Abstract
Peroxiredoxins (prxs) are peroxidases with broad substrate specificity. The seven prx genes expressed in Arabidopsis shoots were analyzed for their expressional response to changing photon fluence rates, oxidative stress, and ascorbate application. The results reveal a highly variable and gene-specific response to reducing and oxidizing conditions. The steady-state transcript amounts of the chloroplast-targeted prxs, namely the two-cysteine (2-Cys) prxs, prx Q and prx II E, decreased upon application of ascorbate. prx Q also responded to peroxides and diamide treatment. prx II B was induced by tertiary butylhydroperoxide, but rather unaffected by ascorbate. The strongest responses were observed for prx II C, which was induced with all treatments. The two Arabidopsis 2-Cys Prxs and four Prx II proteins were expressed heterologously in Escherichia coli. In an in vitro test system, they all showed peroxidase activity, but could be distinguished by their ability to accept dithiothreitol and thioredoxin as electron donor in the regeneration reaction. The midpoint redox potentials (Em′) of Prx II B, Prx II C, and Prx II E were around −290 mV and, thus, less negative than Em′ of Prx II F, 2-Cys Prx A, and 2-Cys Prx B (−307 to −322 mV). The data characterize expression and function of the mitochondrial Prx II F and the chloroplast Prx II E for the first time, to our knowledge. Antibodies directed against 2-Cys Prx and Prx II C showed a slight up-regulation of Prx II protein in strong light and of 2-Cys Prx upon transfer both to high and low light. The results are discussed in context with the subcellular localization of the Prx gene products.
Peroxiredoxins (Prxs) are enzymes that reduce hydrogen peroxide (H2O2) and alkyl hydroperoxides. They are grouped in four classes: (a) 2-Cys Prx; (b) Prx Q; (c) Prx II, which all contain two catalytic Cys residues in distinct sequence environment; and (d) 1-Cys Prx with one conserved Cys residue only (Dietz, 2003). A phylogenetic distance analysis suggests that 2-Cys Prx, Prx Q, and 1-Cys Prx are related proteins, whereas the group of Prx II is likely to have evolved independently (Verdoucq et al., 1999; Horling et al., 2002). The catalytic Cys residues undergo oxidation during the peroxide reduction reaction and need to be reduced by electron donors such as glutaredoxins, thioredoxins, or cyclophilins before the next catalytic cycle (Lee et al., 2001; Rouhier et al., 2001; König et al., 2002). For the bacterial and animal homologs, a broad substrate specificity has been described (Nogoceke et al., 1997; Bryk et al., 2000; Hillas et al., 2000). In in vitro tests, these Prx proteins reduced H2O2, lipid peroxides, such as butyl hydroperoxide, phospholipid peroxides and cumene hydroperoxide, and peroxynitrite. For plant Prxs, the catalytic properties have only poorly been investigated.
The Arabidopsis genome encodes 10 open reading frames (ORFs) for peroxiredoxins. Based on sequence similarities, they can be assigned to the four subgroups of peroxiredoxins: two ORFs code for 2-Cys Prx, one for 1-Cys Prx, one for Prx Q, and six ORFs for Prx II (Dietz et al., 2002; Horling et al., 2002). Expression activity has not been observed for Prx II A and D (Horling et al., 2002), indicating that the two ORFs might be pseudogenes. In Arabidopsis, four Prxs are predicted to be targeted into chloroplasts and one into mitochondria (Horling et al., 2002). The remaining two Prx proteins contain no apparent targeting address and their localization is unknown.
Reactive oxygen species (ROS) and peroxides play a dual role in metabolism. On the one hand, they are highly toxic and must be kept under tight control (Noctor and Foyer, 1998). On the other hand, ROS and peroxides serve as substrates in metabolism and as signals for regulation (Foyer and Noctor, 2000). A complicated multifactorial antioxidant network composed of low-molecular mass antioxidants and enzymes, such as catalase, ascorbate peroxidase (APX), and glutathione peroxidase, decompose ROS and lipid peroxides, and quench radicals. Peroxiredoxins are part of the antioxidant defense. They decompose ROS and lipid peroxides and tune ROS and peroxide levels in signaling events. For 2-Cys Prx, activities below wild-type levels have been shown to disturb early shoot development of Arabidopsis seedlings and photosynthesis (Baier and Dietz, 1999). In animal cells, stimulated or reduced expression of prx genes altered redox-dependent signal transduction; for example, via the nuclear transcription factor NF-κB or in the p53-mediated activation of apoptosis (Jin et al., 1997; Kang et al., 1998; Zhou et al., 2000), suggesting a general role of Prxs not only in detoxification of ROS, but also in balancing signaling cascades involving ROS.
The identification of the prx gene family in the Arabidopsis genome, with its likely function in antioxidant defense and redox regulation prompted us to investigate prx gene expression in context of cellular redox and light conditions. It will be shown that the light-, ascorbate-, and oxidative stress-dependent regulations of gene expression exhibit distinct patterns for the various prx genes that appear to be related to their proposed subcellular location.
RESULTS
Expression of Arabidopsis prx Genes under Changing Light Conditions
Seven of 10 predicted prx genes are expressed in Arabidopsis leaves (Horling et al., 2002). Table I summarizes the key characteristics of these members of the Prx family in Arabidopsis. The first set of experiments was conducted to analyze the expressional behavior of the two 2-Cys prxs, four prx II genes (prx II B, C, E, and F), and prx Q in response to redox and light changes. The 1-Cys prx was excluded from the analysis because it has been shown to be expressed in the embryo and aleuron layer of barley (Hordeum vulgare) caryopses and Arabidopsis seeds (Stacy et al., 1996; Haslekas et al., 1998), thus, in seeds only.
Table I.
