Abstract
The gibberellin (GA)-deficient dwarf na mutant in pea (Pisum sativum) has severely reduced internode elongation, reduced root growth, and decreased leaflet size. However, the seeds develop normally. Two genes, PsKAO1 and PsKAO2, encoding cytochrome P450 monooxygenases of the subfamily CYP88A were isolated. Both PsKAO1 and PsKAO2 had ent-kaurenoic acid oxidase (KAO) activity, catalyzing the three steps of the GA biosynthetic pathway from ent-kaurenoic acid to GA12 when expressed in yeast (Saccharomyces cerevisiae). In addition to the intermediates ent-7α-hydroxykaurenoic acid and GA12-aldehyde, some additional products of the pea KAO activity were detected, including ent-6α,7α-dihydroxykaurenoic acid and 7β-hydroxykaurenolide. The NA gene encodes PsKAO1, because in two independent mutant alleles, na-1 and na-2, PsKAO1 had altered sequences and the five-base deletion in PsKAO1 associated with the na-1 allele cosegregated with the dwarf na phenotype. PsKAO1 was expressed in the stem, apical bud, leaf, pod, and root, organs in which GA levels have previously been shown to be reduced in na plants. PsKAO2 was expressed only in seeds and this may explain the normal seed development and normal GA biosynthesis in seeds of na plants.
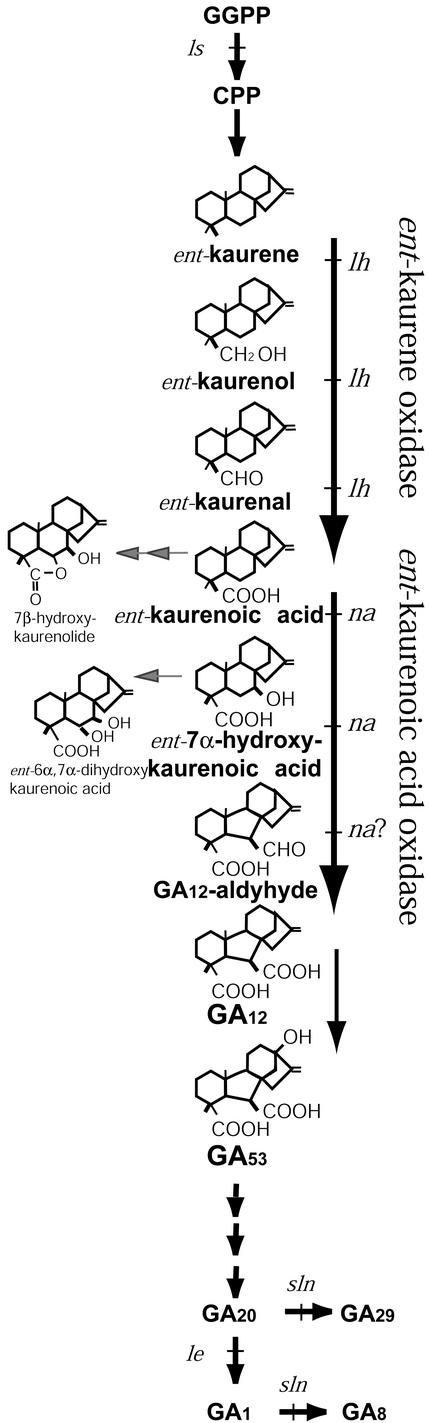
GAs are important plant growth hormones that regulate many aspects of plant growth including stem and petiole elongation, leaf expansion, and the growth of seeds and fruit (Reid and Ross, 1993; Hooley, 1994). They are also involved in seed germination and seed development (Hooley, 1994; Swain et al., 1997). Mutants have been useful in the elucidation of these actions and of the GA biosynthetic pathway (Ross et al., 1997; Hedden and Proebsting, 1999). In pea (Pisum sativum), most of the genes associated with the GA biosynthetic mutants have been cloned, including LS (copalyl diphosphate synthase; Ait-Ali et al., 1997), LE (3-oxidase; Lester et al., 1997; Martin et al., 1997), and SLN (2-oxidase; Lester et al., 1999b; Martin et al., 1999; Fig. 1).
Figure 1.
The GA biosynthetic pathway in pea. Product structures are represented for the cytochrome P450 monooxygenase-mediated steps showing the steps catalyzed by ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO). The steps blocked by the pea mutants (ls, lh, na, le, and sln) are indicated. Putative side products of PsKAO1 and PsKAO2 activity are indicated.
However, this is not the case for the GA-responsive mutant na, which has an extreme dwarf or “nana” phenotype (Fig. 2; Potts and Reid, 1983). It has extremely short internodes with a dramatic decrease in cell length of the epidermal and outer cortical cells as well as a reduction in the total number of these cells in the internode (Reid et al., 1983). The na mutants have small, darkly colored foliage with reduced area of individual leaflets, decreased stipule size, and petiole length (Reid and Ross, 1993). The root growth is also altered with taproot length reduced by 50% (Yaxley et al., 2001). The vegetative part of the na pea plant is severely deficient in endogenous GAs. It was not possible to show the presence of any C-19 GAs by dilution of [13C3H]GA20 metabolites by endogenous [12C]GAs using gas chromatography-mass spectrometry (GC-MS) techniques (Ingram et al., 1984). The na mutation markedly reduces the production of GAs, including the predominant bioactive GA1, in shoots and stems (Potts and Reid, 1983), leaves (Reid and Ross, 1993), roots (Yaxley et al., 2001), and pods (Potts and Reid, 1983; Potts, 1986). However, the effect of the na mutation is tissue specific and this was among the earliest information suggesting that alternative enzymes (or gene families) may be involved in GA biosynthesis (Reid, 1986a). In contrast to the vegetative part of the na plant, where the level of GA1 in expanding tissue was reduced to 2% of the wild type (Proebsting et al., 1992), the developing seeds contain similar GA levels to those found in the seeds of wild-type NA plants (Potts and Reid, 1983; Potts, 1986). In addition, the seeds of na plants develop normally (Potts and Reid, 1983).
Figure 2.
The phenotype of 21-d-old seedlings of wild-type NA (WL1769) and two independent mutants, na-1 (WL1766) and na-2 (L81).
The na mutation appears to block GA biosynthesis before GA12-aldehyde. There are two lines of evidence to support this supposition. The na plants did not respond to the application of precursors before GA12-aldehyde such as ent-kaurene, ent-kaurenoic acid (KA), and ent-7α-hydroxy-kaurenoic acid, but showed marked stem elongation in response to GA12-aldehyde. Also, [2H]GA12-aldehyde was metabolized to C-19 GAs such as GA20, GA29, GA1, and GA8 by na plants, but these plants do not metabolize ent-[3H2]kaurenoic acid to these GAs even though wild-type plants appear to do so (Ingram and Reid, 1987).
The GA biosynthetic pathway (Fig. 1) can be divided into three sections (Hedden and Kamiya, 1997; Hedden and Phillips, 2000). The first section, catalyzed by terpene cyclases, involves the cyclization of geranylgeranyl diphosphate to ent-kaurene. In the second section, the hydrophobic ent-kaurene is oxidized to GA12 or GA53 by membrane-bound cytochrome P450 monooxygenases. The third section consists of further oxidation to form the bioactive GA1 or GA4 by soluble 2-oxoglutarate dependent dioxygenases. The early sections of the pathway are common to all the plant species investigated so far (Hedden and Phillips, 2000). However, the wide range of dioxygenases allows variation in the pathway after GA12 in different species and tissues. The early 13-hydroxylase pathway predominates in vegetative tissue of pea, producing bioactive GA1 (Ingram et al., 1986; Reid and Ross, 1993; Poole et al., 1995). However, GA4 appears to be the main active product in Arabidopsis (Talon et al., 1990; Sponsel et al., 1997).
