Abstract
Nitrogen (N) fixation and assimilation in pea (Pisum sativum) root nodules were studied by in vivo 15N nuclear magnetic resonance (NMR) by exposing detached nodules to 15N2 via a perfusion medium, while recording a time course of spectra. In vivo 31P NMR spectroscopy was used to monitor the physiological state of the metabolically active nodules. The nodules were extracted after the NMR studies and analyzed for total soluble amino acid pools and 15N labeling of individual amino acids by liquid chromatography-mass spectrometry. A substantial pool of free ammonium was observed by 15N NMR to be present in metabolically active, intact nodules. The ammonium ions were located in an intracellular environment that caused a remarkable change in the in vivo 15N chemical shift. Alkalinity of the ammonium-containing compartment may explain the unusual chemical shift; thus, the observations could indicate that ammonium is located in the bacteroids. The observed 15N-labeled amino acids, glutamine/glutamate and asparagine (Asn), apparently reside in a different compartment, presumably the plant cytoplasm, because no changes in the expected in vivo 15N chemical shifts were observed. Extensive 15N labeling of Asn was observed by liquid chromatography-mass spectrometry, which is consistent with the generally accepted role of Asn as the end product of primary N assimilation in pea nodules. However, the Asn 15N amino signal was absent in in vivo 15N NMR spectra, which could be because of an unfavorable nuclear Overhauser effect. γ-Aminobutyric acid accumulated in the nodules during incubation, but newly synthesized 15N γ-aminobutyric acid seemed to be immobilized in metabolically active pea nodules, which made it NMR invisible.
Symbiotic nitrogen (N) fixation, the process whereby N2-fixing bacteria enter into associations with plants, provides the major source of N for the biosphere. Nitrogenase, a bacterial enzyme, catalyzes the reduction of atmospheric dinitrogen to ammonium. In rhizobia-leguminous plant symbioses, a widely accepted and simple model of N transfer from the symbiotic form of the bacterium, called a bacteroid, to the plant implies that nitrogenase-generated ammonia diffuses across the bacteroid membrane and is assimilated into amino acids in the plant compartment of the nodule tissue. However, the transport of symbiotically fixed N across the membranes surrounding the bacteroid and the form in which this occurs has been a matter of controversy.
Until recently, it has been generally accepted that Rhizobium bacteroids do not assimilate ammonium into amino acids to a great extent during symbiosis, but more recent results challenge this view and suggest a possible involvement of amino acids as the form of fixed N delivered from the bacteroid to the plant (for review, see Poole and Allaway, 2000; Day et al., 2001). At present, consensus has not been reached as to whether substantial N assimilation takes place in the nodule bacteroid compartment, and new investigations of nodule N metabolism are required.
The size of the free ammonium pool in the bacteroid and plant cytoplasm during symbiosis is an open question. It has been suggested that ammonium synthesis and subsequent assimilation by Ala dehydrogenase are so tightly coupled in the bacteroid cytoplasm that very little free ammonium is released (Waters et al., 1998). On the other hand, Allaway et al. (2000) observed a plastic partitioning of ammonium and Ala excretion from isolated bacteroids and suggested that the ammonium concentration inside the bacteroids was one of the key variables governing the rate of Ala synthesis. Streeter (1989) estimated the free ammonium concentration in bacteroids to be as high as 12 mm, but the estimate was based on an extrapolation of time-dependent experimental data to time zero and numerous other assumptions, and as a consequence, there is a need for a more direct determination of the concentration of free ammonium inside the bacteroid.
An overall problem in the study of nodule metabolism is the extrapolation from in vitro to in vivo. Information on the microenvironment of the different compartments in the nodule is lacking, and the in vivo significance of in vitro findings, therefore, is difficult to predict. Although determinations of enzyme activities in crude extracts may give a first clue as to which ammonium assimilation pathways are active, they cannot be used to predict the in vivo flux distribution over competing enzyme systems such as Glu dehydrogenase and Gln synthetase/Glu synthase. In vivo NMR spectroscopy, especially when used in combination with stable isotope labeling such as 15N, does allow the characterization of metabolic activities in the living cell.
A crucial factor in studies of bacteroid metabolism and functioning in vitro is the degree to which bacteroid preparations are free from contaminating plant organelles, enzymes, and metabolites. Waters et al. (1998) suggested that the presence of plant enzymes in earlier in vitro studies of bacteroids had caused significant artifacts in the results concerning metabolic products released from bacteroids, but this was subsequently contradicted by Li et al. (2000). Likewise, major errors might occur in the determination of metabolite levels because of reallocation or degradation of metabolites during extraction, processing of extracts, or separation of compartments (Streeter, 1987, 1989; Miller et al., 1991). Metabolic studies by in vivo NMR spectroscopy have the advantage of avoiding artifacts caused by the breakdown of labile compounds during extraction and it also eliminates errors because of incomplete recovery of solutes from the tissue.
15N NMR spectroscopy has, to our knowledge, not been used previously for studying living, N2-fixing organisms. The only reported 15N NMR investigation of 15N2 fixation is by Belay et al. (1988), who incubated methanogenic bacteria in 15N2 and subsequently analyzed harvested, dead cells by 15N NMR. The literature contains a few in vivo 31P NMR spectroscopic studies of symbiotic soybean (Glycine max; Mitsumori et al., 1985; Rolin et al., 1989a, 1989b) and alfalfa (Medicago sativa) root nodules (Nikolaev et al., 1994). Results from these investigations demonstrate that it is possible to maintain nodules in a metabolically active state, while recording NMR spectra.
In conclusion, accurate estimation of the assimilation and translocation of fixed N in nodules would benefit from noninvasive and nondestructive measurements of the fate of the fixed N. Thus, the objective of the present study was to investigate N fixation and assimilation in 15N2-fixing root nodules by in vivo 15N NMR spectroscopy. This included optimization of experimental conditions for maximal 15N incorporation. The investigation presents in vivo 15N NMR spectra of 15N2-fixing root nodules, which in combination with liquid chromatography (LC)-mass spectrometry (MS) results, demonstrate the intracellular buildup of 15N-ammonium and 15N-amino acids.
RESULTS
Optimization of Experimental Conditions for Maximal 15N Incorporation
Factors influencing the 15N incorporation were optimized to make the in vivo signal-to-noise ratio of 15N-labeled metabolites exceed the 15N NMR detection threshold. The 15N enrichment of the root nodules depends primarily on the level of nitrogenase activity, which again depends on inherent properties of the biological material. The proportion of N2-fixing nodule tissue varies with age, and nodules from pea (Pisum sativum) plants of different ages were tested. However, no major differences could be observed in in vivo 15N NMR spectra of nodules from plants aged between 4 and 7 weeks, and 6- to 7-week-old plants were used throughout the study. The root system of a given plant contains nodules of different sizes and developmental stages. The size of the NMR tube limits the amount of nodule tissue that can be included in an experiment, and because smaller nodules may be packed more densely in the tube, they might give rise to a higher total 15N2-fixing rate per volume unit, although more mature nodules are known to contain a larger zone of N2-fixing tissue. We tested whether small nodules from the younger part of the root system or large, older nodules from the top of the tap root gave better 15N NMR spectra and it was found that large nodules gave rise to a higher total 15N2 fixation rate per volume unit. In addition, different pea varieties in combination with different bacterial strains were tested and the most efficient combination (see “Materials and Methods”) was used for all experiments described.
It was tested whether nodules from Lotus japonicum plants gave rise to more informative in vivo 15N NMR spectra, but the obtained spectra displayed less intense signals than spectra from pea nodules, and further experiments with L. japonicum nodules, therefore, were abandoned. However, the L. japonicum 15N spectra provided valuable information that aided the assignment of 15N resonances as described below.