Characteristics and activities of Prxs that are expressed in Arabidopsis leaves
| prx | MATDB Entry | Targeting | No. of Amino Acid Residues | Molecular Mass | pI | Activity with DTT | Activity with Trx |
|---|---|---|---|---|---|---|---|
| kD | mol H2O2 mol Prx min−1 | ||||||
| 2-Cys Prx A | At3g11630 | Chloroplasts | 266 | 29.1 | 4.91 | 1 ± 1 | 6.5 ± 0.5 |
| 2-Cys Prx B | At5g06290 | Chloroplasts | 271 | 29.6 | 4.71 | 6 ± 1 | 7.8 ± 0.6 |
| Prx II B | At1g65980 | Cytosol | 162 | 17.4 | 5.17 | 150 ± 12 | – |
| Prx II C | At1g65970 | Cytosol | 162 | 17.4 | 5.33 | 156 ± 5 | 0.8 ± 0.3 |
| Prx II E | At3g52960 | Chloroplasts | 234 | 24.7 | 5.03 | 57 ± 5 | – |
| Prx II F | At3g06050 | Mitochondria | 199 | 21.2 | 6.29 | 70 ± 5 | – |
| prx Q | At3g26060 | Chloroplasts | 215 | 23.6 | 5.53 | – | – |
The H2O2 reducing activities of heterologously expressed 2-Cys Prx A and B, and Prx II B, C, E and F were determined in a non-physiological activity assay using dithiothreitol (DTT) as electron donor for regeneration of reduced Prx protein. A decrease in H2O2 concentration was measured after formation of colored Fe(III)isothiocyanate at 480 nm. In an enzymatic, Trx-dependent activity assay, NADPH was used as electron donor and the decrease in absorbance at 340 nm was monitored (mean ± se of n ≥ 7 determinations). MATDB, MIPS Arabidopsis thaliana database.
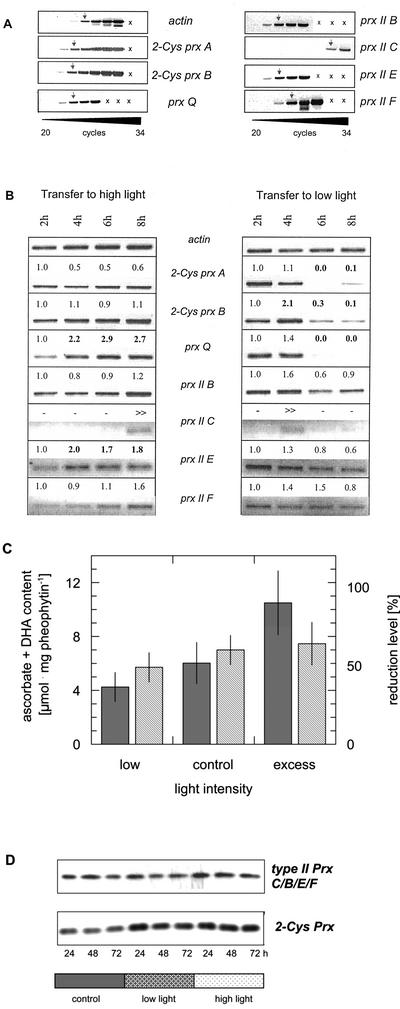
The comparability of the amplification conditions in the PCR reaction was tested with 100 ng of PCR product as template. For all prx cDNAs, similar amounts of products were detected after 12 and 14 cycles (data not shown). As a consequence, similar cycle numbers in the comparative reverse transcription (RT)-PCR analysis (Fig. 1A) indicated similar transcript amounts for 2-Cys prx, prx Q, and prx II B and E and a slightly lower mRNA level for prx II F. The transcript amounts for prx II C were considerably lower. In the PCR reaction, six to eight cycles more were needed to reach similar product amounts (Fig. 1A).
Figure 1.
Semiquantitative RT-PCR with gene-specific primers for 2-Cys prx A, 2-Cys prx B, prx Q, prx II B, prx II C, prx II E, and prx II F. A, Cycle optimization. The arrows indicate the cycle number resulting in a similarly intense amplification product. B, Effect of transfer from adequate light conditions (120 μmol quanta m−2 s−1) to high (1,000 μmol quanta m−2 s−1) and low photon fluence rates (10 μmol quanta m−2 s−1), respectively. The numbers give the normalized results of a semiquantitative band density analysis as related to actin. All experiments were performed three to five times with replicates and showed similar changes in transcript levels in each case. Major changes are indicated by bold letters. C, Ascorbate plus dehydroascorbate levels of leaves in dependence of low, normal, and excessive photon fluence rates as used in A and B. The data are means of n = 8 determinations ± sd (dark bars). The redox state of the ascorbate system is shown with striped bars. D, Western blots of plants maintained at adequate light or transferred to high and low light for 24, 48, or 72 h, respectively, using an antibody against 2-Cys Prx of barley and Arabidopsis Prx II C.
Four Prx proteins (2-Cys Pxr A and B and Prx II E and Prx Q) were predicted by TargetP to be targeted into plastids and have been shown to be expressed in leaves and not in roots (Horling et al., 2002). The expression in green tissues prompted us to investigate the possible relationship between prx gene expression and photosynthesis. The steady-state transcript amounts were compared for the various prx genes after a sudden transfer from adequate photon fluence rate of 120 μmol quanta m−2 s−1 to high light (10-fold excess), or low photon fluence rate (10-fold decrease), respectively (Fig. 1B). The steady-state mRNA amounts of the chloroplast 2-Cys prx A and B and the cytosolic prx II B were little affected by transfer to high light. The transcript of prx Q responded with an increased amount as well as that of prx II E (Fig. 1B). The transcript levels of prx II F were little changed. Prx II C mRNA showed a transient increase after increasing the light intensity with some peak variation. In the result presented, the maximum level was observed after 8 h, and in others after 6 h.
In all four experiments conducted, a transient increase of prx II C transcripts was also observed when the plants were transferred to lower light intensities. The steady-state transcript amounts of prx II B and F were unchanged, when the light intensity was decreased. The transcript amount of all other prx genes decreased and in part were barely detectable 8 h after lowering the light intensity. Ascorbate and dehydroascorbate levels were determined in leaves exposed for 8 h to normal, low, and high photon fluence rates similar to the conditions employed for the expressional analysis (Fig. 1C). Ascorbate plus dehydroascorbate levels were positively correlated with light intensity, with a 2.5-fold increase from low to excess light. The redox level of the ascorbate pool was not significantly different between the treatments, although the trend was observed that the relative amount of ascorbate increased with the light intensity.
To test how the individually controlled RNA dynamics is reflected in the overall protein level of Prx II and 2-Cys Prx, antibodies directed against Arabidopsis Prx II C and barley 2-Cys Prx were used to characterize protein levels in leaf extracts after transfer to low or high photon fluence rates (Fig. 1D). The anti-Prx II C antibody recognized Prx II B, C, E, and F, and anti-2-Cys Prx antibody recognized both 2-Cys Prx A and B, respectively, because of the high degree of amino acid-sequence identity. Western-blot analysis showed that Prx II- and 2-Cys-Prx levels increased after transfer to high light. 2-Cys Prx protein amounts were also increased after transition to low light. The Prx II C/B/E/F protein amount was unchanged in low light.