The second section of the GA biosynthetic pathway from ent-kaurene via KA to GA12 or GA53 is generally assumed to involve cytochrome P450 monooxygenases and requires the coenzyme NADPH-cytochrome P450 reductase, NADPH, and oxygen (Hedden, 1997). The CYP88A family of cytochrome P450s have recently been shown to encode KAO, which catalyzes the three steps from KA to GA12 via ent-7α-hydroxy-kaurenoic acid and GA12-aldehyde, in Arabidopsis and barley (Hordeum vulgare; Helliwell et al., 2001; Fig. 1). Other CYP88A cytochrome P450 genes from pumpkin (Cucurbita maxima; Helliwell et al., 2000) and maize (Zea mays) have also been isolated (Winkler and Helentjaris, 1995).
Arabidopsis does not have a mutant affecting KA oxidation presumably because of redundancy because the two Arabidopsis genes, AtKAO1 and AtKAO2, have similar expression patterns throughout the plant (Helliwell et al., 2001). The GA-responsive maize d3 mutants have defects in a CYP88A gene (Winkler and Helentjaris, 1995). The barley GA-responsive dwarf mutant, grd5, accumulates KA in developing grains (Helliwell et al., 2001). Mutations were found in the barley HvKAO1, in each of three independent mutant alleles of the barley dwarf grd5 (Helliwell et al., 2001).
In this paper, we identify genes encoding KAO activity in pea by screening a cDNA library using a maize D3-like expressed sequence tag (EST) probe from soybean (Glycine max). We show that one of these genes is NA and then explain the tissue-specific nature of the na mutation.
RESULTS
Isolation of Two CYP88A Genes from Pea
Two genes encoding cytochrome P450s of the subfamily CYP88A were isolated by library screening using a maize D3-like EST from soybean as probe. A partial cDNA of PsKAO1 was obtained from a pea cv Alaska apical bud cDNA library. The longest clone (500 bp) was extended by 3′- and 5-′ RACE using cDNA prepared from wild-type NA (WL1769) as template (Frohman et al., 1988). A full-length cDNA, PsKAO2, was obtained from a pea cv Torsdag seed library.
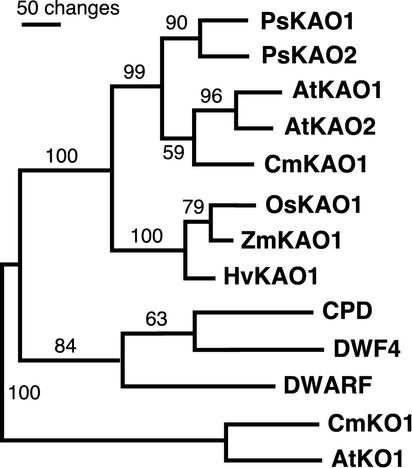
PsKAO1 (CYP88A6, GenBank accession no. AF537321 sequenced from NA WL1769) and PsKAO2 (CYP88A7, GenBank accession no. AF537322) showed close homology at the nucleotide level (60–100-bit score, 82%–93% identities) to AtKAO1 and AtKAO2 (BLASTN; Altschul et al., 1997). At the amino acid level, the full-length putative proteins PsKAO1 and PsKAO2 are similar to AtKAO1, AtKAO2, and CmKAO1 (644–661-bit score, 63%–65% identities, 79%–81% positives; National Center for Biotechnology Information Blast 2 sequences). Over the full-length PsKAO1 is similar to PsKAO2 at the nucleotide (462-bit score, 78% identity) and protein level (756-bit score, 74% identity, 86% positive; National Center for Biotechnology Information Blast 2 sequences). This is comparable with the Arabidopsis KAO putative proteins, where AtKAO1 is 76% identical to AtKAO2 (Fig. 3).
Figure 3.
Inferred phylogenetic relationship of KAOs [CYP88A] and representatives of related cytochrome P450 enzymes. Numbers shown represent the bootstrap support values (%). The phylogram was generated by PAUP 4.8b8 (Swofford, 1999) using putative amino acids of full-length genes (excluding gaps) with CYP701A as the outgroup. KAO proteins used in addition to the pea PsKAO1 and PsKAO2 were from Arabidopsis (AtKAO1 and AtKAO2, Helliwell et al., 2001), pumpkin (CmKAO1, Helliwell et al., 2000), rice (Oryza sativa; OsKAO1, GenBank accession no. AP000616), maize (D3, ZmKAO1, Winkler and Helentjaris, 1995), and barley (Grd5, HvKAO1, Helliwell et al., 2001). The related brassinosteroid biosynthetic enzymes used include Arabidopsis CPD (CYP90A1, Szekeres et al., 1996) and DWF4 (CYP90B1, Choe et al., 1998) and tomato (Lycopersicon esculentum) DWARF (CYP85A1, Bishop et al., 1999). The outgroup consisted of the cytochrome P450 monooxygenase ent-kaurene oxidases (CYP701A) from pumpkin CmKO1 (Helliwell et al., 2000) and Arabidopsis GA3 (AtKO1, Helliwell et al., 1998) of the GA biosynthetic pathway.
Northern-Blot Expression Studies
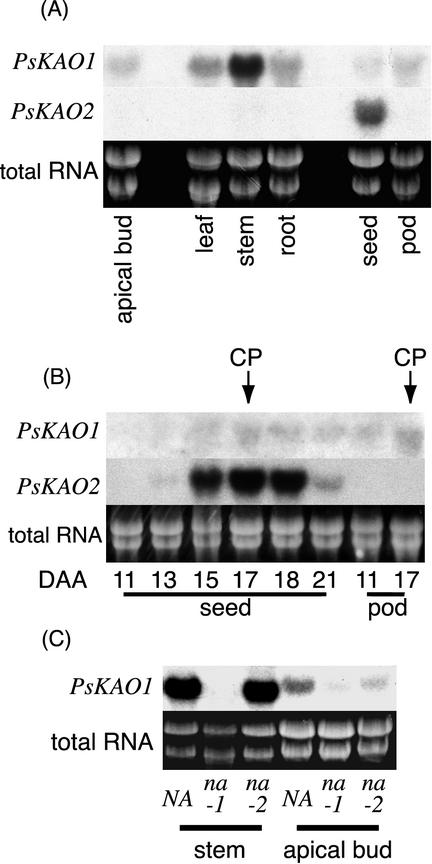
The members of the pea kaurenoic acid oxidase (KAO) gene family are differentially expressed. PsKAO1 is expressed in the stem and to a lesser extent in the leaf, root, apical bud, pod, and seed, whereas PsKAO2 is only expressed in the seed (Fig. 4A). PsKAO2 is expressed most strongly around the time of contact point when the embryo just fills the testa and the liquid endosperm is all consumed (Fig. 4B). This coincides with the rapid buildup of GA levels in maturing seeds (Frydman et al., 1974; Swain et al., 1993).
Figure 4.
