were done in triplicates.
ACKNOWLEDGMENTS
Unknown reviewers are acknowledged for critical comments to the manuscript. Dr. Morten Petersen is thanked for fruitful discussions and Christina Mattson for excellent technical assistance.
Footnotes
This work was supported by a European Molecular Biology Organization long-term fellowship (for E.G.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.011015.
LITERATURE CITED
- Bak S, Feyereisen R. The involvement of two cytochrome P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 2001;127:108–118. doi: 10.1104/pp.127.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R. CYP83B1, a cytochrome P450 at the metabolic branchpoint in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell. 2001;13:101–111. doi: 10.1105/tpc.13.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci USA. 2000;19:14819–14824. doi: 10.1073/pnas.260502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnaryk RP. Effects of wounding on glucosinolates in the cotyledons of oilseed rape and mustard. Phytochemistry. 1992;31:2671–2677. [Google Scholar]
- Bodnaryk RP. Potent effect of jasmonates on indole glucosinolates in oilseed rape and mustard. Phytochemistry. 1994;35:301–305. [Google Scholar]
- Borek V, Elberson LR, McCaffrey JP, Morra JM. Toxicity of isothiocyanates produced by glucosinolates in Brassicaceae species to black vine weevil eggs. J Agric Food Chem. 1998;46:5318–5323. [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner R. Glucosinolates in rapeseeds: analytical aspects. In: Wathelet JP, editor. World Crops: Production, Utilization, Description. Dordrecht, The Netherlands: Martinus Nijhoff Publishers, Klüwer Academic Publishers Group; 1987. pp. 50–58. [Google Scholar]
- Brader G, Tas É, Palva ET. Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiol. 2001;126:849–860. doi: 10.1104/pp.126.2.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Shockey JA. The ethylene-response pathway: signal perception to gene regulation. Curr Opin Plant Biol. 1999;5:352–358. doi: 10.1016/s1369-5266(99)00004-7. [DOI] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang H-S, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA et al. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Quiros HC, Magrath R, McCallum D, Kroymann J, Schnabelrauch D, Mitchell-Olds T, Mithen R. α-Keto acid elongation and glucosinolate biosynthesis in Arabidopsis thaliana. Theor Appl Genet. 2000;101:429–437. [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Doares SH, Narváez-Vásquez J, Conconi A, Ryan CA. Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol. 1995;108:1741–1746. doi: 10.1104/pp.108.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doughty KJ, Kiddle GA, Pye BJ, Wallsgrove RM, Pickett JA. Selective induction of glucosinolates in oilseed rape leaves with methyl jasmonate. Phytochemistry. 1995;38:347–350. [Google Scholar]
- Du L, Lykkesfelt J, Olsen CE, Halkier BA. Involvement of cytochrome P450 in oxime production in glucosinolate biosynthesis as demonstrated by an in vitro microsomal enzyme system isolated from jasmonic acid-induced seedlings of Sinapis alba L. Proc Natl Acad Sci USA. 1995;92:12505–12509. doi: 10.1073/pnas.92.26.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Benedetti CE, Penfold CN, Turner JG. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to bacterial pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Parker JE. Interplay of signalling pathways in plant disease resistance. Trends Genet. 2000;16:449–455. doi: 10.1016/s0168-9525(00)02107-7. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr Opin Plant Biol. 2001;4:301–308. doi: 10.1016/s1369-5266(00)00177-1. [DOI] [PubMed] [Google Scholar]
- Goddijn OJ, de Kam RJ, Zanetti A, Schilperoort RA, Hoge JH. Auxin rapidly down-regulates transcription of the tryptophan decarboxylase gene from Catharanthus roseus. Plant Mol Biol. 1992;18:1113–1120. doi: 10.1007/BF00047714. [DOI] [PubMed] [Google Scholar]
- Halkier BA. Glucosinolates. In: Ikan R, editor. Naturally Occurring Glycosides. New York: John Wiley & Sons; 1999. pp. 193–223. [Google Scholar]
- Hansen CH, Du L, Naur P, Olsen CE, Axelsen KB, Hick AJ, Pickett JA, Halkier BA. CYP83B1 is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. J Biol Chem. 2001a;276:24790–24796. doi: 10.1074/jbc.M102637200. [DOI] [PubMed] [Google Scholar]
- Hansen CH, Wittstock U, Olsen CE, Hick AJ, Pickett JA, Halkier BA. Cytochrome P450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J Biol Chem. 2001b;276:11078–11085. doi: 10.1074/jbc.M010123200. [DOI] [PubMed] [Google Scholar]
- Haughn GW, Davin L, Giblin M, Underhill EW. Biochemical genetics for plant secondary metabolites in Arabidopsis thaliana. Plant Physiol. 1991;97:217–226. doi: 10.1104/pp.97.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull AK, Rekha V, Celenza JL. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PR, Ecker JR. The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet. 1998;32:227–254. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- Kiddle GA, Doughty KJ, Wallsgrove RM. Salicylic acid-induced accumulation of glucosinolates in oilseed rape (Brassica napus) leaves. J Exp Bot. 1994;45:1343–1346. [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T. Gene duplication in the diversification of secondary metabolism: Tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell. 2001;13:681–693. doi: 10.1105/tpc.13.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T. A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol. 2001;127:1077–1088. [PMC free article] [PubMed] [Google Scholar]