Expression of prx Genes under Oxidative Stress
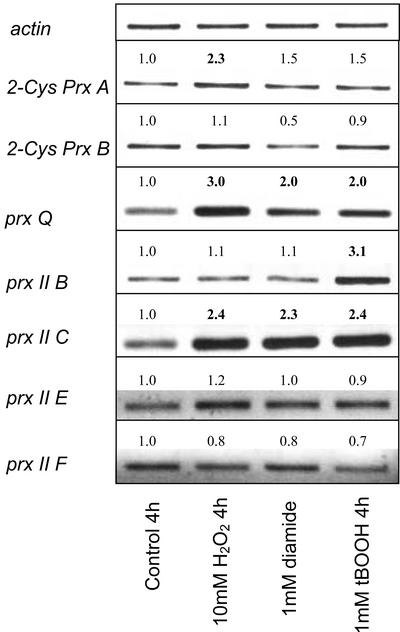
To test whether the steady-state transcript amounts respond to oxidants and pro-oxidants, H2O2, tertiary butyl hydroperoxide, and diamide were used. The effectors were applied to leaf slices by incubation in effector solution after infiltration to ensure fast and homogenous application (Fig. 2). H2O2 and tertiary butyl hydroperoxide are compounds with directly oxidizing properties. Diamide acts indirectly as oxidative stressor by depletion of the cellular thiol pool. Transcript levels of 2-Cys prx A and B and prx II E and F were almost unaffected by the treatments. The steady-state mRNA levels of the 2-Cys prx A only slightly increased after peroxide treatment. Under the same conditions, prx Q and prx II C were induced by all three treatments. prx II C showed a general strong increase in the transcript amount, whereas for prx Q, the increase was stronger with H2O2 than with diamide and tertiary butylhydroperoxide. prx II B was specifically induced by tertiary butylhydroperoxide treatment.
Figure 2.
Effect of oxidative stressors on prx gene expression in leaf slices of Arabidopsis. Leaf slices were incubated in the presence of H2O2 (10 mm), diamide (1 mm), and butyl hydroperoxide (1 mm) as mediators of oxidative stress for 4 h before RNA extraction and prx gene-specific semiquantitative RT-PCR. The figure shows a data set representative for two to four independent experiments, each with replicates. The presented and described trends were seen in each experiment. The numbers indicate the factor of change obtained by a semiquantitative band density analysis as related to the control condition and actin.
Ascorbate Effects on prx Gene Expression
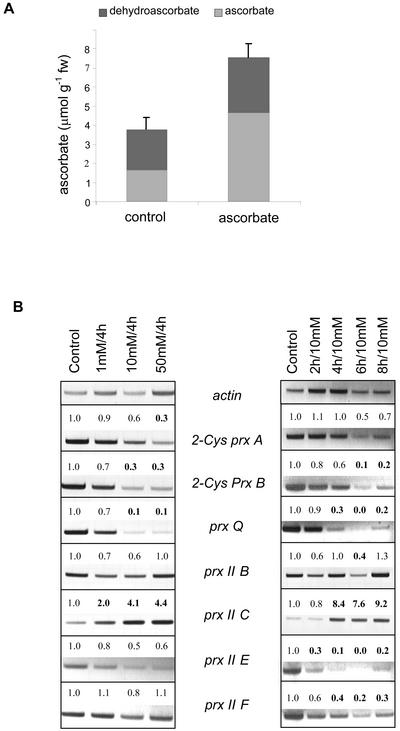
Ascorbate is the major soluble low-molecular mass antioxidant in plants (Noctor and Foyer, 1998). To test its effects on prx gene expression, leaf slices were suspended in media supplemented with ascorbate. The quantum yield of PS II was measured during the incubation period to monitor the photosynthetic performance and proved to be unaffected by ascorbate feeding during the incubation period (not shown). With 10 mm ascorbate in the suspension medium, the ascorbate contents of the leaf slices increased within 4 h by a factor of about 2 from 3.8 ± 0.8 to 7.6 ± 1.4 μmol ascorbate g fresh weight−1 (means ± sd, n = 10; Fig. 3A). The relative oxidation state of the ascorbate pool was high, which might be because of the use of leaf slices. The mRNA amounts of all chloroplast-targeted prx genes, i.e. 2-Cys prx A and B, prx Q and prx II E, decreased upon external application of ascorbate. The response was dependent on incubation time and effector concentration. The two 2-Cys prxs responded slightly differently to ascorbate with 2-Cys prx B being more sensitive to lower ascorbate concentrations than 2-Cys prx A. The time-resolved response to 10 mm ascorbate was similar. prx II B and F were largely unaffected by externally supplied ascorbate. In a converse manner, the transcript level of prx II C was increased upon addition of ascorbate (Fig. 3B).
Figure 3.
Effect of exogenous application of ascorbate on ascorbate and dehydroascorbate contents, and prx gene expression of Arabidopsis leaf slices. A, Ascorbate and dehydroascorbate contents of leaf slices incubated in 0 or 10 mm ascorbate for 4 h. The data are mean values ± sd; n = 10. B, Concentration and time dependency of the changes in prx gene expression. Leaf slices were incubated in the presence of 1, 10, and 50 mm ascorbate for 4 h and for 2, 4, 6, and 8 h exposed to 10 mm ascorbate. Prx transcript abundance was analyzed by gene-specific RT-PCR. The numbers indicate the factor of change obtained by a semiquantitative band density analysis as related to the control condition. All experiments were performed three times and showed similar changes in transcript levels in each case.