A, PsKAO1 and PsKAO2 transcript levels in various parts of wild-type pea (L107). Five micrograms of total RNA from the apical bud (all material above the uppermost fully expanded leaf), leaf (the uppermost fully expanded leaf), stem (internode immediately below the uppermost fully expanded leaf, the internode was 80%–100% fully expanded), and root (50 mm off the end of the tap and lateral roots) of 19-d-old seedlings; also, 5 μg of total RNA from seeds (3 d after contact point) and pods (that originally contained these seeds) from mature plants were loaded on the gel. B, PsKAO1 and PsKAO2 transcript levels in wild-type pea (L107) seeds at various developmental ages. Total RNA was extracted from whole seeds and their pods between 11 and 21 d after anthesis. Contact point (CP, the 1st d that no liquid endosperm remained in seeds) occurred at 17 d after anthesis. C, PsKAO1 transcript levels in wild-type NA (WL1769) and mutants na-1 (WL1766) and na-2 (L81) from the apical bud (all tissue above the uppermost fully expanded leaf) or fully expanded stem tissue of 18-d-old pea seedlings. The ethidium bromide (EtBr)-stained total RNA is included as a loading control for each northern analysis.
PsKAO1 Is Mutated in the na-1 and na-2 Mutants
The PsKAO1 cDNA from the na-1 line WL1766 contained a five-base deletion when compared with NA (WL1769). This would change the reading frame for the encoded protein leading to a premature stop codon. The predicted protein would be 194 amino acids long, which is much smaller than the expected 485-amino acid length of the putative PsKAO1 enzyme. The predicted protein would be truncated before the catalytic domains (Kalb and Loper, 1988), including the active haem-binding site common to all cytochrome P450 enzymes. There was markedly less PsKAO1 mRNA measured in the na-1 mutant tissue than the isogenic wild-type NA (Fig. 4C). This may be because of the instability of mRNA with a premature stop codon (Gutierrez et al., 1999).
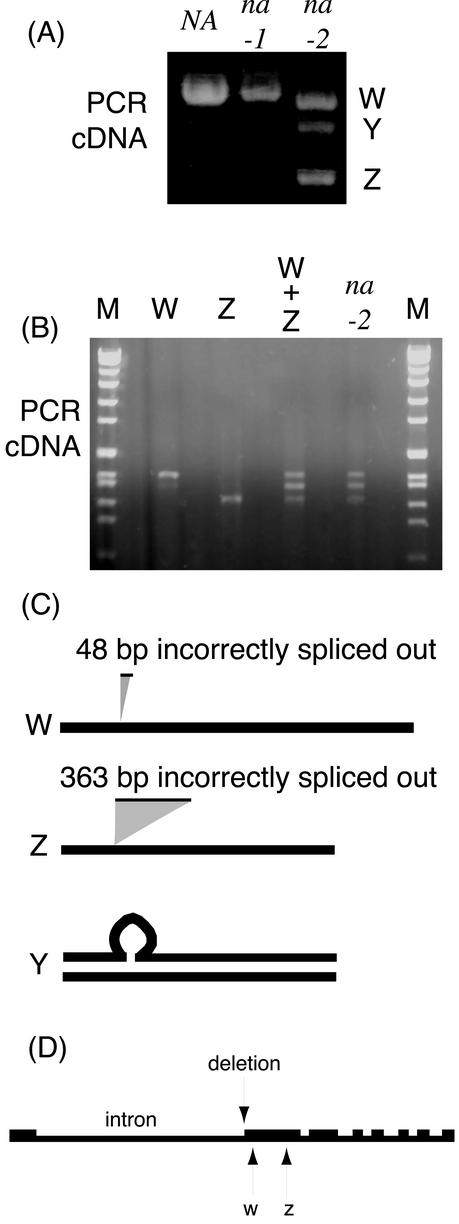
The PsKAO1 from the na-2 line L81 also is altered compared with NA (WL1769). Initially, PCR of the cDNA obtained from RNA of na-2 stems produced three bands (Fig. 5). When gel purified, the largest band (W) was found to have 48 bp incorrectly spliced out and the smallest band (Z) was found to have 364 bp incorrectly spliced out (Fig. 5). The band Y could not be gel purified. However, if the products W and Z were combined, melted, and annealed, the original three-band PCR pattern reappeared. This suggests that the Y band represents a duplex between the W and Z bands (Fig. 5). Genomic DNA sequence data was then used to further define the PsKAO1 na-2 mutation. The genomic sequence of na-2 revealed a 25-bp deletion (5 bp from the 3′ end of a large intron and 20 bp of exon sequence) compared with the wild type. The AG that is required for the positioning of splicing (Brown, 1996) was lost from the intron. Hence, splicing did not occur in the same place as in the wild type. Therefore, this mutation leads to aberrant processing of the resultant pre-RNA. The second and 18th AG after the deletion were used as 3′-splicing points for the RNA that produced the W and Z PCR bands, respectively. Splicing of the following intron was not affected (Fig. 5).
Figure 5.
A, The cDNA PCR products of PsKAO1 from wild-type NA (WL1769) and mutants na-1 (WL1766) and na-2 (L81) using the same specific primers run on 1% (w/v) agarose/Tris-acetate EDTA gel containing EtBr at 80 V for 45 min. The na-2 bands have been labeled W, Y, and Z. B, Lanes W and Z contain previously gel-purified PCR product bands W and Z from na-2 mutant cDNA (see A). Lane W + Z is the product formed when the gel-purified products W and Z were combined, melted, and annealed (three cycles of melting at 95°C and annealing at 55°C then a 70°C extension). Lane na-2 is the PCR product of the na-2 mutant cDNA as seen in A and lane M is the SPPI-EcoRI size marker run on 11% (w/v) agarose/Tris-acetate EDTA gel containing EtBr. C, Schematic diagram explanations of the cDNA bands W, Z, and Y of A from sequence and experimental data (B). D, Schematic diagram of genomic DNA of PsKAO1 from sequence data. The 25-bp deletion site as well as the W and Z cryptic splice sites of the na-2 mutation are indicated.
The Mutation in na-1 Cosegregates with the na Mutant Phenotype
DNA was extracted from individual plants from four segregating F9 families from a cross between NA (WL1769) and na-1 (WL1766) plants (Fig. 2). The hexachloro fluorescein-labeled PCR products (243 bp in the wild type) identified the five-base deletion of the na-1 mutant gene when run on denaturing gel. The five-base deletion of PsKAO1 cosegregates with the dwarf phenotype of na plants (data not shown). Of the 50 individuals in the four families, 12 were homozygous tall (NA NA), 26 were heterozygous tall (NA na), and 12 were homozygous dwarf (na na) in agreement with expected (χ22 = 0.08; P > 0.9). Therefore, the na-1 phenotype cosegregates with the mutation in the PsKAO1 gene.
Yeast (Saccharomyces cerevisiae) Expression
Yeast strains WAT11 and WAT21 were transformed with PsKAO1 and PsKAO2 expression constructs and yeast strains expressing PsKAO1 and PsKAO2 were identified by RNA gel blots. Both yeast strains expressing PsKAO1 and PsKAO2 converted KA through to GA12 at a greater rate than yeast expressing the Arabidopsis AtKAO1 (Table I). The intermediates ent-7α-hydroxykaurenoic acid and GA12-aldehyde, as well as the final product GA12, were detected (Table I) and confirmed to be authentic by comparison to known standards (Table II). The PsKAO2 construct expressed in both yeast strains appeared more effective than the PsKAO1 and converted all the KA substrate provided. Direct comparison may not be possible because the PsKAO1 construct may have four extra amino acids in the 5′-untranslated region because there were two possible start codons in the PsKAO1 sequence. GA53 and GA14 were not detected in any of the samples (Table I). WAT21 yeast transformed with PsKAO1 and PsKAO2 expression constructs fed with intermediates ent-7α hydroxy kaurenoic acid or GA12-aldehyde converted these substrates to GA12 although at a lower rate than expected from the KA feed data (data not shown). A substrate earlier in the GA biosynthetic pathway, ent-kaurene, was not metabolized by yeast expressing PsKAO1 or PsKAO2 (data not shown). Untransformed wild-type yeast did not metabolize KA to intermediates in the GA biosynthetic pathway (Table I).