Root nodules in the perfusion system were immersed in buffer, and this is well known to cause a decrease in nitrogenase activity, which, however, may be partially recovered by increasing the oxygen supply to the nodules (Sprent, 1969). The O2 availability for the nodules in the NMR tube depends on a number of factors such as the composition and pressure of the gas phase, the equilibrium between the perfusion buffer and the gas phase, the perfusion buffer flow rate, and the packing of nodules in the NMR tube. An adequate flow rate must be maintained to prevent significant gradients of oxygen tension across the sample because of both depletion of oxygen by cells nearest the medium inlet, and nonuniform flow over individual pieces of tissue. The most intense in vivo 15N signals occurred when nodules were perfused at a flow rate of 40 to 50 mL min−1 of a buffer in equilibrium with a gas phase containing about 50% (v/v) O2 and a total gas pressure of 1.6 atm. When the O2 concentration was raised to approximately 70% (v/v), less intense 15N NMR signals were observed, which indicated an inhibition of nitrogenase activity.
The total amount of 15N incorporated by pea nodules during an 8-h incubation period in the perfusion system was determined by MS to be 1.97 μmol 15N g−1 fresh weight. Thus, the mean nitrogenase activity could be estimated to be 0.12 μmol 15N2 g−1 fresh weight h−1. The nitrogenase activity was observed to be higher in the beginning of the incubation and decreasing throughout the incubation period in other experiments (data not shown).
Monitoring of Physiological State of Root Nodules
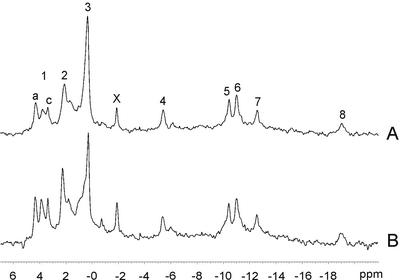
During NMR experiments, the energy status of root nodules and possible changes in intracellular pH, which may indicate hypoxic conditions, were monitored by acquiring 31P NMR spectra. Figure 1 shows representative in vivo 31P spectra obtained at the beginning (0 h) and end (8 h) of the 15N experiment shown in Figure 2. ATP concentrations did not change throughout the experiment, as seen from the unchanged intensity of the well-resolved signals from the β- and γ-phosphate groups of ATP at −19.0 and −5.5 ppm, respectively. At optimal N2-fixation conditions, nodules have ATP to ADP ratios, which are lower than those normally found in fully aerobic plant tissues (Kuzma et al., 1999), and the presence of ADP in the nodule tissue is demonstrated by the weak β-ADP signal at −6.2 ppm. The intracellular pH can be estimated from the chemical shift of the inorganic phosphate (Pi) signal based on calibration curves (see “Materials and Methods”), and we observed that the apparent pH in the cytoplasm as well as in the vacuole remained stable during the course of the experiment. The pH values in the cytoplasm and vacuole were estimated to be 7.2 and 5.2, respectively, from the Pi chemical shifts at 2.1 and 0.3 ppm, respectively. The cytoplasmic compartment at pH 7.2 might include both the plant and the bacteroid cytoplasm. The resonance at 1.6 ppm could represent Pi in a different subcellular compartment with a pH of approximately 6.8, or it could be a signal from one of the phosphate groups of phytate. Another possible signal from phytate was observed at 1.0 ppm, but the exact position of the phytate multiplet is highly sensitive to pH and to the chemical environment, making its correct identification difficult (Saint-Ges et al., 1991). However, it seems likely that the two small peaks represent phytate because 31P spectra of perchloric acid extracts from root nodules also contained phytate resonances of approximately the same intensity (data not shown).
Figure 1.
In vivo 31P NMR spectra of pea root nodules showing the unchanged metabolic status during an 8-h incubation period. The 30-min spectra were recorded before the addition of 15N2 (A) and at the end of the treatment period (B) after acquisition of the in vivo 15N spectra shown in Figure 2. The numbered peaks may be assigned to: 1, several phosphomonoesters including Glc 6-phosphate (1a) and phosphocholine (1c); 2, cytoplasmic Pi; 3, vacuolar Pi; X, unidentified compound; 4, 5, and 8, the γ-, α- and β-phosphates of nucleoside triphosphate; 6, UDP-Glc and NAD(P)(H); and 7, UDP-Glc. Chemical shifts are quoted relative to 85% (w/v) H3PO4 at 0 ppm.
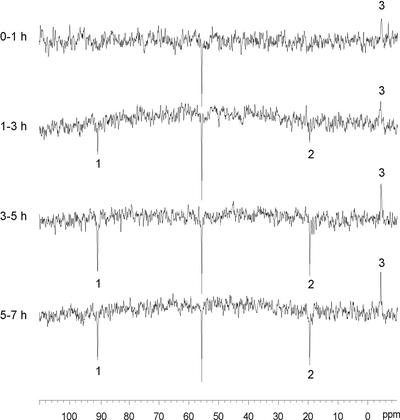
Figure 2.
In vivo 15N NMR spectra showing a time course of N assimilation in 15N2-fixing pea root nodules. The numbered peaks may be assigned to: 1, Asn amide-N; 2, Gln/Glu amino-N; and 3, ammonium. The signal at 55.8 ppm is from the reference compound 15N-urea.
Ammonium in Metabolically Active Nodules
Incubation of metabolically active root nodules in 15N2 in the perfusion system while recording 15N NMR spectra resulted in the observation of intracellular 15N ammonium in experiments with both pea and L. japonicum (see Table I; Fig. 2). The resonance frequency showed some variability in the range from −4.2 to −3.9 ppm. This is an unusually low ammonium frequency compared with in vitro measurements and previous in vivo 15N NMR studies, which all have demonstrated ammonium resonances at around 0 ppm in prokaryotic as well as in eukaryotic organisms (Legerton et al., 1981; Fox et al., 1992; Aarnes et al., 1995; Tesch et al., 1999). A spectrum from an extract of 15N-enriched pea root nodules showed a small signal at −0.14 ppm, which was assigned to ammonium (Table I).
Table I.
Assignments of amino acid and ammonium 15N chemical shifts
| Sample | D2O | Gln Amide | Asn Amide | Ala | Gln Amino | Glu | Asn Amino | Asp | γ-Aminobutyric Acid (GABA) | NH4+ |
|---|---|---|---|---|---|---|---|---|---|---|
| % | ppm | |||||||||
| Authentic 15N standards (pH 7) | 10 | 90.86 | 90.51 | – | 19.85 | 19.65 | 18.90 | 18.34 | 11.75 | −0.26 |
| Dead 15N-enriched pea nodules immersed in buffer (pH 6) | 20 | – | 90.6 | – | – | – | 18.8 | – | 11.6 | – |
| Extract from dead nodules (pH 5.8) | 4 | – | 90.48 | 22.06 | – | – | 19.07 | – | 11.79 | −0.14 |
| Metabolically active, 15N2-fixing pea nodules | 10 | – | 90.7 | – | 19.6 | 19.6 | – | – | – | −4.2 |
| Metabolically active, 15N2-fixing L. japonicum nodules | 10 | – | 90.7 | – | 19.6 | 19.6 | 18.4 | 18.4 | 11.3 | −4.1 |
The in vivo signal at around −4 ppm was assigned to ammonium based on several indications. First, no other well-known 15N metabolite resonates anywhere near −4 ppm. Second, the signal at −4 ppm represents the first detectable 15N metabolite in 15N2-fixing nodules (see Fig. 2), which is in accordance with the well-established fact that the product of nitrogenase activity is ammonium. Third, the nuclear Overhauser effect (NOE) factor is between −1 and 0, which is characteristic for ammonium in vivo (Martin, 1985; Robinson et al., 1991; Fox et al., 1992; Joy et al., 1997) and gives rise to spectra with oppositely phased signals for amino acids and ammonium, respectively. Finally, the signal at −4 ppm is intense despite being attenuated by NOE under the applied acquisition parameters; thus, it must originate from a metabolite that is present in a large amount, which is what should be expected from ammonium in the bacteroid compartment.