H2O2-Reducing Activities and Midpoint Redox Potentials of Six Prx Proteins
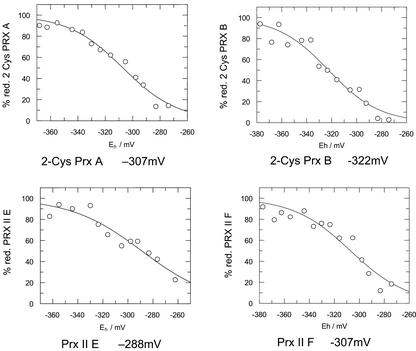
For in vitro analysis of the biochemical properties of the peroxiredoxins, six Prx proteins were heterologously expressed as His-tagged proteins in Escherichia coli. In kinetic measurements with a DTT-based nonenzymatic regeneration system, the Prx II proteins showed H2O2 activities between 57 and 156 mol H2O2 mol Prx min−1 (Table I). The mean initial rates (Vo) from seven determinations can be ordered: Vo(Prx II C) > Vo(Prx II B) ≫ Vo(Prx II F) ≫ Vo(Prx II E). The 2-Cys prx proteins were less active. 2-Cys prx B showed a specific activity of 6.5 mol H2O2 mol Prx min−1, whereas 2-Cys prx A activity had no activity distinguishable from the background in the DTT-assay. In the NADPH/E. coli thioredoxin reductase/E. coli thioredoxin assay system (König et al., 2002), which gave a similar activity for 2-Cys Prx B and 2-Cys prx A, the Prx II only had a trace activity.
The midpoint redox potentials (Em′) of the Prx proteins was determined in a fluorimetric test. The Prx proteins were incubated in a defined redox buffer of oxidized and reduced DTT, followed by labeling with excess monobromobimane. The preliminary midpoint potentials were between −287 and −289 mV for Prx II B, Prx II C, and Prx II E (Fig. 4). Prx II F and 2-Cys Prx A had a midpoint potential of −307 mV. 2-Cys Prx B had the most negative Em′ with −322 mV.
Figure 4.
Midpoint redox potentials of 2-Cys Prx A and B, and Prx II B and F. The redox potential of the samples was adjusted by varying the ratio of DTToxidized to DTTreduced. After reacting reduced thiol groups with monobromobimane, the samples were analyzed for bound fluorophore.
DISCUSSION
Prx proteins are found in all organisms. So far, the physiological role of Prxs in antioxidant defense of photosynthesizing cells has only been worked out for 2-Cys Prx. By analyzing antisense Arabidopsis plants with decreased 2-Cys Prx levels (Baier and Dietz, 1999; Baier et al., 2000) and knockout mutants of the cyanobacterium Synechocystis PPC 6803 (Klughammer et al., 1998; Nishiyama et al., 2001), a protective function of 2-Cys Prx was established with importance for maintaining photosynthesis in a functional state. Because of the high sequence similarity, the antisense approach influenced both 2-Cys Prx isoforms of Arabidopsis. The partial loss of 2-Cys Prx function could not fully be compensated by other components of the antioxidant network (Baier et al., 2000). The induction of ascorbate-regenerating enzymes and the shift in the ascorbate redox poise indicated that a high burden was put on the ascorbate system. The enzymatic properties of the barley homolog were recently analyzed in vitro, demonstrating that the plant 2-Cys Prx reduces H2O2 and alkyl hydroperoxides (König et al., 2002).
Putative homologs of Prx Q and Prx II B/C were described from Sedum lineare (Kong et al., 2000), Brassica rapa (Choi et al., 1999), and poplar (Populus trichocarpa; Rouhier et al., 2001), respectively. The poplar Prx II B/C homolog is expressed in phloem cells (Rouhier et al., 2001) and can use thioredoxin and glutaredoxin with similar efficiency as the electron donor in the regeneration reaction (Rouhier et al., 2002).
The data presented here compare for the first time, to our knowledge, the expressional behavior and biochemical characteristics of all Prxs expressed in green tissues of one plant, Arabidopsis. This includes the first description, to our knowledge, of genetic and biochemical data on Prx II E and F.
The Catalytic Activity of Prx
All Prx proteins showed peroxidase activity, however, with strong quantitative differences and different specificity for the regenerating electron donor. Despite the use of His-tagged proteins, the Prx activities are assumed to be similar to those of native Prxs as shown in the comparative work with Prxs of Crithidia fasciculata by Montemartini et al. (1998), in which N-terminally His-tagged Prx was shown to be as active as the purified Prx protein. Nevertheless, the use of His-tagged proteins should be kept in mind and the enzymic data interpreted with caution.
For the first time in plants, to our knowledge, H2O2-reducing activity is shown for the mitochondrial and chloroplastic isoforms of Prx II. In the antioxidative system of mitochondria, the presence and function of H2O2-scavenging enzymes are discussed controversially (Foyer and Noctor, 2000). Prx II F, which is expressed in green and nongreen tissues (Horling et al., 2002), is likely to be an important component of the mitochondrial defense system against peroxide stress. Recently, Laloi et al. (2001) identified a functional mitochondrial thioredoxin system in Arabidopsis that could act as an electron donor of Prx II F. In the chloroplast, Prx II E is present in addition to Prx Q and both 2-Cys Prx. In comparison with 2-Cys Prx, Prx II E shows a more positive midpoint potential indicating a distinct physiological activation of this antioxidative enzyme within the redox hierarchy of the chloroplast (König et al., 2002).
In an enzymic assay using DTT as an electron donor, the Arabidopsis 2-Cys Prx B showed the same activity as in the presence of thioredoxin. 2-Cys Prx A was not active in the DTT system, whereas all Prx IIs were highly active in the DTT-based assay. Considering the concentration and the negative redox potential of DTT (Em′ = −0.336 mV), the differences in activity are unlikely to be caused by insufficient reduction potential of DTT. It is more likely that the different responses may reflect the accessibilities of the oxidized catalytic centers in the Prx proteins or different reaction mechanisms. For Prx II, Rouhier et al. (2002) suggested a reaction mechanism, in which only one of the two Cys of Prx II is involved in the catalytic mechanism. The sulfenic acid intermediate formed in the active site of Prx II by peroxide reduction is directly reduced by the sulfhydryl group of an electron donor as DTT. In 2-Cys Prx, the sulfenic acid intermediate first reacts with the other conserved Cys residue in the second subunit of the 2-Cys Prx dimer. Because of sterical hindrance, the sulfenic acid side chain might not be accessible for the sulfhydryl group of DTT. The 2-Cys Prx proteins were more active in the thioredoxin system than Prx II B and Prx II C, which were most active in the DTT system. It is assumed that in the interaction with thioredoxin, the disulfide structure of the oxidized 2-Cys Prx is a better target for the bi-thiol/disulfide transition of thioredoxin than the monoreduction reaction of Prx II regeneration. It should be noted that the method of determination of redox potentials does not allow one to distinguish between disulfide bridges formed within Prx molecules and mixed DTT-Prx complexes. Therefore, the redox potentials of Prx IIs with yet unclear reaction mechanism should be considered as preliminary trends.