Table I.
Putative products from WAT21 and WAT11 yeast strains expressing CYP88A cytochrome P450s
| Enzyme | Yeast | KA | ent-7α-OH KA | GA12-Aldehyde | GA12 | GA53 or GA14 | 7α-OH Kaurenolide | ent-6α,7α-diOH KA |
|---|---|---|---|---|---|---|---|---|
| TIC (×106) | ||||||||
| Wild-type yeast | W21 | 1,131 | n.d.a | n.d. | n.d. | n.d. | n.d. | n.d. |
| AtKAO1 | W21 | 1,204 | 49 | 15 | ≈2.5b | n.d. | n.d. | 18 |
| PsKAO1 | W21 | 702 | 414 | 22 | 84 | n.d. | ≈5b | 39 |
| PsKAO2 | W21 | n.d. | 520 | 108 | 265 | n.d. | ≈54b | 96 |
| PsKAO1 | W11 | 921 | 437 | 23 | 95 | n.d. | ≈5b | 42 |
| PsKAO2 | W11 | n.d. | 480 | 131 | 265 | n.d. | ≈49b | 79 |
The GC-MS total ion current (TIC) areas were measured for ent-7α-hydroxykaurenoic acid, GA12-aldehyde, GA12, GA53, GA14, 7β-hydroxy kaurenolide, and ent-6α,7α-dihydroxykaurenoic acid after KA feeds.
n.d., Not detected.
Approximate TIC area value where baseline is too high or peak is contaminated, calculated from the uncontaminated major ion and a conversion factor based on the appropriate standard.
Table II.
Authentication of GA biosynthetic intermediates and additional products in yeast extracts
| Reference or Putative Compounds | KRI | Characteristic Ions– Relative Abundance as % of Base Peak | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 404(M+) | 389 | 314 | 299 | 255 | 254 | 239 | 223 | 199 | ||
| ent-7α-hydroxy KA METMS standard | 2,459 | 22 | 8 | 100 | 32 | 69 | 59 | 43 | 19 | 22 |
| ent-7α-hydroxy KA METMS | 2,460 | 35 | 13 | 100 | 33 | 67 | 60 | 45 | 19 | 23 |
| 330(M+) | 287 | 270 | 255 | 242 | 241 | 239 | 227 | 199 | ||
| GA12 aldehyde ME standard | 2,437 | 12 | 14 | 40 | 14 | 24 | 100 | 24 | 14 | 10 |
| GA12 aldehyde ME | 2,436 | 11 | 14 | 40 | 15 | 25 | 100 | 24 | 15 | 11 |
| 360(M+) | 328 | 300 | 285 | 241 | 240 | 225 | 185 | |||
| GA12 ME standard | 2,415 | 1 | 17 | 100 | 24 | 35 | 29 | 20 | 12 | |
| GA12 ME | 2,416 | 1 | 17 | 100 | 22 | 35 | 31 | 26 | 15 | |
| 388(M+) | 345 | 298 | 283 | 270 | 255 | 227 | 163 | 137 | ||
| 7β-Hydroxykaurenolide TMS standard | 2,557 | 2 | 9 | 100 | 15 | 17 | 12 | 10 | 12 | 58 |
| 7β-Hydroxykaurenolide TMS | 2,552 | 1 | 10 | 100 | 17 | 22 | 14 | 13 | 27 | 95 |
| 492(M+) | 477 | 402 | 387 | 343 | 327 | 269 | 253 | 209 | ||
| ent-6α,7α-dihydroxy KA METMSa | 2,593 | <1 | 92 | 16 | 5 | 4 | 8 | 100 | 16 | 50 |
GC-MS relative ion abundances and Kovats' retention index (KRI) were compared with authentic standards. Samples are methyl ester (ME), trimethyl silyl (TMS), or methyl ester trimethyl silyl (METMS) derivatives.
Tentative identification based on comparison with published spectra (Gaskin and MacMillan, 1991).
The compound 7β-hydroxy-kaurenolide was detected and confirmed against authentic standard (Table II) in the samples with PsKAO1 and PsKAO2 activity after KA feeds but not when the intermediates ent-7α-hydroxykaurenoic acid or GA12-aldehyde were used as substrates. ent-6α,7α-dihydroxy-kaurenoic acid was detected (Table I) and tentatively identified based on comparison with published spectra (Gaskin and MacMillan, 1991; Table II) in yeast with PsKAO1, PsKAO2, or AtKAO1 activity when fed with KA. However, this product was also present after feeds of ent-7α-hydroxykaurenoic acid (data not shown). Further conversion to fujenoic acid was not observed. Neither 7β-hydroxy-kaurenolide nor ent-6α,7α-dihydroxy-kaurenoic acid were detected in the wild-type untransformed yeast samples and appeared to be a result of the KAO activity. However, in the wild-type untransformed yeast and yeast expressing AtKAO1 and PsKAO1 samples, the C/D ring-rearranged compounds, stachenoic acid and trachylobanic acid, were present in significant amounts. However, neither compound was present in the more active PsKAO2-expressing samples that appeared to metabolize these compounds to the ent-7α hydroxy and ent-6α,7α-dihydroxy derivatives and also through to GA12-like derivatives (Table III).
Table III.
Putative C/D ring rearranged products in untransformed yeast and yeasts expressing KAOs fed with KA
| Putative Compound | Present following Feed of KA to | KRI | Characteristic Ions (Relative Abundance as % of Base Peak) |
|---|---|---|---|
| Stachenoic acid ME | Wild-type yeast, AtKAO1, and PsKAO1 | 2,224 | 316 (75), 301 (8), 273 (5), 257 (22), 256 (11), 241 (11), 194 (27), 181 (24), 159 (20), 148 (39), 135 (100) |
| Trachylobanic acid ME | Wild-type yeast, AtKAO1, and PsKAO1 | 2,278 | 316 (77),301 (27), 273 (5), 260 (100), 257 (58), 256 (19), 245 (45), 241 (46), 201 (30), 200 (27) |
| ent-7α-hydroxy stachenoic acid METMSa | PsKAO1 and PsKA02 | 2,344 | 404 (25), 389 (7), 314 (51), 301 (18), 299 (15), 286 (76), 283 (63), 255 (54), 254 (44), 239 (32), 223 (42), 193 (24), 181 (46), 133 (97), 73 (100) |
| ent-7α-hydroxy trachylobanic acid METMSa | PsKAO1 and PsKA02 | 2,399 | 404 (2), 389 (4), 314 (100), 299 (17), 255 (55), 254 (60), 239 (41), 209 (31), 185 (43), 157 (65) |
| ent-6α,7α-dihydroxy stachenoic acid METMS | PsKA02 | 2,471 | 492 (<1), 477 (57), 402 (18), 387 (4), 343 (4), 327 (11), 269 (100), 253 (15), 209 (45) |
| ent-6α,7α-dihydroxy Trachylobanic acid METMS | PsKA02 | 2,531 | 492 (<1), 477 (18), 402 (12), 387 (3), 343 (2), 327 (4), 269 (100), 253 (17), 209 (40) |
| Stacheno-GA12 MEb | PsKA02 | 2,299 | 360 (4), 328 (47), 300 (63), 285 (22), 240 (34), 225 (53), 119 (100) |
| Trachylo-GA12 ME | PsKA02 | 2,360 | 360 (4), 328 (21), 300 (100), 285 (84), 241 (73), 240 (29), 225 (31), 164 (68), 119 (24) |
GC-MS relative ion abundances and KRI are indicated.