- Manici LM, Lazzeri L, Palmieri S. In vitro fungitoxic activity of some glucosinolates and their enzyme-derived products toward plant pathogenic fungi. J Agric Food Chem. 1997;45:2768–2773. [Google Scholar]
- Mari M, Iori R, Leoni O, Marchi A. In vitro activity of glucosinolate-derived isothiocyanates against post harvest fruit pathogens. Ann Appl Biol. 1993;123:155–164. [Google Scholar]
- Marillia E-F, MacPherson JM, Tsang EWT, Audenhove KV, Keller WA, Groot Wassink JWD. Molecular cloning of a Brassica napus thiohydroximate S-glucosyltransferase and its expression in Escherichia coli. Physiol Plantarum. 2001;113:176–184. doi: 10.1034/j.1399-3054.2001.1130204.x. [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem. 2000;275:33712–33717. doi: 10.1074/jbc.M001667200. [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Petersen BL, Olsen CE, Halkier BA. Biosynthesis and metabolic engineering of glucosinolates. Amino Acids. 2002;22:279–295. doi: 10.1007/s007260200014. [DOI] [PubMed] [Google Scholar]
- Moran PJ, Thompson GA. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001;125:1074–1085. doi: 10.1104/pp.125.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman-Setterblad C, Vidal S, Palva ET. Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol Plant-Microb Interact. 2000;13:430–438. doi: 10.1094/MPMI.2000.13.4.430. [DOI] [PubMed] [Google Scholar]
- Peña-Cortés H, Albrecht T, Prat S, Weiler EW, Willmitzer L. Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta. 1993;191:123–128. [Google Scholar]
- Penninckx IAMA, Eggermont K, Terras FRG, Thomma BPHJ, De Samblax GW, Buchala A, Metraux J-P, Manners JM, Broekeart WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchala A, Metraux J-P, Broekeart WF. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;10:2103–2113. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen BL, Andreasson E, Bak S, Agerbirk N, Halkier BA. Characterization of transgenic Arabidopsis thaliana with metabolically engineered high levels of p-hydroxybenzylglucosinolate. Planta. 2001;212:612–618. doi: 10.1007/s004250000429. [DOI] [PubMed] [Google Scholar]
- Petersen BL, Chen S, Hansen CH, Olsen CE, Halkier BA. Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta. 2002;214:562–571. doi: 10.1007/s004250100659. [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE et al. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, van Loon LC. Salicylic acid-dependent plant defense pathways. Trends Plant Sci. 1999;4:52–58. doi: 10.1016/s1360-1385(98)01364-8. [DOI] [PubMed] [Google Scholar]
- Rask L, Andreasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J. Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol. 2000;42:93–113. [PubMed] [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalczyk M, Sandberg G, Godde M, Uhl R, Palme K. bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell. 2001;13:351–367. doi: 10.1105/tpc.13.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Farmer EE. Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol. 1998;1:404–411. doi: 10.1016/s1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- Rojo E, Leon J, Sanchez-Serrano JJ. Cross-talk between wound signaling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J. 1999;20:135–142. doi: 10.1046/j.1365-313x.1999.00570.x. [DOI] [PubMed] [Google Scholar]
- Rojo E, Titarenko E, Leon J, Berger S, Vancanneyt G, Sanchez-Serrano JJ. Reversible protein phosphorylation regulates jasmonic acid-dependent and -independent wound signal transduction pathways in Arabidopsis thaliana. Plant J. 1998;13:153–165. doi: 10.1046/j.1365-313x.1998.00020.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Stotz HU, Pittendrigh BR, Kroymann J, Weniger K, Fritsche J, Bauke A, Mitchell-Olds T. Induced plant defense responses against chewing insects: Ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm but not diamondback moth. Plant Physiol. 2000;124:1007–1017. doi: 10.1104/pp.124.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantikanjana T, Yong JW, Letham DS, Griffith M, Hussain M, Ljung K, Sandberg G, Sundaresan V. Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev. 2001;15:1577–1588. doi: 10.1101/gad.887301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Penninckx IAMA, Broekaert WF, Cammue BPA. The complexity of disease signaling in Arabidopsis. Curr Opin Immunol. 2001;13:63–68. doi: 10.1016/s0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Acquired resistance in Arabidopsis. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA. Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. catalyzes the conversion of l-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolate. J Biol Chem. 2000;275:14659–14666. doi: 10.1074/jbc.275.19.14659. [DOI] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA. Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 2002;7:263–270. doi: 10.1016/s1360-1385(02)02273-2. [DOI] [PubMed] [Google Scholar]
- Xie D-X, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Xu Y, Chang P-FL, Liu D, Narasimhan ML, Raghothama KG, Hasegawa PM, Bressan RA. Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell. 1994;6:1077–1085. doi: 10.1105/tpc.6.8.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]