The difference between the two 2-Cys Prx proteins is more difficult to explain. The sequences of the mature proteins differ in seven amino acid residues, six positions of which are substituted by similar amino acids. A possible functional exchange may be the substitution of the His-130 in 2-Cys Prx A by a Pro residue in 2-Cys Prx B. This position is highly variable in the 2-Cys Prx family and often replaced by charged amino acid residues in animal, bacterial, and fungal 2-Cys Prx. Pro may increase rigidity of the protein and enable 2-Cys Prx B to accept DTT as a reductant in the active site.
The natural electron donor is still unknown. For Prx proteins of yeast, man, plants, and other sources, thioredoxins (Chae et al., 1994; König et al., 2002), glutaredoxins (Rouhier et al., 2001), and the redox active cyclophilin hCyp-A (Lee et al., 2001) are discussed and have been described either as interacting partners or as electron donors in vitro. In the light of more than 30 genes encoding thioredoxins and thioredoxin-like proteins, about 25 genes for glutaredoxins and glutaredoxin-like proteins and 20 genes coding for cyclophilins (with at least one or more conserved Cys-residues) in the Arabidopsis genome (data not shown), the complementary pairs of Prx protein and optimum electron donor still need to be identified.
The antioxidant capacity of Prxs has to be compared with that of other antioxidant enzymes. Rate constants are as follows: 2-Cys Prx, 105 m−1 s−1; Prx II C, 1.6 × 106 m−1 s−1; selenium-free HGPx × 106 m−1 s−1, and Apx, about 107 m−1 s−1 (Asada et al., 1996; Hofmann et al., 2002; this work). Thus, Prxs belong to the less active antioxidant enzymes, which may be suggested to either function in antioxidant defense at specific sites (König et al., 2002) or in antioxidant signaling (Dietz, 2003).
The Subcellular Localization of Prx Proteins in Arabidopsis
Four of the 10 putative Prx gene products are predicted to be targeted to the chloroplast (Horling et al., 2002). 2-Cys Prx A and B and Prx Q and Prx II have N-terminal extensions with defined properties of plastid-targeting addresses, which is experimentally confirmed for the 2-Cys Prx and Prx II E (Baier and Dietz, 1997; König et al., 2002; Peltier et al., 2002). In mitochondria, a Prx-like protein was found in a proteomic approach (Kruft et al., 2001), which is Prx II F. Prx II B and C are likely to be retained in the cytosol (Horling et al., 2002). Immunocytochemistry localized a poplar Prx that is most homologous to Arabidopsis Prx II B/C to the plastids in sieve elements (Rouhier et al., 2001). The poplar Prx lacks an apparent targeting address for plastid import. The contradiction may be solved on the basis of immunological cross-reactivity of Prx II isoforms. With an antibody raised against Prx II C protein, heterologously expressed Prx II B, E, and F proteins were detected in western blots in addition to Prx II C and with similar affinity. In immunocytochemical studies using that antibody, signals were observed in the plastids and in the cytosol of mesophyll cells (not shown). The cytosolic signal might either indicate the presence of Prx II B, Prx II C, or both.
Redox Regulation of Prx Gene Expression
With the exception of the nuclear 1-Cys Prx (Stacy et al., 1996, 1999), all functional Arabidopsis prx genes are expressed in leaves (Horling et al., 2002). In plant cells, the cellular redox poise is tightly linked to photosynthesis. Thus, the influence of light intensity changes on the transcript level was analyzed for all seven leaf-expressed prxs.
Excess light triggers various acclimation responses at distinct metabolic and genetic levels with different kinetics. Fast responses are state transition and violaxanthin synthesis for redirecting excitation energy and safe energy dissipation (Dietz et al., 2001). Increasingly excessive photon fluence rates induce changes in plastidic and nuclear gene expression, which include light-harvesting proteins and reaction center proteins (Escoubas et al., 1995; Pfannschmidt et al., 1999), and finally up-regulation of antioxidant defense genes both locally and systemically (Mullineaux et al., 2000). Ten-fold excess light had only a small stimulatory effect on prx expression. In a converse manner, a 10-fold drop in photon fluence rate suppressed expression of all plastidic prx genes within 4 to 8 h. A regulatory scenario could be as follows: The chloroplast prxs are expressed at almost maximum level under normal conditions of photosynthesis. Thus, only a moderate up-regulation is possible in excess light. Conversely, chloroplast prx expression is down-regulated when the excitation pressure is substantially decreased, i.e. when the activity of the photosynthesis-related oxidative metabolism is low. The 2-Cys Prx protein amount appeared slightly up-regulated both at low and excess light. This seemingly contradicting result may hint at strongly decreased 2-Cys Prx turnover under low light. The signaling events involved in low-light-induced down-regulation of chloroplast prx expression are unknown. The low light effect on prx gene expression was paralleled by the response to exogenous application of ascorbate: The transcript levels of chloroplastic prx genes declined after addition of ascorbate in a time- and concentration-dependent manner. For the time being, increased ascorbate or decreased dehydroascorbate may be considered as candidate signaling elements transducing a low burden on antioxidant metabolism from the chloroplast to the nucleus (Horling et al., 2001). An apparent contradiction is seen in the high light experiment where the ascorbate pool increased during an 8-h exposure to 10-fold increased photon fluence rates but had no major effect on prx gene expression. Ascorbate is directly involved in the Apx-mediated detoxification of H2O2 and indirectly of lipid peroxides. Lipid peroxides and lipid radicals are quenched by tocopherol. Ascorbate is the electron donor in the reduction of oxidized tocopherol (Noctor and Foyer, 1998). The data on light-dependent adjustment of prx transcript levels show that the regulation of prx gene expression cannot be exclusively explained by changing ascorbate or monodehydroascorbate concentrations. Prx proteins are alternative enzymes in H2O2 and lipid peroxide reduction. In this context, ascorbate could act as a negative, or dehydroascorbate as a positive, regulator of chloroplast prx expression.
In contrast to 2-Cys prx A and B and prx II E, expression of prx Q increased upon exposure of leaf slices to H2O2. Apparently, another signaling pathway interferes with prx Q gene expression independent of light and ascorbate. The regulatory redox linkage to ascorbate is suggested on the basis of the differential response to thiols and ascorbate of 2-Cys prx in Riccia fluitans (Horling et al., 2001) and barley (Baier and Dietz, 1996) and the response of plants with decreased levels of 2-Cys Prx (Baier and Dietz, 1999; Baier et al., 2000). Additional experiments will have to dissect the potential role of thiols in the ascorbate-induced regulation of the various prx genes.