Tentatively identified as ent-7α-hydroxy stachenoic acid METMS and ent-7α-hydroxy trachylobanic acid METMS based on the similarities in their mass spectra with ent-7α-hydroxy KA METMS, and similar relative KRI values of ent-7α-stachenoic (putative)/ent-7α-trachylobanic (putative)/ent-7α-kaurenoic acid METMS compared with the stachenoic/trachylobanic/kaurenoic acid ME KRI values.
Tentatively identified as “stacheno-GA12 ME” because of the similarity of its spectra to GA12 ME and trachylo-GA12 ME, and similar relative KRI values to the two groups of three above.
DISCUSSION
PsKAO1 and PsKAO2 Encode CYP88A Cytochrome P450 Monooxygenases
Two genes, PsKAO1 and PsKAO2, encoding cytochrome P450 monooxygenases from the subfamily CYP88A were identified in pea. They have high similarity to the recently cloned genes AtKAO1 and AtKAO2 of Arabidopsis (Helliwell et al., 2001) and CmKAO1 of pumpkin (Helliwell et al., 2000). They are grouped with the GA biosynthetic KAO enzymes in the subfamily CYP88A as defined by the maize D3 enzyme (Winkler and Helentjaris, 1995). The KAOs from the dicotelydons (pea, Arabidopsis, and pumpkin) are grouped separately from those of the monocotyledons (rice, maize, and barley; Fig. 3). In addition, the two enzymes of pea are grouped, as are those of Arabidopsis, suggesting that gene duplication occurred late in the evolutionary process. The KAO-deduced proteins contain the four catalytic domains (A–D) common to eukaryotic cytochrome P450s (Kalb and Loper, 1988). However, a critical conserved Thr of the A domain that was shown in the crystal structure of P450cam to form a hydrogen bond with Gly to produce an oxygen-binding pocket (Poulos et al., 1985), and may also be involved in oxygen transfer (Imai et al., 1989), is replaced by Ser in all the KAOs sequenced so far. In P450cam, this Thr could be replaced by Ser in site-directed mutagenesis without altering the monooxygenase activity, whereas replacement with amino acids without the hydroxyl group uncoupled the oxygen consumption of the enzyme (Imai et al., 1989).
Pea NA Encodes PsKAO1
The evidence shows that the pea NA gene encodes PsKAO1. First, PsKAO1 from both the na-1 and na-2 GA-responsive dwarf mutants had altered sequences. The mutations na-1 and na-2 are allelic and produced by independent mutational events (Reid et al., 1983). Furthermore, the five-base deletion in PsKAO1 associated with the na-1 mutation cosegregated with the dwarf phenotype. PsKAO1 is expressed in the tissues where the na mutant phenotype is expressed. The mutant na plant has dwarfed stature, reduced taproot length, and reduced leaflet area (Reid et al., 1983; Reid and Ross, 1993; Yaxley et al., 2001). The GA1 levels are reduced in the shoot, roots, leaves, and pods (Potts and Reid, 1983; Ingram et al., 1984; Potts, 1986; Yaxley et al., 2001). PsKAO1 was expressed in stems, roots, leaves, apical buds, pods, and seeds (Fig. 4). However, the other pea CYP88A gene, PsKAO2, was only expressed in developing seeds and not any of the other tissues tested (Fig. 4). Because PsKAO2 was expressed in seeds, KAO activity could be expected in the seeds of the na mutant. This would explain the observation that mutant na plants have normal seed development and the same GA content in their seeds as wild-type pea seeds (Potts and Reid, 1983). In contrast, the two Arabidopsis genes have similar expression patterns. Probably because of this redundancy, no mutants have been found in Arabidopsis (Helliwell et al., 2001).
The na mutants are dwarfs with extreme reduction in internode lengths. The differences in internode lengths between WL1766 and L81 (Fig. 2) arise from differences in their genetic background (Reid et al., 1983). The putative proteins from the mutants (na-1 and na-2) are severely altered from wild-type PsKAO1. However, the double recessive with other GA biosynthetic mutants (na lh and na ls) are shorter than the single na mutant (Reid, 1986b). This could be because of limited activity by other KAO enzymes in pea shoots because extra bands were observed on genomic southern blots at low stringency (data not shown). Alternatively, there may be movement of intermediates from other tissues where the PsKAO2 gene is expressed. The latter is possible because the effect of the NA (PsKAO1) gene is graft transmissible (Reid et al., 1983; Proebsting et al., 1992) and mature pea seeds at sowing contain GA20 (Ross et al., 1993). The other KAO gene (PsKAO2) is expressed in the seed (Fig. 4, A and B) and the seeds of na mutant plants develop normally (Potts and Reid, 1983; Potts, 1986). This may provide a limited amount of GA precursors in the stem of the mutant na seedlings allowing the epicotyl to develop almost normally before the extreme dwarfism sets in (Reid et al., 1983). Furthermore, the phenotype of the double mutant seedlings appears to reflect the accumulation of GAs in the seed (na sln, Reid et al., 1992; Ross et al., 1995; na le, Reid et al., 1983; Lester et al., 1999a; and na ls and na lh, Swain et al., 1995).
PsKAO1 and PsKAO2 Catalyze KA to GA12
PsKAO1 and PsKAO2, when expressed in yeast, catalyzed the three steps from KA to ent-7α-hydroxykaurenoic acid to GA12-aldehyde to GA12 (Table I; Fig. 1). This was observed previously with AtKAO1 and AtKAO2 from Arabidopsis and HvKAO1 of barley (Helliwell et al., 2001). Many of the enzymes in the GA biosynthetic pathway are multifunctional (Hedden, 1997). This occurs normally where the enzyme catalyzes multiple oxidations at the same carbon position (Phillips et al., 1995; Xu et al., 1995; Helliwell et al., 1999) as occurs with all three oxidations from KA to GA12 at the C-7 position (Hedden, 1997; MacMillan, 1997; Helliwell et al., 2001).
The na Mutation Blocks GA Biosynthesis before GA12-Aldehyde
To see if the three-step oxidation of KA observed in the yeast expression studies is demonstrated in the plant, we can look at the corresponding mutants. The pea na-1 and na-2 mutants, which have a defective PsKAO1 gene, do not metabolize [3H] kaurenoic acid to substances co-eluting with GA20, GA1, or GA8, even though NA plants do carry out this conversion (Ingram and Reid, 1987). This supports the yeast expression data (Table I) and the finding that the barley mutant (grd5) accumulates KA in its seed (Helliwell et al., 2001). However, metabolism studies show that na plants can convert [2H] GA12-aldehyde to C19-GAs such as GA20, GA29, GA1, and GA8. In addition, application studies show that although na plants do not respond to precursors before GA12-aldehyde, including KA and ent-7α-hydroxy-kaurenoic acid, they do respond to GA12-aldehyde and readily convert labeled GA12-aldehyde to GA1 (Ingram and Reid, 1987). The na mutation, therefore, appears to block the first two biosynthetic steps but not the final GA12-aldehyde to GA12 step in the plant. This could occur if the nature of the na mutation altered the specificity of the PsKAO1 enzyme for the substrates. However, this is not likely because na-1 (WL1766) is a null mutation.