Acquisition of 15N NMR spectra from the perfusion buffer after the incubation was ended demonstrated that no extracellular 15N-labeled ammonium was present (data not shown).
Total Amino Acid Pools and 15N Labeling
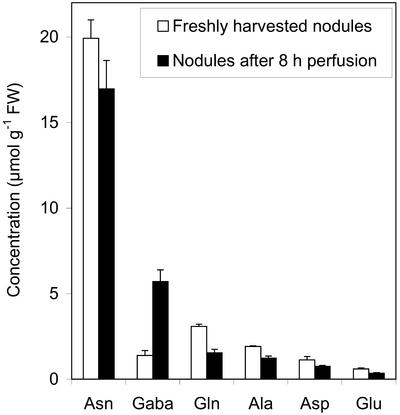
Pea root nodules contained Asn as the dominating free amino acid, and its concentration amounted to as much as 19.9 μmol g−1 fresh weight in freshly harvested nodules and 16.9 μmol g−1 fresh weight in nodules that had been perfused for 8 h during the NMR experiment (Fig. 3). This is an order of magnitude higher than each of the other major amino acids (Gln, Ala, Asp, and Glu). In general, the concentrations of free α-amino acids were slightly lower in perfused nodules than in freshly harvested nodules. GABA constituted a substantial part of the soluble pool of N-containing metabolites in freshly harvested nodules (1.4 μmol g−1 fresh weight) and the pool size apparently increased substantially during perfusion (5.7 μmol g−1 fresh weight).
Figure 3.
Concentrations of soluble amino acids in freshly harvested pea root nodules and nodules that have been subjected to perfusion for 8 h. Error bars = ses (n = 3).
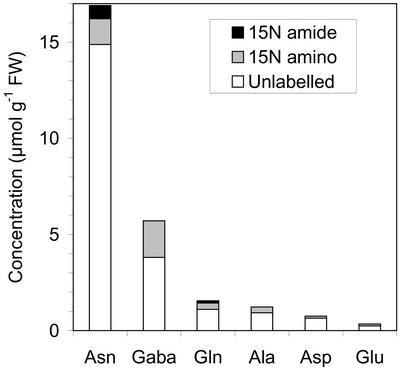
A new mass spectrometric technique involving separation of amino acids by LC made it possible to analyze the position of 15N in monolabeled Gln and Asn by using MS/MS and MS/MS/MS, respectively (A.M. Scharff, C. Schou, and H. Egsgaard, unpublished data). The analysis can be carried out on-line in a single experiment without any previous derivatization procedures, which is a major simplification and improvement compared with GC-MS procedures, which are usually used for analysis of positional labeling. GABA, Glu, and Ala were the most highly 15N-enriched compounds after the 8-h incubation period with 15N in excess of 33.4, 27.9, and 25.0 atom%, respectively, but all examined compounds were found to be significantly 15N-labeled (Table II). When the pool sizes were taken into account, Asn and GABA were found to contain most of the 15N label in soluble amino acids in the nodule tissue, namely 2.03 and 1.91 μmol 15N g−1 fresh weight, respectively (Table II; Fig. 4). Ala and Glu contained a minor part of the total 15N label (0.31 and 0.10 μmol 15N g−1 fresh weight, respectively), which may be ascribed to their smaller pool sizes. Asn contained more 15N label in the amino group (1.33 μmol 15N g−1 fresh weight) than in the amide group (0.70 μmol 15N g−1 fresh weight). Gln was also more labeled in the amino group (0.34 μmol 15N g−1 fresh weight) than in the amide group (0.11 μmol 15N g−1 fresh weight).
Table II.
15N labeling of soluble amino acids in pea root nodules after 8 h of incubation in 15N2 in the perfusion system
| Amino Acid | Total Amino Acid Concentration
|
15N Enrichment
|
Position of Mono-15N Label
|
Total 15N Concentrationa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Meanb | se | Mean | se | n | Mean | se | n | Mean | ||||
| μmol g−1 fresh wt | atom% excess | % | μmol g−1 fresh wt | |||||||||
| Asn | 16.91 | 1.65 | Mono-15N | 8.17 | 0.27 | 11 | Amino | 73 | 6 | 6 | Amino | 1.33 |
| Amide | 27 | 6 | 6 | Amide | 0.70 | |||||||
| Double 15N | 1.91 | 0.12 | 12 | |||||||||
| GABA | 5.72 | 0.67 | 33.39 | 2 | 1.91 | |||||||
| Gln | 1.55 | 0.20 | Mono-15N | 15.74 | 2.08 | 5 | Amino | 98 | 2 | 3 | Amino | 0.34 |
| Amide | 2 | 2 | 3 | Amide | 0.11 | |||||||
| Double 15N | 6.59 | 1.10 | 4 | |||||||||
| Ala | 1.24 | 0.12 | 25.02 | 0.80 | 7 | 0.31 | ||||||
| Asp | 0.76 | 0.05 | 14.22 | 1.27 | 10 | 0.11 | ||||||
| Glu | 0.35 | 0.04 | 27.86 | 0.98 | 5 | 0.10 | ||||||
The values given for amino and amide group total 15N concentration include both mono- and double-labeled molecules.
n = 3.
Figure 4.
Concentrations of unlabeled, 15N-amino-labeled, and 15N-amide-labeled soluble amino acids in pea root nodules after an 8-h incubation period in 15N2 in the perfusion system and acquisition of the in vivo 15N NMR spectra shown in Figure 2.
Assignment of Amino Acid 15N NMR Resonances in Metabolically Active Nodules
Considering the very unusual chemical shift of the ammonium ions in root nodules, the assignments of the in vivo NMR resonances of amino acid 15N amino groups required some attention. The amino acid amide resonances are much less likely to be severely perturbed because they are less prone to complexation and virtually independent of pH in the physiologically relevant range. In vivo 15N NMR spectra from 15N2-fixing pea root nodules showed two amino acid 15N resonances at 90.7 and 19.6 ppm (Fig. 2), which correspond to an amide and an α-amino group, respectively. This was observed in several experiments. The amide resonance at 90.7 ppm could, in comparison with in vitro measurements and previously observed values, unequivocally be assigned to the amide resonance of Asn. The α-amino resonance at 19.6 ppm,, which was also observed in in vivo spectra from L. japonicum nodules, was assigned to Gln and Glu according to in vitro measurements as well as previously observed values. The Gln and Glu amino groups resonate in a 0.2 ppm interval around 19.7 ppm and are difficult to resolve in vivo, so the resonance at 19.6 ppm is likely to contain contributions from both.
However, analyses by LC-MS of the 15N labeling in pea nodules that were incubated in 15N2 in the perfusion system showed that the most abundant 15N-labeled amino group was the one of Asn (Table II; Fig. 4). Analyses of dead nodules and extracts (see Table I) confirmed that the dominating 15N-labeled α-amino group was the one of Asn and that it resonated at the expected 18.8 to 19.1 ppm when inside dead nodules. An α-amino signal at 18.4 ppm was observed in an in vivo 15N NMR spectrum from L. japonicum nodules and assigned to Asn (Table I). Thus, it was surprising that the amino group of Asn did not give rise to a large NMR signal in living pea root nodules, but further investigations indicated that differences in NOE factors could provide an explanation (see below).
GABA was observed at the expected 11.6, 11.8, and 11.3 ppm in 15N NMR spectra from dead pea root nodules, an extract from nodules, and metabolically active, 15N2-fixing L. japonicum nodules, respectively (Table I). The GABA 15N resonances in spectra from nodules were in several experiments observed to be broader than resonances from other amino acids. However, GABA was never observed in in vivo 15N NMR spectra from 15N2-fixing pea nodules (Fig. 2), although LC-MS analyses demonstrated very high 15N labeling of GABA in these nodules (see Table II; Fig. 4). Possible reasons for this apparent inconsistency are discussed below.