Prx II C Expression as Indicator of Metabolic Imbalances
Prx II B and C have a putative location in the cytosol. Together with the mitochondrial prx II F, they frequently showed regulation in response to redox and stress conditions distinct from chloroplast-targeted prx. In general, prx II F transcript amounts were unaffected by the light conditions. A stable steady-state transcript level was also described in dependence of leaf age and under NaCl stress (Horling et al., 2002) and may be interpreted as constitutive requirement and controlled turnover rate of prx II F transcripts. In the case of prx II B, the high sensitivity of expression to externally added butyl hydroperoxide seems noteworthy. Butyl hydroperoxide is lipohilic and likely to initiate its oxidative activity in the plasma membrane upon application through the external medium. It will have to be investigated whether Prx II B functions in protecting the plasma membrane.
Among all prx genes, prx II C showed the most peculiar expression pattern. Its expression was stimulated upon an increase and decrease of photon fluence rate, after addition of ascorbate and under oxidative stress, mediated by H2O2, butyl hydroperoxide, or diamide, and as shown previously under salt stress (Horling et al., 2002). The sensitive response of prx II C expression to all kind of changing conditions suggests that any deviation from the steady state activates the promoter. Expression of cytosolic apx is a frequently employed marker of stress (Mullineaux et al., 2000). It will be interesting to compare the promoters of apxcytosol and prx II C because both gene products are involved in antioxidant defense.
The description of the rather surprising divergence of the light-, ascorbate-, and oxidative stress-dependent expressional regulation of the peroxiredoxin gene family in Arabidopsis provides an essential basis for the elucidation of Prx function in future work.
MATERIALS AND METHODS
Plant Growth and Harvesting
Arabidopsis (Columbia) was grown in soil culture with a 14-h light period at 22°C and a photon fluence rate of 120 μmol quanta m−2 s−1. For the light transition experiment, the whole plants were transferred to 1,000 μmol quanta m−2 s−1 or 10 μmol quanta m−2 s−1 for the time periods as indicated. The youngest fully expanded leaves were harvested by rapid freezing in liquid nitrogen and extracted for total RNA. For effector studies, 10 to 20 leaves from 4-week-old plants were pooled and cut into 1-mm-diameter leaf slices. After vacuum infiltration with distilled water and suspension in effector solution, the leaf slices were incubated at 120 μmol quanta m−2 s−1 for time periods as indicated. The ascorbate solution was adjusted to pH 4 for increased stability.
Measurement of Reduced and Total Ascorbate
Ascorbate contents were determined according to Foyer et al. (1983). Leaves were ground to a fine powder in liquid N2. For extraction, 1 mL of ice-cold 1 m HClO4 was added to the frozen plant material. After centrifugation at 13,000 rpm (5 min at 4°C), 400 μL of the supernatant was transferred to 200 μL of 1 m HEPES/KOH buffer (pH 7.0). The pH of the solution was adjusted to about pH 5.0 with 5 m K2CO3. The samples were spun at 13,000 rpm for 5 min at 4°C to remove the precipitates. The supernatant was used for measuring the contents of reduced and total ascorbate spectrophotometrically.
Reduced ascorbate was measured after addition of 100 μL of the supernatant to 900 μL of 0.1 m sodium phosphate buffer (pH 5.6) by monitoring a decrease in A265 in the presence of 5 units of ascorbate oxidase (Sigma, Deisenhofen, Germany). For the measurement of total ascorbate, the ascorbate pool was reduced with 50 mm DTT in four volumes of 0.1 m sodium phosphate (pH 7.0) during 30 min of incubation on ice and analyzed as described above for reduced ascorbate. DHA was calculated as the difference of ascorbate contents determined in the presence and absence of DTT according to authentically treated ascorbate and dehydroascorbate standards (Foyer et al., 1983).
Semiquantitative RT-PCR
Total RNA was isolated from 100 mg of plant material using TRIZOL Reagent (Gibco-BRL, Cleveland) according to the instruction manual. cDNA synthesis and RT-PCR were performed as described earlier (Horling et al., 2001, 2002). For expression studies of the different prx isotypes, primers were designed from the 5′-untranslated region and 3′-untranslated region and gene-specific amplification was verified by sequencing (MWG-Biotech, Eberswalde, Germany).
Heterologous Expression and Purification of Prx
prx II E was cloned using the TA cloning kit (Invitrogen, Carlsbad, CA). All other prx cDNAs were cloned using the BamHI recognition site flanking the appropriate forward primer and the KpnI recognition site flanking the reverse primer. The gene-specific cDNA was amplified by PCR using the proof-reading Pfu polymerase (Stratagene, La Jolla, CA), eluted from a 1% (w/v) agarose gel and purified using the Easy Pure Kit (Biozym, Hessisch Oldendorf, Germany). After restriction digestion with BamHI and KpnI and further purification, the cDNAs were ligated into the pQE30 vector using T4 ligase at 15°C over night and transformed into TOPO10-competent Escherichia coli cells (Invitrogen). The pQE30 vector allows expression and purification of an N-terminally 6× His-tagged recombinant protein. The prx genes were expressed without putative signal peptides. Clones from overnight Luria-Bertani (LB)-ampicillin plates were verified by PCR and restriction analysis. Plasmids were transformed into CaCl2-competent M15 E. coli cells and plated on LB medium supplemented with ampicillin (50 μg mL−1). Single clones were picked for expression in a 1-L culture (LB medium, 50 μg mL−1) inoculated at a 1:100 (v/v) ratio with noninduced overnight culture grown to an OD600 of 0.6 to 0.8. The expression was induced by adding isopropylthio-β-galactoside (0.4 mm final concentration) to the medium. Cells were harvested after 4 h by centrifugation at 5,000 rpm for 30 min at 4°C. The cell pellet was frozen at −80°C and stored over night.