Alternatively, there may be other specific or nonspecific enzymes present in the plant that can catalyze the last step but not the earlier steps catalyzed by PsKAO1 and PsKAO2 when expressed in yeast. A GA 7-oxidase dioxygenase has been found in pumpkin (Lange, 1997) in addition to the monooxygenase 7-oxygenase activity (Hedden et al., 1984) but has not yet been found in other species (Hedden and Phillips, 2000). Perhaps the most likely explanation is that nonspecific activity may be involved because multigene families of aldehyde oxidases have been cloned from maize and Arabidopsis (Sekimoto et al., 1997, 1998) and some of these oxidize a wide range of aldehydes (Seo et al., 1998).
PsKAO1 and PsKAO2, when expressed in yeast, did not produce GA53 from any of the precursors provided, although the early 13-hydroxylation GA biosynthetic pathway predominates in pea (Ingram et al., 1986; Reid and Ross, 1993; Poole et al., 1995). In immature pea and barley embryos, the formation of GA53 from GA12 was associated with the microsomal fraction and required NADPH and oxygen (Kamiya and Graebe, 1983; Groβelindemann et al., 1992), suggesting that there may be another membrane-bound cytochrome P450 monooxygenase in pea catalyzing GA12 to GA53.
Additional Products of PsKAO Activity
In addition to ent-7α hydroxykaurenoic acid, GA12-aldehyde, and GA12, several side products of PsKAO1 and PsKAO2 activity were identified in the yeast expression studies. After either KA or ent-7α-hydroxykaurenoic acid feeds, the byproduct ent-6α,7α-dihydroxykaurenoic acid was detected (Fig. 1). We also detected this compound as a product of AtKAO1 activity. The compound, ent-6α,7α-dihydroxykaurenoic acid, was noted in pumpkin (Hedden, 1997; MacMillan, 1997) and related products were previously detected in pea (Ingram and Reid, 1987). The product 7β-hydroxykaurenolide was detected after KA (but not ent-7α-hydroxykaurenoic acid) feeds in yeast expressing PsKAOs and is presumably a side product of the formation of the double bond and epoxide from KA via ent-kauradienoic acid (Hedden, 1997). The P450–1 enzyme of Gibberella fujikuroi that catalyzes the four steps from KA to GA14 also produced 7β-hydroxykaurenolide and ent-6α,7α-dihydroxykaurenoic acid (Rojas et al., 2001). Fungi and higher plants appear to have evolved their GA biosynthetic pathway independently and P450–1 belongs to a different subfamily (CYP68) with low sequence homology to higher plant KAOs of subfamily CYP88A (Hedden et al., 2002). Because the additional products are common to both enzymes, they may be inevitable consequences of the reactions rather than specific products of the respective enzymes. In line with the expected difference between the fungi and higher plant KAO enzymes, the compound GA14 was not detected from PsKAO activity. It was interesting to note, however, that the C/D ring-rearranged stachenoic acid and trachylobanic acid can act as substrates for the PsKAO activity.
CONCLUSION
We have cloned two CYP88A genes in pea, PsKAO1 and PsKAO2. Both of these genes catalyze the three steps from KA to GA12 when expressed in yeast. The genes have distinct expression patterns. PsKAO1 is the pea NA gene and is expressed in the stem, apical bud, root, leaf, pod, and seed. Mutation in the PsKAO1 gene results in the extreme dwarf na phenotype. PsKAO2 is expressed in developing seeds, explaining the normal seed GA levels and seed development of na plants.
MATERIALS AND METHODS
Plant Material and Growing Conditions
Two independent mutational events in pea (Pisum sativum) resulted in the alleles na-1 and na-2 (Reid et al., 1983). The na-1 fast neutron induced recessive mutation is in the Weibullsholm line WL1766 (genotype na-1 LE LH LS) and the na-2 mutation is in the Hobart line L81 (genotype na-2 le LH LS). The tall NA WL1769 (genotype NA LE LH LS) was used as wild type and contains the progenitor sequence for the na-1 mutation. The na-1 and NA plants used for the cosegregation analysis were isogenic as a result of eight generations of single plant selection after a cross between lines WL1766 (na-1) and WL1769 (NA). Another wild-type Hobart line L107 (genotype NA LE LH LS) derived from pea cv Torsdag was used for some northern analyses.
Plants were grown two per pot in a heated greenhouse under an 18-h photoperiod (Beveridge and Murfet, 1996).
Library Screening
Both seed and shoot cDNA libraries were screened. The seed cDNA library was constructed in Lambda ZAPII (Stratagene, La Jolla, CA) with cDNA prepared from L107 pea cv Torsdag seeds at “contact point” (Ait-Ali et al., 1997). The library screening and the isolation of clones were according to methods recommended by the manufacturer (Stratagene). The shoot cDNA library was in Lambda gt11 prepared from pea cv Alaska apical buds (CLONTECH Laboratories, Palo Alto, CA). The library screening method was similar to above; however, inserts were obtained directly by PCR with nested vector primers from original pure clones. The probe used was a 339-nucleotide fragment from a maize (Zea mays) D3-like EST from soybean (Glycine max). The conserved 3′ end of soybean gi9483278 (BE657386), nearly identical to gi6915567 (AW397097), was 32P labeled using the Decalabel DNA labeling kit (MBI Fermentas, Burlington, ON, Canada).
Northern-Blot Analysis
Total RNA was extracted using either the Phenol/SDS Method (Ausubel et al., 1994; Fig. 4A) or the RNeasy Plant Kit (Qiagen USA, Valencia, CA; Fig. 4, B and C) consistent within the blot. The RNA (5 μg per lane) was fractionated in 1.5% (w/v) agarose gel containing formaldehyde and transferred to Genescreen Plus hybridization transfer membrane (PerkinElmer Life Sciences, Boston) using 10× SSC. The membrane was hybridized with a 32P-labeled cDNA fragment of PsKAO1 or PsKAO2 at 42°C in 5× SSC, 5× Denhardts, 50% (w/v) formamide, 1% (w/v) SDS, and 200 μg mL−1 salmon sperm. The membrane was washed in 2× SSC and 0.1% (w/v) SDS, then 0.2× SSC and 0.1% (w/v) SDS at 65°C and exposed to Biomax x-ray film (Eastman-Kodak, Rochester, NY) at −70°C.
Cosegregation Analysis
DNA was extracted from the leaves of 50 individuals of four segregating families from the F9 generation of cross WL1766 (na-1) × WL1769 (NA). The genomic PsKAO1 PCR products (243 bp in the wild type) were visualized using a 5′ primer labeled with the fluorescent dye, hexachloro fluorescein. The PCR fragment encompassed the five-base deletion of the na-1 mutant gene. The PCR products were denatured (94°C for 3 min) in loading buffer containing deionized formamide and bromphenol blue, then placed on ice before loading on a denaturing gel (5% [w/v] acrylamide gel in 0.6× Tris-borate/EDTA buffer containing 7 m urea) in the Gel-Scan 2000 (Corbett Research, Sydney).