In Vivo 15N NMR Time Course of 15N Fixation and Assimilation
Figure 2 shows a representative series of consecutive in vivo 15N NMR spectra that were started immediately after addition of 15N2 at time zero. An ammonium signal at −4.2 ppm was evident during the 1st h of incubation and increased in intensity throughout the first 5 h, after which steady state seemed to occur. Two additional 15N signals representing the amide group of Asn and the amino groups of Glu/Gln, respectively, emerged in the spectrum period from 1 to 3 h and likewise increased in intensity in the next spectrum from 3 to 5 h. The intensities of these two signals were unchanged in the last spectrum from 5 to 7 h, indicating steady state.
NOE Effects
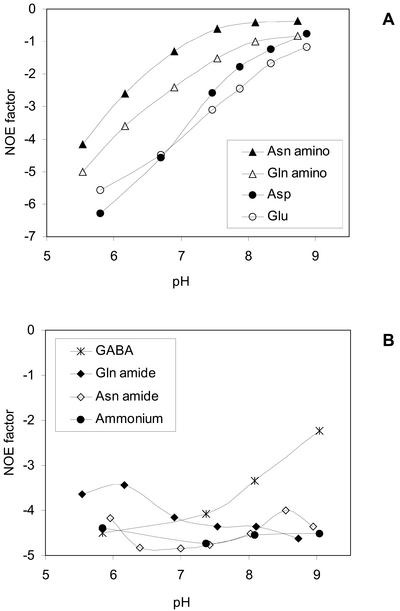
To observe quantitative results, in vivo 15N spectra should ideally be recorded without NOE effects. This turned out not to be feasible in the present study. However, the NOE effects led to variation in intensities for the various amino acids resonances. This was investigated in vitro as a function of pH in standard solutions as shown in Figure 5, and as seen, the effects are related to the acid dissociation constant (pKa) values of the compounds. The amino group of Asn was observed to have a NOE factor of −1 at around pH 7.1 in vitro, whereas NOE factors of the Gln and Glu amino groups were between −1.5 and −3.5 in the physiologically relevant pH range of 7 to 7.5. If the NOE factors exhibit the same pH dependency in vivo as was demonstrated in vitro, it would be expected that the signal from the α-amino group of Asn would be lost, whereas the signals from the amino groups of Gln and Glu would be enhanced.
Figure 5.
Amino acid and ammonium NOE factors at different pH values. A, α-Amino groups; B, amide groups, ammonium, and GABA. NOE factors were determined as I/I0 − 1, where I and I0 represent signal intensities in spectra with full decoupling and spectra with inverse gated decoupling, respectively.
DISCUSSION
15N Ammonium in Living Root Nodules
Ammonium ions in root nodules appear from the present results to reside in an intracellular environment, which is remarkably different from all other plant systems that have been studied by in vivo 15N NMR. The 15N NMR ammonium signal in living pea and L. japonicum root nodules was in several experiments observed at around −4 ppm, which is an unusually low ammonium frequency. Previous in vivo 15N NMR studies have demonstrated ammonium resonances at around 0 ppm in a diversity of biological systems such as maize (Zea mays) root tips (Amancio and Santos, 1992), carrot (Daucus carota) cells (Fox et al., 1992), Corynebacterium glutamicum (Tesch et al., 1999), and ectomycorrhizal mycelium (Martin et al., 1994). The observed enhanced shielding of ammonium can be because of many different factors such as pH effects, bonding to anions, macromolecules, or paramagnetic ions. We investigated whether the observed unusual chemical shift could be explained by the presence of one of the more common anions, phosphate, and found no significant effects of physiologically relevant phosphate concentrations on the ammonium chemical shift (data not shown).
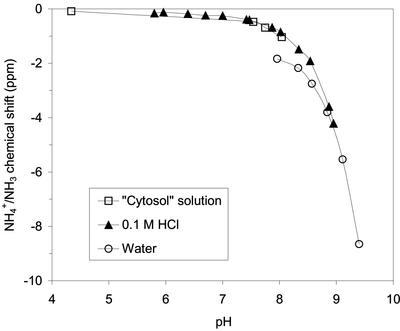
A location of ammonium in an alkaline compartment in the root nodules would cause a change of the chemical shift in the observed direction, and the bacteroids may constitute such a compartment. Base titration of ammonium in vitro demonstrated, as expected, large pH effects on the chemical shifts, and an ammonium chemical shift of −4 ppm was observed at pH 8.9 (see Fig. 6). It has previously been estimated that free ammonium in nodules is primarily confined to bacteroids (Streeter, 1989), and the bacteroid cytoplasm is anticipated to be slightly alkaline (Day et al., 2001) because of the proton pumping activity of the electron transport chain in the bacteroid inner membrane as well as the proton-consuming nitrogenase activity. The peribacteroid space, which is the compartment between the bacteroid membrane and the symbiosome membrane, is, on the contrary, assumed to be acidic (Day et al., 2001), and is therefore unlikely to contain the observed free ammonium. It does not seem reasonable that the pH could be as high as 8.9 in a tissue that is metabolically active, but it is possible that a minor contribution to the shielding of the observed 15N NMR ammonium signal is because of a higher pH in the bacteroids. It should be noted that as pH is raised, more and more of the ammonium would be converted to ammonia. This will cause a change in the 15N chemical shift but will also imply that the ammonium-ammonia couple, because of the latter, will complex metal ions much better. This, in turn, may lead to a further change in the chemical shift. The 31P spectra (Fig. 1) do not show a phosphate signal from an alkaline compartment. However, this does not rule out the possibility of the existence of such a compartment because the absence of the signal could merely reflect a very low phosphate concentration.
Figure 6.
15N ammonium chemical shift as a function of pH. Chemical shifts are referenced to urea at 55.8 ppm, which was present as an aqueous solution in a capillary.
Our results may suggest that the ammonium ions are located in the bacteroid, whereas the labeled Gln/Glu and Asn are located in the plant cytoplasm. If the observed large shift of the in vivo ammonium NMR signal is caused by pH and binding to paramagnetic sites, this would be likely to have an impact on both ammonium ions and the amino acids. Chemical shifts of Gln/Glu amino groups and Asn amide groups change dramatically at pH values above 8 (data not shown), and complexation constants of amino groups with physiologically relevant metal ions are in general orders of magnitude larger than ammonium complexation constants (Martell and Smith, 1974, 1976). However, no changes in the in vivo chemical shifts of amino acids were observed, which may suggest that the labeled amino acids reside outside the bacteroids.
Further indication that free 15N-labeled ammonium and amino acids are located in different compartments is provided by the observation that the phase of the ammonium resonance during decoupling was opposite of the phase of all amino acid 15N signals (see Fig. 2). This means that the ammonium NOE factor is between −1 and 0, whereas NOE factors of the observed 15N amino acids are numerically larger than −1. The reason for the difference in NOE factors is not known, but Altenburger et al. (1991) observed a similar effect and suggested that it was caused by the ammonium ions being bound to ion-exchange sites in the cell wall. It has been observed previously that addition of cell debris to an ammonium solution reduced the NOE factor to between 0 and −1, whereas no change of the NOE factor was seen for Glu (Fox et al., 1992). It should be noticed that neither a change in the chemical shifts nor any apparent line broadening occurred.