For protein purification, the cells were suspended in 20 mL of lysis buffer (50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazol, 10 mm ascorbate, and 0.5 mg mL−1 lysozyme at pH 8.0). The solution was shaken for 1 h on ice and sonicated using six 10 s-cycles at 200 W. The supernatant obtained from centrifugation at 15,000 rpm for 30 min at 4°C was loaded onto a nickel-nitrilotriacetic acid agarose column (2 mL L−1 cell culture, Qiagen, Hilden, Germany) previously equilibrated in washing buffer (50 mm NaH2PO4, 300 mm NaCl, and 10 mm imidazol, pH 8.0 using NaOH). The loaded resin was washed with 20 column volumes of the same buffer and 20 column volumes of this buffer supplemented with 20% (v/v) glycerol. The His-tagged protein was eluted using washing buffer containing 250 mm imidazol. The protein-containing fractions, as measured at 280 nm, were pooled and dialyzed against 40 mm K phosphate (Pi) buffer at pH 7.0.
Western-Blot Analyses
For western-blot analysis, frozen plant material was ground to a fine powder in liquid N2 and proteins were extracted in a buffer containing 250 mm Tris-Cl (pH 6.8). The protein contents of the aqueous extracts were quantified spectrophotometrically using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, CA) according to the supplier's manual. SDS-PAGE and western-blot analyses were performed as described before (Horling et al., 2002). For detection of peroxiredoxins, antibodies were produced against proteins heterologously expressed in E. coli (barley [Hordeum vulgare] 2-Cys Prx and Arabidopsis Prx II C).
Peroxiredoxin Activity Assay and Determination of Midpoint Potential
Reduction of H2O2 by Arabidopsis peroxiredoxins was analyzed in vitro using a nonenzymatic, DTT-dependent activity assay and an enzymatic thioredoxin-dependent activity assay, respectively. The nonenzymatic test was performed by measuring the decrease in H2O2 concentration in the assay solution. The assay contained 100 mm K-Pi buffer (pH 7.0), 0.3 to 3 μm Prx, 10 mm DTT, and 100 μm H2O2 in a total volume of 1,000 μL. The reaction was initiated with H2O2 and stopped with 800 μL of trichloroacetic acid (12.5% [w/v]) to an aliquot of 50 μL of assay solution. After adding 200 μL of 10 mm Fe(NH4)2(SO4)2 and 100 μL of 2.5 m KSCN, the A480 was measured to quantify the H2O2 contents of the solution, and H2O2 reduction rates were calculated. The enzymatic, thioredoxin-dependent assay was performed as described before (König et al., 2002). The assay typically contained 50 μm peroxide, 100 mm K-Pi buffer (pH 7.0), 1 mm EDTA, 0.1 mm NADPH, 1 μm Prx, 7.5 μm Trx, and 2.5 μm TR.
For determination of the oxidation reduction midpoint potential, recombinant Prx protein was titrated at pH 7.0. Prx (100 μg) was incubated in MOPS buffer (100 mm) containing 2 mm total DTT in a volume of 500 μL. After 3 h at ambient temperature, monobromobimane was added at a final concentration of 10 mm. The samples were prepared for fluorescence analysis as described by Hirasawa et al. (1999).
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft within the Special Research Focus FOR 387 (TP 3: redox regulation of nuclear gene expression) and Di 346/6 (2-Cys Prx).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010017.
LITERATURE CITED
- Asada K, Miyake C, Ogawa K, Hossain MA. Microcompartmentation of ascorbate peroxidase and regeneration of ascorbate from ascorbate radical: its dual role in chloroplasts. In: Obinger C, Burner U, Ebermann R, Penel C, Greppin H, editors. Plant Peroxidases: Biochemistry and Physiology. Université de Genève. 1996. pp. 163–167. [Google Scholar]
- Baier M, Dietz KJ. Primary structure and expression of plant homologues of animal and fungal thioredoxin-dependent peroxide reductases and bacterial alkyl hydroperoxide reductases. Plant Mol Biol. 1996;31:553–564. doi: 10.1007/BF00042228. [DOI] [PubMed] [Google Scholar]
- Baier M, Dietz KJ. The plant 2-Cys peroxiredoxin BAS1 is a nuclear encoded chloroplast protein. Its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J. 1997;12:179–190. doi: 10.1046/j.1365-313x.1997.12010179.x. [DOI] [PubMed] [Google Scholar]
- Baier M, Dietz KJ. Protective function of chloroplast 2-Cys peroxiredoxin in photosynthesis. Evidence from transgenic Arabidopsis. Plant Physiol. 1999;119:1407–1414. doi: 10.1104/pp.119.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M, Noctor G, Foyer CH, Dietz KJ. Antisense suppression of 2-Cys peroxiredoxin in Arabidopsis thaliana specifically enhances the activities and expression of enzymes associated with ascorbate metabolism, but not glutathione metabolism. Plant Physiol. 2000;124:823–832. doi: 10.1104/pp.124.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk R, Griffin P, Nathan C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature. 2000;407:211–215. doi: 10.1038/35025109. [DOI] [PubMed] [Google Scholar]
- Chae HZ, Chung SJ, Rhee SG. Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem. 1994;269:27670–276784. [PubMed] [Google Scholar]
- Choi YO, Cheong NE, Lee KO, Jung BG, Hong CH, Jeong JH, Chi YM, Kim K, Cho MJ, Lee SY. Cloning and expression of a new isotype of the peroxiredoxin gene of Chinese cabbage and its comparison to 2 Cys-peroxiredoxin isolated from the same plant. Biochem Biophys Res Commun. 1999;258:768–771. doi: 10.1006/bbrc.1999.0714. [DOI] [PubMed] [Google Scholar]
- Dietz KJ (2003) Plant peroxiredoxins. Annu Rev Plant Biol (in press) [DOI] [PubMed]
- Dietz KJ, Horling F, König J, Baier M. The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. J Exp Bot. 2002;53:1321–1329. [PubMed] [Google Scholar]
- Dietz KJ, Link G, Pistorius EK, Scheibe R. Redox regulation in oxygenic photosynthesis. Prog Bot. 2001;63:207–245. [Google Scholar]
- Escoubas J-M, Lomas M, La Roche J, Falkowski PG. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Tansley review no. 112, oxygen processing in photosynthesis: regulation and signaling. New Phytol. 2000;146:359–388. [Google Scholar]
- Foyer CH, Rowell J, Walker D. Measurements of the ascorbate content of spinach leaf protoplasts and chloroplasts during illumination. Planta. 1983;157:239–244. doi: 10.1007/BF00405188. [DOI] [PubMed] [Google Scholar]