Yeast (Saccharomyces cerevisiae) Expression
The constructs were prepared in the pYEDP60 plasmid vector (Pompon et al., 1996). Oligonucleotide primers with restriction sites incorporated at the 5′ end were designed and checked with the aid of the Oligo Primer Analysis Software (version 6.74, Molecular Biology Insights, Cascade, CO). The PsKAO1 and PsKAO2 cDNA were prepared from RNA extracted from WL1769 stems or L107 seeds, respectively. The cDNA PCR products encompassed the putative protein-coding sequence with the 5′-untranslated region as short as possible and were amplified using Pfu Turbo DNA polymerase (Stratagene). These PCR products were cloned into pGEM-T vector (Promega, Madison, WI) and sequenced to check for PCR-generated mutations. Selected clones were digested using restriction enzymes corresponding to the sites introduced in the PCR primers and ligated into pYEDP60 vector in the sense orientation with reference to the GAL10-CYC1 promoter (Pompon et al., 1996). An Arabidopsis AtKAO1 construct was used for comparison (Helliwell et al., 2001). The WAT11 and WAT21 yeast lines that are modified to express Arabidopsis NADPH-cytochrome P450 reductases, ATR1 and ATR2-1, respectively (Pompon et al., 1996; Urban et al., 1997), were transformed with the construct plasmids (Cullin and Pompon, 1988). The transformed yeasts and untransformed yeast as a control were incubated with 10 μg of the substrates (KA, ent-7α hydroxy kaurenoic acid, GA12-aldehyde, or ent-kaurene) for 2 h at 28°C (Helliwell et al., 1999). In preparation for GC-MS analysis, methylation or trimethylsilylation was required. Extracts in about 2 mL of hexane/EtOAc were dried almost completely by speed vacuum, then to completion under nitrogen. Methylation was in the same test tubes by addition of 50 μL of MeOH and 400 μL of ethereal diazomethane. Samples were left for 15 min, dried as before, then transferred to reactivials using 4 × 50 μL EtOAc. These were dried and then trimethylsilylated using 5 μL of pyridine and 5 μL of bis(TMS) trifluoroacetamide + 1% (w/v) trimethylchlorosilane, which was heated at 90°C for 30 min (Helliwell et al., 1999). Injections were 1-μL samples with 0.1-μL parafilm standard. The KRIs were calculated using hydrocarbon peaks from the co-injected parafilm standard. Identities of products were confirmed by GC-MS comparison of spectra and KRI with authentic standards where possible. Alternatively, some side products were tentatively identified based on comparison with published spectra (Gaskin and MacMillan, 1991) and relative KRI values.
ACKNOWLEDGMENTS
We thank Jenny Smith and Adam J. Smolenski (University of Tasmania, Hobart, Australia) for technical assistance in the molecular laboratory, Ian Cummings and Tracey Jackson (University of Tasmania) for greenhouse assistance, Denis Pompon (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France) for the WAT11 and WAT21 yeast strains, Bruce Twitchin and Professor Lewis Mander (Australian National University, Canberra) for the provision of authentic GA standards. We would also like to thank Dr. John Ross and Dr. L. Huub Kerckhoffs (University of Tasmania) for helpful discussions.
Footnotes
This work was supported by the Australian Research Council (grants to J.B.R.). S.E.D. was the recipient of an Australian Postgraduate Award.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.012963.
LITERATURE CITED
- Ait-Ali T, Swain SM, Reid JB, Sun TP, Kamiya Y. The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J. 1997;11:443–454. doi: 10.1046/j.1365-313x.1997.11030443.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JG, Struhl K. Current Protocols in Molecular Biology 1. New York: Wiley Interscience; 1994. [Google Scholar]
- Beveridge CA, Murfet IC. The gigas mutant in pea is deficient in the floral stimulus. Physiol Plant. 1996;96:637–645. [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JDG, Kamiya Y. The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc Natl Acad Sci USA. 1999;96:1761–1766. doi: 10.1073/pnas.96.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JWS. Arabidopsis intron mutations and pre-mRNA splicing. Plant J. 1996;10:771–780. doi: 10.1046/j.1365-313x.1996.10050771.x. [DOI] [PubMed] [Google Scholar]
- Choe SW, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22 alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullin C, Pompon D. Synthesis of functional mouse cytochromes P-450 P1 and chimeric P-450 P3–1 in the yeast Saccharomyces cerevisiae. Gene. 1988;65:203–217. doi: 10.1016/0378-1119(88)90457-x. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman VM, Gaskin P, MacMillan J. Qualitative and quantitative analyses of gibberellins throughout seed maturation in Pisum sativum cv. Progress no.9. Planta. 1974;118:123–132. doi: 10.1007/BF00388388. [DOI] [PubMed] [Google Scholar]
- Gaskin P, MacMillan J. GC-MS of the gibberellins and related compounds: methodology and a library of spectra. Bristol, UK: University of Bristol, Cantock's Enterprises; 1991. [Google Scholar]
- Groβelindemann E, Lewis MJ, Hedden P, Graebe JE. Gibberellin biosynthesis from gibberellin A12-aldehyde in a cell-free system from germinating barley (Hordeum vulgare L, Cv Himalaya) embryos. Planta. 1992;188:252–257. doi: 10.1007/BF00216821. [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, MacIntosh GC, Green PJ. Current perspectives on mRNA stability in plants: multiple levels and mechanisms of control. Trends Plant Sci. 1999;4:429–438. doi: 10.1016/s1360-1385(99)01484-3. [DOI] [PubMed] [Google Scholar]
- Hedden P. The oxidases of gibberellin biosynthesis: their function and mechanism. Physiol Plant. 1997;101:709–719. [Google Scholar]
- Hedden P, Graebe JE, Beale MH, Gaskin P, MacMillan J. The biosynthesis of 12α-hydroxylated gibberellins in a cell-free system from Cucurbita maxima endosperm. Phytochemistry. 1984;23:569–574. [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B. Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J Plant Growth Regul. 2002;20:319–331. doi: 10.1007/s003440010037. [DOI] [PubMed] [Google Scholar]
- Hedden P, Proebsting WM. Genetic analysis of gibberellin biosynthesis. Plant Physiol. 1999;119:365–370. doi: 10.1104/pp.119.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA. 2001;98:2065–2070. doi: 10.1073/pnas.041588998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Olive MR, Gebbie L, Forster R, Peacock WJ, Dennis ES. Isolation of an ent-kaurene oxidase cDNA from Cucurbita maxima. Aust J Plant Physiol. 2000;27:1141–1149. [Google Scholar]
- Helliwell CA, Poole A, Peacock WJ, Dennis ES. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 1999;119:507–510. doi: 10.1104/pp.119.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Sheldon CC, Olive MR, Walker AR, Zeevaart JAD, Peacock WJ, Dennis ES. Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc Natl Acad Sci USA. 1998;95:9019–9024. doi: 10.1073/pnas.95.15.9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooley R. Gibberellins: perception, transduction and responses. Plant Mol Biol. 1994;26:1529–1555. doi: 10.1007/BF00016489. [DOI] [PubMed] [Google Scholar]