The subcellular distribution of ammonium in living maize root tissue has previously been elucidated by studying the pH dependence of line widths of proton-coupled ammonium signals (Lee and Ratcliffe, 1991). The proton exchange rate is, however, fast on the NMR time scale at pH values of approximately 7 and above, and ammonium and amino groups give narrow resonances (Lee and Ratcliffe, 1991). We have measured line widths of all 15N signals in proton-coupled 15N in vivo spectra (data not shown), but because these line widths are all narrow (15–20 Hz with 8-Hz line broadening) and of similar size as line widths in uncoupled spectra, they only demonstrate that both ammonium and the amino acids are present in compartments with pH above 7. Therefore, these measurements do not give any supplementary information on a possible location of ammonium in an alkaline compartment.
The concentration of free 15N ammonium in metabolically active pea root nodules may be roughly estimated to be at least as large as that of 15N-labeled Asn amide, which was determined to constitute 0.7 μmol g−1 fresh weight nodule based on LC-MS measurements. The intensity of the 15N ammonium in in vivo NMR signal was of a comparable size to the signal from the Asn amide group after an 8-h incubation in 15N2 (see Fig. 2). The NMR signal intensities are influenced by both T1 and NOE, which were not quantified under in vivo conditions in the present study, but from other studies it can be ascertained that both processes would cause an attenuation of the ammonium signal relative to the Asn amide signal. In vivo NOE factors for amino acid amide groups have been shown previously to be of the order of −4.5 (Kanamori et al., 1982; Thorpe et al., 1989; Joy et al., 1997), whereas the ammonium NOE factor is between 0 and −1 in the present study. Amino acid amide group T1s of 3 to 4 s in plant tissue have been reported (Kanamori et al., 1982; Thorpe et al., 1989; Joy et al., 1997), whereas very long ammonium T1s of up to 50 s are commonly observed in vivo (Tesch et al., 1999).
Our suggestion that ammonium is primarily localized in the bacteroid compartment and a free ammonium concentration of minimum 0.7 μmol g−1 fresh weight nodule is in accordance with earlier estimates. A direct analysis of the concentration and compartmentation of ammonium in symbiotic root nodules is lacking, but Streeter (1989) estimated, based on extractions and extrapolations, that soybean bacteroids contained 1.6 μmol ammonium g−1 fresh weight nodule, whereas the concentration of ammonium in the plant cytoplasm was essentially nil. The uneven distribution of ammonium in the nodule tissue is believed to be caused by a slow, restricted ammonium transport across the bacteroid membrane because passive diffusion of ammonia is the only known transport mechanism across this membrane. It is evident from the present study that substantial, although not quantifiable, 15N labeling of ammonium took place in the metabolically active pea root nodules and that ammonium was the first detectable 15N-labeled compound in the intact symbiosis. This is in accordance with many previous reports (for review, see Day et al., 2001), but in contrast to recent results by Waters et al. (1998). The latter detected no 15N labeling of ammonium in isolated 15N2-fixing soybean bacteroids, when these were incubated at high densities under microaerobic conditions. In the experimental setup used by Waters et al. (1998), Ala was demonstrated to contain 98 atom% excess 15N, and to be the primary product excreted by the bacteroids.
N Assimilation in Pea Nodules
Asn was observed by LC-MS to be 15N labeled in both the amino and the amide groups when nodules were incubated in 15N2, although the amino resonance was invisible in 15N NMR spectra with full decoupling. The amide group for the biosynthesis of Asn from Asp may either come from direct incorporation of ammonium or from the Gln amide group, and previous results have indicated that both pathways occur in alfalfa nodules (Snapp and Vance, 1986; Ta et al., 1986). The observed high total 15N labeling of Asn is consistent with its generally accepted role as the end product of the primary N assimilation processes taking place in indeterminate root nodules.
An in vivo 15N NMR signal from the Asn amino group was never observed in pea nodules (see Fig. 2), although LC-MS analyses demonstrated substantial labeling of the amino group (see Table II and Fig. 4). This apparent inconsistency could, however, be explained by an unfavorable NOE effect if the NOE factors exhibit the same pH dependency in vivo as was demonstrated in vitro (Fig. 5). The variation in the amino group NOE factors with pH has been debated intensely over the years (Cooper et al., 1973; Leipert and Noggle, 1975; Irving and Lapidot, 1975), and the reason for the variation is still not entirely clear (Neuhaus and Williamson, 2000). From our in vitro experiments (see Fig. 5), it seems obvious that the NOE effect is related to the pKa values. If it is correct that the in vivo NOE factor of the Asn amino group is of an unfavorable size, which causes elimination of the signal in spectra recorded with full decoupling, it is surprising that this has not been reported previously because the majority of in vivo 15N NMR studies applies full decoupling. On the other hand, very few in vivo 15N NMR studies have observed a signal from the Asn amino group. To our knowledge, only a single 15N NMR study, namely the investigation of 15N ammonium metabolism in duckweeds by Monselise and Kost (1993), demonstrated the presence of 15N labeling in the amino group of Asn. Amancio and Santos (1992) assigned an 15N signal at 18.6 ppm to the amino groups of both Asn and Asp, but in our opinion, only Asp would resonate at this low frequency. The absence of reported Asn 15N amino signals in the in vivo NMR literature may reflect that no labeling occurs or that the Asn pool of the studied organisms was below the detection limit, but it may also be caused by NOE signal elimination. Very few investigations, apart from the present, have included parallel analyses by in vivo 15N NMR and other analytical methods that would reveal a possible NMR invisible Asn amino signal.
A substantial amount of GABA (labeled as well as unlabeled) was shown to accumulate in root nodules during incubation in the NMR tube (see Fig. 3), but the newly synthesized 15N GABA seemed to be immobilized in some way in metabolically active pea nodules, which made it NMR invisible. The accumulation of 15N labeling in the GABA pool could indicate that GABA is a biosynthetic end point under the given experimental conditions. The results concerning the size of the GABA pool in the nodule tissue do not allow for a discrimination of GABA localized in the bacteroid and in the plant cytoplasm. It is, however, evident from the high 15N labeling that the GABA is formed from a newly synthesized pool of Glu, presumably catalyzed by Glu α-decarboxylase, which is well known in plant tissue (for review, see Bown and Shelp, 1997) and has also been reported to be present in symbiotic rhizobia bacteroids (Fitzmaurice and O'Gara, 1991; Miller et al., 1991). The GABA shunt pathway has been suggested previously to play a prominent role in R. meliloti bacteroids (Fitzmaurice and O'Gara, 1988; Miller et al., 1991), although the function of GABA remains unclear. Some experimental evidence indicates that the locations of GABA production and accumulation are not identical, and that accumulated GABA is sequestered within organelles, possibly in the vacuoles (for review, see Shelp et al., 1999). It has also been demonstrated that nodules from many legume species accumulate bound forms of GABA amounting to as much as 20% of the total N content of the nodule (Larher et al., 1983). The apparent NMR invisibility of 15N-labeled GABA in pea root nodules in in vivo experiments may result from GABA being immobilized in vivo. Immobilized GABA may be released from the tissue by the applied extraction procedures and/or enzymatic reactions and, therefore, detected by LC-MS. A support for such a suggestion is the observation of broad resonances of GABA in dead nodules, indicating that GABA is less tightly bound under such conditions but still not free, e.g. in the cytoplasm.
Concluding Remarks
The present work represents the first study of N2-fixation and assimilation in 15N2-fixing root nodules by in vivo 15N NMR spectroscopy. The in vivo approach has generated new information, which has not been made available by other techniques previously used for studying N assimilation in legume nodules. A substantial pool of free ammonium was observed to be present in the metabolically active, intact symbiosis. The ammonium ions were located in an unusual intracellular environment, which caused a remarkable change in the in vivo 15N chemical shift. Alkalinity of the ammonium-containing compartment is suggested as a partial explanation for the unusual chemical shift; thus, the observations point to the bacteroids as a probable location of the free ammonium pool in root nodules. Furthermore, the observed 15N-labeled amino acids, Gln/Glu and Asn, apparently reside in a different compartment, presumably the plant cytoplasm, because no changes in the expected in vivo 15N chemical shifts were observed. GABA accumulated in the root nodules during incubation, but newly synthesized 15N GABA seemed to be immobilized in metabolically active pea root nodules, which made it NMR invisible.