- Haslekas C, Stacy RA, Nygaard V, Culianez-Macia FA, Aalen RB. the expression of a peroxiredoxin antioxidant gene AtPer1, in Arabidopsis thaliana is seed-specific and related to dormancy. Plant Mol Biol. 1998;36:833–845. doi: 10.1023/a:1005900832440. [DOI] [PubMed] [Google Scholar]
- Hillas PJ, del Alba FS, Oyarzabal J, Wilks A, de Montellano PRO. The AhpC and AhpD antioxidant defense system of Mycobacterium tuberculosis. J Biol Chem. 2000;275:18801–18809. doi: 10.1074/jbc.M001001200. [DOI] [PubMed] [Google Scholar]
- Hirasawa M, Schürmann P, Jacquot JP, Manieri W, Jacquot P, Keryer E, Hartman FC, Knaff DB. Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin:thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry. 1999;38:5200–5205. doi: 10.1021/bi982783v. [DOI] [PubMed] [Google Scholar]
- Hofmann B, Hecht H-J, Flohé L. Peroxiredoxins. Biol Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- Horling F, Baier M, Dietz K-J. The cellular redox poise regulates expression of the peroxide detoxifying chloroplast 2-Cys peroxiredoxin in the liverwort Riccia fluitans. Planta. 2001;214:283–287. doi: 10.1007/s004250100623. [DOI] [PubMed] [Google Scholar]
- Horling F, König J, Dietz KJ. Type II peroxiredoxin C, a member of the peroxiredoxin family of Arabidopsis thaliana: its expression and activity in comparison with other peroxiredoxins. Plant Physiol Biochem. 2002;40:491–499. [Google Scholar]
- Jin DY, Chae HZ, Rhee SG, Jeang KT. Regulatory role for a novel human thioredoxin peroxidase in NF-kB activation. J Biol Chem. 1997;272:30952–30961. doi: 10.1074/jbc.272.49.30952. [DOI] [PubMed] [Google Scholar]
- Kang SW, Baines IC, Rhee SG. Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J Biol Chem. 1998;273:6303–6311. doi: 10.1074/jbc.273.11.6303. [DOI] [PubMed] [Google Scholar]
- Klughammer B, Baier M, Dietz KJ. Inactivation by gene disruption of 2-cysteine-peroxiredoxin in Synechocystis sp. PCC 6803 leads to increased stress sensitivity. Physiol Plant. 1998;104:699–706. [Google Scholar]
- Kong W, Shiota S, Shi Y, Nakayama H, Nakayama K. A novel peroxiredoxin of the plant Sedum lineare is a homologue of Escherichia coli bacterioferritin co-migratory protein (Bcp) Biochem J. 2000;351:107–114. doi: 10.1042/0264-6021:3510107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Baier M, Horling F, Kahmann U, Harris G, Schürmann P, Dietz KJ. The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc Natl Acad Sci USA. 2002;99:5738–5743. doi: 10.1073/pnas.072644999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruft V, Eubel H, Jänsch L, Werhahn W, Braun H-P. Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 2001;127:1694–1710. [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Rayapuram N, Chartier Y, Grienenberger JM, Bonnard G. Identification and characterization of a mitochondrial thioredoxin system in plants. Proc Natl Acad Sci USA. 2001;98:14144–14149. doi: 10.1073/pnas.241340898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SP, Hwang YS, Kim YJ, Kwon KS, Kim HJ, Kim K, Chae HZ. Cyclophilin a binds to peroxiredoxins and activates its peroxidase activity. J Biol Chem. 2001;276:29826–29832. doi: 10.1074/jbc.M101822200. [DOI] [PubMed] [Google Scholar]
- Montemartini M, Nogoceke E, Singh M, Steinert P, Flohé L, Kalisz HM. Sequence analysis of the tryparedoxin peroxidase gene from Crithidia fasciculata and its functional expression in Escherichia coli. J Biol Chem. 1998;273:4864–4871. doi: 10.1074/jbc.273.9.4864. [DOI] [PubMed] [Google Scholar]
- Mullineaux P, Ball L, Escobar C, Karpinski B, Creissen G, Karpinski S. Are diverse signaling pathways integrated in the regulation of Arabidopsis antioxidant defence gene expression in response to excess excitation energy. Philos Trans R Soc Lond. 2000;355:1531–1540. doi: 10.1098/rstb.2000.0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Yamamoto H, Allakhverdiev S, Inaba M, Yokota A, Murata N. Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J. 2001;20:5587–5594. doi: 10.1093/emboj/20.20.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Nogoceke E, Gommel DU, Kiess M, Kalisz HM, Flohé L. A unique cascade of oxidoreductases catalyses trypanothionine-mediated peroxide metabolism in Crithidia fasciculata. Biol Chem. 1997;378:827–836. doi: 10.1515/bchm.1997.378.8.827. [DOI] [PubMed] [Google Scholar]
- Peltier J-B, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Söderberg L, Roepstorff P, von Heijne G et al. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimental and genome-wide prediction. Plant Cell. 2002;14:211–236. doi: 10.1105/tpc.010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- Rouhier N, Gelhaye E, Jacquot JP. Glutaredoxin-dependent peroxiredoxin from poplar. J Biol Chem. 2002;277:13609–13614. doi: 10.1074/jbc.M111489200. [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Sautiere PE, Brun A, Laurent P, Tagu D, Gerard J, de Fa E, Meyer Y, Jacquot JP. Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiol. 2001;127:1299–1309. [PMC free article] [PubMed] [Google Scholar]
- Stacy RAP, Munthe E, Steinum T, Sharma B, Aalen RB. A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Mol Biol. 1996;31:1205–1216. doi: 10.1007/BF00040837. [DOI] [PubMed] [Google Scholar]
- Stacy RAP, Nordeng TW, Culianez-Macia FA, Aalen RB. The dormancy-related peroxiredoxin anti-oxidant, PER1, is localized to the nucleus of barley embryo and aleurone cells. Plant J. 1999;19:1–8. doi: 10.1046/j.1365-313x.1999.00488.x. [DOI] [PubMed] [Google Scholar]
- Verdoucq L, Vignol F, Jacquot JP, Chartier Y, Meyer Y. In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J Biol Chem. 1999;274:19714–19722. doi: 10.1074/jbc.274.28.19714. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Kok KH, Chun ACS, Wong C-M, Wu HW, Lin MCM, Fung PCW, Kung H-F, Jin D-Y. Mouse peroxiredoxin V is a thioredoxin peroxidase that inhibits p53-induced apoptosis. Biochem Biophys Res Commun. 2000;268:921–927. doi: 10.1006/bbrc.2000.2231. [DOI] [PubMed] [Google Scholar]