- Imai M, Shimada H, Watanabe Y, Matsushima-Hibiya Y, Makino R, Koga H, Horiuchi T, Ishimura Y. Uncoupling of the cytochrome P-450cam monooxygenase reaction by a single mutation, threonine-252 to alanine or valine: possible role of the hydroxy amino acid in oxygen activation. Proc Natl Acad Sci USA. 1989;86:7823–7827. doi: 10.1073/pnas.86.20.7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram TJ, Reid JB. Internode length in Pisum1. Gene na may block gibberellin synthesis between ent-7-alpha-hydroxykaurenoic acid and gibberellin A12-aldehyde. Plant Physiol. 1987;83:1048–1053. doi: 10.1104/pp.83.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram TJ, Reid JB, MacMillian J. The quantitative relationship between gibberellin A1 and internode growth in Pisum sativum L. Planta. 1986;168:414–420. doi: 10.1007/BF00392370. [DOI] [PubMed] [Google Scholar]
- Ingram TJ, Reid JB, Murfet IC, Gaskin P, Willis CL, MacMillian J. Internode length in Pisum. Planta. 1984;160:455–463. doi: 10.1007/BF00429763. [DOI] [PubMed] [Google Scholar]
- Kalb VF, Loper JC. Proteins from eight eukaryotic cytochrome P-450 families share a segmented region of sequence similarity. Proc Natl Acad Sci USA. 1988;85:7221–7225. doi: 10.1073/pnas.85.19.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya Y, Graebe JE. The biosynthesis of all major pea gibberellins in a cell-free system from Pisum sativum. Phytochemistry. 1983;22:681–689. [Google Scholar]
- Lange T. Cloning gibberellin dioxygenase genes from pumpkin endosperm by heterologous expression off enzyme activities in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:6553–6558. doi: 10.1073/pnas.94.12.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, MacKenzie-Hose AK, Davies PJ, Ross JJ, Reid JB. The influence of the null le-2 mutation on gibberellin levels in developing pea seeds. Plant Growth Regul. 1999a;27:83–89. [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB. Mendels stem length gene (Le) encodes a gibberellin 3-beta-hydroxylase. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB. Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J. 1999b;19:65–73. doi: 10.1046/j.1365-313x.1999.00501.x. [DOI] [PubMed] [Google Scholar]
- MacMillan J. Biosynthesis of the gibberellin plant hormones. Nat Prod Rep. 1997;14:221–243. [Google Scholar]
- Martin DN, Proebsting WM, Hedden P. Mendel's dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc Natl Acad Sci USA. 1997;94:8907–8911. doi: 10.1073/pnas.94.16.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P. The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol. 1999;121:775–781. doi: 10.1104/pp.121.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NEJ, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of 3 gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Poole AT, Ross JJ, Lawrence NL, Reid JB. Identification of gibberellin A4 in Pisum sativum L. and the effects of applied gibberellins A9, A4, A5 and A3 on the le mutant. Plant Growth Regul. 1995;16:257–262. [Google Scholar]
- Potts WC. Gibberellins in light-grown shoots of Pisum sativum L. and the influence of reproductive development. Plant Cell Physiol. 1986;27:997–1003. [Google Scholar]
- Potts WC, Reid JB. Internode length in Pisum: III. The effect and interaction of the Na/na and Le/le gene differences on endogenous gibberellin-like substances. Physiol Plant. 1983;57:448–454. [Google Scholar]
- Poulos TL, Finzel BC, Gunsalus IC, Wagner GC, Kraut J. The 2.6-A crystal structure of Pseudomonas putida cytochrome P-450. J Biol Chem. 1985;260:16122–16130. [PubMed] [Google Scholar]
- Proebsting WM, Hedden P, Lewis MJ, Croker SJ, Proebsting LN. Gibberellin concentration and transport in genetic lines of pea: effects of grafting. Plant Physiol. 1992;100:1354–1360. doi: 10.1104/pp.100.3.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB. Gibberellin mutants. In: King PJ, Blonstein AD, editors. Plant Gene Research: A Genetic Approach to Plant Biochemistry. Vol. 3. New York: Springer-Verlag; 1986a. pp. 1–34. [Google Scholar]
- Reid JB. Internode length in Pisum. Three further loci, lh, ls and lk. Ann Bot. 1986b;57:577–592. [Google Scholar]
- Reid JB, Murfet IC, Potts WC. Internode Length in Pisum: II. Additional information on the relationship and action of loci Le, La, Cry, Na, and Lm. J Exp Bot. 1983;34:349–364. [Google Scholar]
- Reid JB, Ross JJ. A mutant based approach, using Pisum sativum, to understand plant growth. Int J Plant Sci. 1993;154:22–34. [Google Scholar]
- Reid JB, Ross JJ, Swain SM. Internode length in Pisum. A new, slender mutant with elevated levels of C19 gibberellins. Planta. 1992;188:462–467. doi: 10.1007/BF00197036. [DOI] [PubMed] [Google Scholar]
- Rojas MC, Hedden P, Gaskin P, Tudzynki B. The P450–1 gene of Gibberella fujikuroi encodes a multifunctional enzyme in gibberellin biosynthesis. Proc Natl Acad Sci USA. 2001;98:5838–5843. doi: 10.1073/pnas.091096298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, Murfet IC, Reid JB. Gibberellin mutants. Physiol Plant. 1997;100:550–560. [Google Scholar]
- Ross JJ, Reid JB, Swain SM. Control of stem elongation by gibberellin A1: evidence from genetic studies including the slender mutant sln. Aust J Plant Physiol. 1993;20:585–599. [Google Scholar]
- Ross JJ, Reid JB, Swain SM, Hasan O, Poole AT, Hedden P, Willis CL. Genetic regulation of gibberellin deactivation in Pisum. Plant J. 1995;7:513–523. [Google Scholar]
- Sekimoto H, Seo M, Dohmae N, Takio K, Kamiya Y, Koshiba T. Cloning and molecular characterization of plant aldehyde oxidase. J Biol Chem. 1997;272:15280–15285. doi: 10.1074/jbc.272.24.15280. [DOI] [PubMed] [Google Scholar]
- Sekimoto H, Seo M, Kawakami N, Komano T, Desloire S, Liotenberg S, Marion-Poll A, Caboche M, Kamiya Y, Koshiba T. Molecular cloning and characterization of aldehyde oxidases in Arabidopsis thaliana. Plant Cell Physiol. 1998;39:433–442. doi: 10.1093/oxfordjournals.pcp.a029387. [DOI] [PubMed] [Google Scholar]
- Seo M, Akaba S, Oritani T, Delarue M, Bellini C, Caboche M, Koshiba T. Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana. Plant Physiol. 1998;116:687–693. doi: 10.1104/pp.116.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponsel VM, Schmidt FW, Porter SG, Nakayama M, Kohlstruk S, Estelle M. Characterization of new gibberellin-responsive semidwarf mutants of Arabidopsis. Plant Physiol. 1997;115:1009–1020. doi: 10.1104/pp.115.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Reid JB, Kamiya Y. Gibberellins are required for embryo growth and seed development in pea. Plant J. 1997;12:1329–1338. [Google Scholar]
- Swain SM, Reid JB, Ross JJ. Seed development in Pisum: The lhi allele reduces gibberellin levels in developing seeds, and increases seed abortion. Planta. 1993;191:482–488. [Google Scholar]
- Swain SM, Ross JJ, Reid JB, Kamiya Y. Gibberellins and pea seed development: expression of the lhi, ls and le5839 mutations. Planta. 1995;195:426–433. [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, MA: Sinauer Associates; 1999. [Google Scholar]
- Szekeres M, Nemeth K, KonczKalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Talon M, Koornneef M, Zeevaart JAD. Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc Natl Acad Sci USA. 1990;87:7983–7987. doi: 10.1073/pnas.87.20.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem. 1997;272:19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- Winkler RG, Helentjaris T. The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in gibberellin biosynthesis. Plant Cell. 1995;7:1307–1317. doi: 10.1105/tpc.7.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Li L, Wu KQ, Peeters AJM, Gage DA, Zeevaart JAD. The Ga5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaxley JR, Ross JJ, Sherriff LJ, Reid JB. Gibberellin biosynthesis mutations and root development in pea. Plant Physiol. 2001;125:627–633. doi: 10.1104/pp.125.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]