MATERIALS AND METHODS
Plant Material
Surface-sterilized seeds of pea (Pisum sativum L. cv Solara) were germinated in humid vermiculite inoculated with a 3-d-old yeast broth suspension culture of Rhizobium leguminosarum bv viceae strain Risø 18a. After 6 d, the seedlings were transferred to an aeroponic system consisting of a large plastic caisson tank equipped with a mist generator circulating 12 L of nutrient solution from the bottom of the tank. The nutrient solution was prepared as described previously (Rosendahl and Jakobsen, 1987) with the addition of 8 mm MES to provide some pH buffering. pH was kept at 6 to 6.5 by adjusting with 5 m KOH when necessary. The nutrient solution was inoculated once a week with 50 mL of the R. leguminosarum suspension culture. The roots of seedlings were pushed through holes in the lid of the tank and, thus, exposed to the mist. The lid was carefully tightened around the stems of the plants to avoid salt deposition on the stems, which may result in plant death. Pea nodules were harvested from this system with minimal physical disturbance and were observed to retain a higher nitrogenase activity than pea nodules harvested from the below-mentioned vermiculite growth system (data not shown). Nodules grown in the aeroponic system maintained a higher nitrogenase activity when immersed in water than vermiculite-grown nodules (data not shown). This may be attributed to the fact that nodules are covered with a water film in the aeroponic system and, thus, adapted to taking up O2 and N2 under these circumstances.
Nodules from Lotus japonicum inoculated with Mesorhizobium loti strain R7A (Sullivan et al., 1995) were used for a few experiments. Plants were grown in vermiculite-filled pots in an experimental setup described by Rosendahl and Jakobsen (1987).
All plants were cultured in a growth chamber under a 16-h-light/8-h-dark cycle at 20°C/16°C and a photosynthetically active photon flux density of 600 μmol m−2 s−1. Nodules were excised from the roots immediately before NMR experiments, when plants were 6 to 7 weeks old (early pod-filling stage) and nitrogenase activity is maximal.
Experimental Design of in Vivo Studies
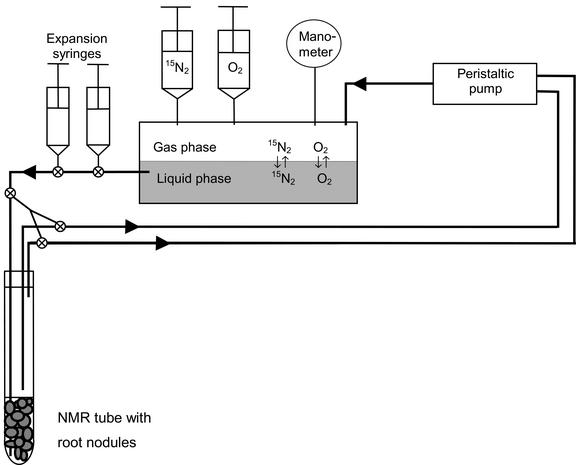
In vivo NMR spectra were recorded from detached root nodules that were incubated in 15N2 in a perfusion system (Fig. 7). After perfusion, the nodules were quickly removed from the NMR tube, gently rinsed with water, and immediately frozen in liquid N2. The nodules were kept at −80°C until further analysis. Subsamples were later taken for determination of fresh weight to dry weight ratios, total 15N incorporation, soluble amino acid pools, and 15N labeling of individual amino acids. Spectra and matching analyses from a single experiment are presented in this paper, but similar results were obtained in several experiments.
Figure 7.
Schematic drawing (not proportional) of the perfusion system used for studying N2-fixation and assimilation in living pea root nodules by 15N NMR spectroscopy. Approximately 1.5 g fresh weight root nodules could be contained within the NMR tube, and about 1 g fresh weight was within the volume of the NMR detection coil. ⊗, Three-way stopcock.
Perfusion and 15N2 Incubation of Nodules for in Vivo NMR
Approximately 1.5 g fresh weight root nodules were placed in a 10-mm NMR tube, which was connected to a perfusion system (Fig. 7). Nodules were maintained in a physiologically viable and controllable state by perfusion with an oxygenated nutrient buffer consisting of 25 mm Glc, 25 mm malate, 0.1 mm CaSO4, 10 mm MES (pH 6.0), and 10% (v/v) D2O at a temperature of 21°C to 22°C. A teflon inlet tube delivered the buffer to the bottom of the NMR tube below the nodules and the buffer left through two output tubes above the nodules. The tubes and nodules were kept in place by a plastic rod insert fixed to the screw cap top of the threaded NMR tube. All tubing outside the NMR tube was made of Tygon (R-3603, 1.6-mm wall, Cole-Parmer Instrument Co., Vernon Hills, IL), which is only slightly permeable to N2 and O2. A peristaltic pump attached to the two output tubes circulated the 700-mL nutrient buffer at 45 mL min−1 through the nodule-filled volume, and recycled it via a closed gas-tight reservoir. The reservoir consisted of a 500-mL rubber stopped serum bottle that contained a liquid phase and an 80-mL gas phase. Equilibrium between the phases was ensured by continuous stirring of the liquid phase and by spraying the perfusate returning from the NMR tube through the gas phase. To achieve a high enrichment of 15N2 in the perfusion system, the reservoir was first flushed for 30 min with an O2:Ar mixture (1:1 [v/v]), while circulating the nutrient buffer to drive off all 14N2. Then, the reservoir was filled with O2, and 15N2 (99.95 atom% 15N, Isotec Inc., Miamisburg, OH) was introduced in the system by injecting it into the reservoir. The composition and 15N enrichment of the final gas mixture in the perfusion system was 49% (v/v) N2 (92 atom% 15N), 48% (v/v) O2, and 3% (v/v) Ar as determined by MS of a subsample. The gas mixture in the perfusion system was compressed to a total pressure of 1.6 atm during the entire experiment to dissolve more gas in the circulating liquid phase and to reduce intrusion of atmospheric air. At the end of the experiment, after 8 h of perfusion of the nodules, the composition of the gas phase was 56% (v/v) N2 (73 atom% 15N), 39% (v/v) O2, 4% (v/v) Ar, and 0.4% (v/v) CO2. The pH value of the perfusate was measured after the experiment to ensure that pH was unchanged. The perfusate was also examined by 15N NMR spectroscopy to ensure that all observed 15N resonances originated from intracellular 15N metabolites.
Total 15N Incorporation
Nodules were dried and homogenized, and the 15N enrichment was determined on an isotope ratio mass spectrometer (Finnigan MAT Delta E, ThermoFinnigan MAT, Bremen, Germany) coupled in continuous flow mode to an EA 1110 elemental analyzer (ThermoFinnigan Italia, Milan) as described previously (Egsgaard et al., 1989).
Preparation of 15N-Enriched Biological Material and Solutions
To aid the initial assignments of 15N metabolites, highly 15N-enriched pea nodules were prepared in a separate experiment and studied by 15N NMR spectroscopy after quenching of all metabolic activity. These pea plants were grown with the entire root system in a gas-tight chamber (Rosendahl and Jakobsen, 1988). The root chamber of six-week-old plants was flushed with an Ar:O2 mixture (70%:30% [v/v]), and 15N2 was subsequently added to a final gas composition of: 59% (v/v) Ar, 24% (v/v) O2, 10% (v/v) 15N2, and 7% (v/v) 14N2. After 4 h of incubation in the 15N-enriched atmosphere, root nodules were quickly harvested on ice, immediately frozen in liquid N2, and kept at −80°C until NMR analysis. After recording of NMR spectra, nodules and the medium bathing the nodules during NMR analysis were extracted with MeOH:CHCl3:water (12:5:3 [v/v]). The extract was subsequently purified as described below, redissolved in 0.1 m HCl, and the pH adjusted to 5.8 before NMR analysis. Identification of 15N-labeled metabolites was obtained by spiking the extract with authentic 15N-labeled amino acid standards followed by 15N NMR analysis.
Standard solutions of 15N-enriched ammonium and amino acids were prepared by dissolving authentic 99 atom% 15N-enriched compounds (Icon Services Inc., Summit, NJ) in 0.1 m HCl to a concentration of about 30 mm. The solutions were titrated to different pH values with KOH and HCl for the study of pH dependence of NOE factors. For the study of the pH dependence of the ammonium chemical shift (Fig. 6), 99 atom% 15N-enriched ammonium chloride was dissolved in three different solutions: (a) a “cytosol” solution consisting of 100 mm KCl, 5 mm MgSO4, and 5 mm NaH2PO4 (Spickett et al., 1993), (b) 0.1 m HCl, and (c) water. The ammonium concentrations were 5, 30, and 5 mm, respectively, in the three solutions, and the titrations to different pH values were done with KOH and HCl.
Extraction of Amino Acids from Nodules
Frozen nodules (approximately 0.4 g fresh weight) were homogenized in a mortar on liquid N2, and α-aminobutyric acid was added as an internal standard. The homogenate was transferred to a tube and kept on ice, and amino acids were extracted as described by Johansen et al. (1996) by three subsequent additions of 4 mL of MeOH:CHCl3:water (12:5:3 [v/v]), vortex mixing for 1 min, and centrifugation (2,000g, 5 min, 5°C). All subsequent purification of extracts was performed on ice. The supernatants were pooled and CHCl3 (14 mL) and water (3 mL) were added. The tube was vortexed and centrifuged (2,000g, 2 min, 5°C) to facilitate phase separation. The methanol-water phase containing the amino acids was evaporated in a Speed-Vac concentrator (Maxidry Lyo Heto-Holten Als, Allerød, Denmark), and the amino acids were finally taken up in 1 mL of 0.1 m HCl. An amino acid standard solution was subjected to the extraction procedure and analyzed in parallel with nodule extracts to ensure that Gln and Asn, in particular, were not degraded during the applied procedures.
Determination of Soluble Amino Acid Pools
Nodule extracts were deproteinized by addition of sulfosalicylic acid to a final concentration of 0.1 m, and nor-Leu was added as an internal standard. The samples were left for 15 min at room temperature before they were centrifuged (4,000 rpm, 30 min, 5°C), and the supernatant was applied to an HPLC system (Waters, Milford, MA) modified as an amino acid analyzer with cooled autosampler and fluorometer. The instrument used cation exchange chromatography for separation of the amino acids and post-column reaction with ortho-phthaldialdehyde for quantification. Millennium software (Waters) was used for control of the instrument and for integration of the amino acid peaks.
Analysis of 15N Labeling of Amino Acids by LC-MS
Underivatized amino acids were analyzed using the ion-pair reverse-phase chromatographic strategy as pioneered by Petritis et al. (2000). Asp, Asn, Gln, Glu, and Ala were baseline separated using a Purospher RP-18e column (125 × 4 mm, 5 μm, VWR International, West Chester, PA) with 0.5 mm pentadecafluoroctanoic acid as mobile phase (0.5 mL min−1). The rather difficult separation of GABA and α-aminobutyric acid was achieved using 10 mm nonafluorpentanoic acid as mobile phase. In all cases, a sample size of 50 μL was used. The isotope analyses were carried out on-line using an LCQ (Classic) MSn system (ThemoFinnigan, San Jose, CA) bundled with an electrospray ionization source and a complete TSP HPLC system (Thermo Separation Products, San Jose, CA). Specific scan events were designed to meet the analysis of the individual amino acids. The 15N content was determined using the single ion traces of the MH+ and [M + 1]H+ ions. The 15N content was calculated directly from the measured ratios and corrected for natural abundances.
The positional labeling of Gln was quantified by monitoring the daughter ions formed by the loss of NH3 from the [M + 1]H+ ions using MS/MS experiments. Thus, the daughter ions ([[M + 1] − NH3]H+ and [[M +1] − 15NH3]H+) were monitored using two different MS/MS experiments. The positional labeling of Asn was quantified using the granddaughter ions arising from the consecutive loss of [CO + H2O] and HNCO from the[M +1]H+ ions by MS/MS/MS experiments. Thus, the granddaughter ions ([[M + 1] − [CO + H2O] − HNCO]H+ and [[M + 1] − [CO + H2O] − 15HNCO]H+) were quantified using two different MS experiments. The isolation widths in the MSn experiments were carefully optimized with respect to sensitivity and accuracy. The positional labeling was calculated on the principles of isotope dilution (A.M. Scharff, C. Schon, and H. Egsgaard, unpublished data).
NMR Spectroscopy
31P and 15N spectra were recorded at 242.8 and 60.79 MHz, respectively, on a Unity Inova 600 spectrometer (Varian, Palo Alto, CA) using a broadband 10-mm-diameter probe head.
In vivo 31P spectra were acquired with a 30° pulse angle (12 μs), 0.125-s acquisition time, proton decoupling by WALTZ-16 composite pulse sequence, 11.2-kHz sweep width, 13,824 transients, and 20-Hz line broadening. 31P chemical shifts were measured relative to the signal from methylene diphosphonic acid (pH 8.9 in Tris buffer) contained in a capillary included in the NMR tube and are quoted relative to the resonance of 85% (w/v) phosphoric acid at 0 ppm. Assignments of 31P signals were based on literature reports (Saint-Ges et al., 1991) and 31P NMR analyses (data not shown) of neutralized perchloric acid extracts performed as described by Roby et al. (1987). Estimates of intracellular pH were based on cytoplasmic and vacuolar calibration curves, respectively, that were made as suggested by Spickett et al. (1993).
15N spectra of metabolically active, 15N2-fixing nodules as well as of dead 15N-enriched nodules were acquired with a 60° pulse angle, 0.25-s acquisition time, recycle delay of 1.75 s, proton decoupling at high power during the acquisition and at low power during the delay by WALTZ-16 composite pulse sequence, 9.3-kHz sweep width, and 1,792 or 3,584 transients leading to total acquisition times of 1 or 2 h, respectively. Line broadening (8 Hz) was applied.
15N spectra of nodule extracts were acquired with a 90° pulse angle, 2-s acquisition time, 3-s delay 1, 5-s delay 2, proton decoupling at high power during the acquisition and at low power during delay 2 by WALTZ-16 composite pulse sequence, 18-kHz sweep width, and 5,800 transients leading to a total acquisition time of 16 h. Line broadening (5 Hz) was applied.
15N NMR spectra of standard solutions of 15N-enriched amino acids were acquired with a 90° pulse angle, 2-s acquisition time, 10-s delay 1, 10-s delay 2, and 9.3-kHz sweep width. Spectra of 15N-enriched ammonium and GABA were acquired with the same parameters, except for delay 1 and delay 2, which were 200 and 150 s, respectively. No line broadening was applied. NOE factors were determined as the ratio between signal intensities in: (a) spectra where NOE was applied by WALTZ-16-modulated proton decoupling during delay 2 as well as during the acquisition, and (b) spectra with inverse gated decoupling. Line widths were similar in the two types of spectra.
15N chemical shifts were measured relative to the signal at 55.8 ppm from 0.25 m aqueous 15N-urea. For 15N NMR analyses of nodules, urea was contained in a capillary included in the NMR tube, whereas urea was added directly to extracts and 15N standard solutions.
ACKNOWLEDGMENTS
Ina B. Hansen (Risoe National Laboratory) is thanked for skilled technical assistance and Per Ambus and Merete Brink Jensen (Risoe National Laboratory) are thanked for the mass spectrometric determinations of total 15N incorporation in nodules. We are grateful to Christian Schou (Risoe National Laboratory) for assistance with developing and performing the LC-MS analyses. Dr. Ernst Christensen (Copenhagen University Hospital) is acknowledged for the analyses of soluble amino acid pools. Finally, Dr. George Ratcliffe (University of Oxford) is thanked for inspiring discussions as well as comments and suggestions for the manuscript.
Footnotes
This work was supported by the Danish Research Agency and by the Danish Research Foundation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.015156.
LITERATURE CITED
- Aarnes H, Eriksen B, Southon TE. Metabolism of nitrate and ammonium in seedlings of Norway spruce (Picea abies) measured by in vivo N-14 and N-15 NMR spectroscopy. Physiol Plant. 1995;94:384–390. [Google Scholar]
- Allaway D, Lodwig EM, Crompton LA, Wood M, Parsons R, Wheeler TR, Poole PS. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol Microbiol. 2000;36:508–515. doi: 10.1046/j.1365-2958.2000.01884.x. [DOI] [PubMed] [Google Scholar]
- Altenburger R, Abarzua S, Callies R, Grimme LH, Mayer A, Leibfritz D. Ammonia rhythm in Microcystis firma studied by in vivo 15N and 31P NMR spectroscopy. Arch Microbiol. 1991;156:471–476. [Google Scholar]
- Amancio S, Santos H. Nitrate and ammonium assimilation by roots of maize (Zea mays L.) seedlings as investigated by in vivo 15N-NMR. J Exp Bot. 1992;43:633–639. [Google Scholar]
- Belay N, Sparling R, Choi BS, Roberts M, Roberts JE, Daniels L. Physiological and 15N-NMR analysis of molecular nitrogen fixation by Methanococcus thermolithotrophicus, Methanobacterium bryantii and Methanospirillum hungatei. Biochim Biophys Acta. 1988;971:233–245. doi: 10.1016/0167-4889(88)90138-3. [DOI] [PubMed] [Google Scholar]
- Bown AW, Shelp BJ. The metabolism and functions of gamma-aminobutyric acid. Plant Physiol. 1997;115:1–5. doi: 10.1104/pp.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RA, Lichter RL, Roberts JD. pH-dependent nuclear magnetic resonance spectra of 15N-enriched glycine. Line shape and relaxation studies. J Am Chem Soc. 1973;95:3724–3729. doi: 10.1021/ja00792a042. [DOI] [PubMed] [Google Scholar]
- Day DA, Poole P, Tyerman SD, Rosendahl L. Ammonia and amino acid transport across symbiotic membranes in nitrogen-fixing legume nodules. Cell Mol Life Sci. 2001;58:61–71. doi: 10.1007/PL00000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egsgaard H, Larsen E, Jensen ES. Evaluation of automated analysis of 15N using on-line Dumas combustion. Anal Chem Acta. 1989;226:345–349. [Google Scholar]
- Fitzmaurice AM, O'Gara F. Involvement of glutamate as a carbon source in supporting nitrogen fixation activity in R. meliloti. In: Bothe G, de Bruijn FJ, Newton WE, editors. Nitrogen Fixation: Hundred Years After. Stuttgart: Gustav Fischer; 1988. p. 558. [Google Scholar]
- Fitzmaurice AM, O'Gara F. Glutamate catabolism in Rhizobium meliloti. Arch Microbiol. 1991;155:422–427. [Google Scholar]
- Fox GG, Ratcliffe RG, Robinson SA, Slade AP, Stewart GR. Detection of 15N-labeled ammonium. In vivo 15N NMR versus mass spectrometry. J Magn Reson. 1992;96:146–153. [Google Scholar]
- Irving CS, Lapidot A. Origin of the anomalous pH dependence of the 15-[1H] Nuclear Overhauser effect of amino and amide nitrogens. J Am Chem Soc. 1975;97:5945–5946. [Google Scholar]
- Johansen A, Finlay RD, Olsson PA. Nitrogen metabolism of external hyphae of the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 1996;133:705–712. [Google Scholar]
- Joy RW, Vogel HJ, Thorpe TA. Inorganic nitrogen metabolism in embryogenic white spruce cultures: a nitrogen 14/15 NMR study. J Plant Physiol. 1997;151:306–315. [Google Scholar]
- Kanamori K, Legerton TL, Weiss RL, Roberts JD. Nitrogen-15 spin-lattice relaxation times of amino acids in Neurospora crassa as a probe of intracellular environment. Biochemistry. 1982;21:4916–4920. doi: 10.1021/bi00263a013. [DOI] [PubMed] [Google Scholar]
- Kuzma MM, Winter H, Storer P, Oresnik I, Atkins CA, Layzell DB. The site of oxygen limitation in soybean nodules. Plant Physiol. 1999;119:399–407. doi: 10.1104/pp.119.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larher F, Goas G, le Rudulier D, Gerard J, Hamelin J. Bound 4-aminobutyric acid in root nodules of Medicago sativa and other nitrogen fixing plants. Plant Sci Lett. 1983;29:315–326. [Google Scholar]
- Lee RB, Ratcliffe RG. Observations on the subcellular distribution of the ammonium ion in maize root tissue using in vivo N-14-nuclear-magnetic-resonance spectroscopy. Planta. 1991;183:359–367. doi: 10.1007/BF00197734. [DOI] [PubMed] [Google Scholar]
- Legerton TL, Kanamori K, Weiss RL, Roberts JD. 15N NMR studies of nitrogen metabolism in intact mycelia of Neurospora crassa. Proc Natl Acad Sci USA. 1981;78:1495–1498. doi: 10.1073/pnas.78.3.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipert TK, Noggle HN. The nitrogen-15 magnetic resonance of glycine. J Am Chem Soc. 1975;97:269–272. doi: 10.1021/ja00835a006. [DOI] [PubMed] [Google Scholar]
- Li YZ, Green LS, Day DA, Bergersen FJ. Ammonia and alanine efflux from nitrogen-fixing soybean bacteroids. In: Pedrosa FO, Hungria M, Yates MG, Newton WE, editors. Nitrogen Fixation: From Molecules to Crop Productivity. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. p. 391. [Google Scholar]
- Martell AE, Smith RM. Critical Stability Constants. Vol. 1. New York: Plenum Press; 1974. Amino acids; pp. 25–27. , 40–41. [Google Scholar]
- Martell AE, Smith RM. Critical Stability Constants. Vol. 4. New York: Plenum Press; 1976. Inorganic complexes; pp. 40–41. [Google Scholar]
- Martin F. 15N-NMR studies of nitrogen assimilation and amino acid biosynthesis in the ectomycorrhizal fungus Cenococcum graniforme. FEBS Lett. 1985;182:350–354. [Google Scholar]
- Martin F, Cote R, Canet D. NH4+ assimilation in the ectomycorrhizal basidiomycete Laccaria bicolor (Maire) Orton, a 15N-NMR study. New Phytol. 1994;128:479–485. doi: 10.1111/j.1469-8137.1994.tb02994.x. [DOI] [PubMed] [Google Scholar]
- Miller RW, McRae DG, Joy KW. Glutamate and gamma-aminobutyrate metabolism in isolated Rhizobium meliloti bacteroids. Mol Plant-Microbe Interact. 1991;4:37–45. [Google Scholar]
- Mitsumori F, Yoneyama T, Ito O. Phosphorus-31 nuclear magnetic resonance studies on higher plant tissue. Plant Sci. 1985;38:87–92. [Google Scholar]
- Monselise EB-I, Kost D. Different ammonium-ion uptake, metabolism and detoxification efficiencies in two Lemnaceae. A 15N-nuclear magnetic resonance study. Planta. 1993;189:167–173. [Google Scholar]
- Neuhaus D, Williamson MP. The Nuclear Overhauser Effect in Structural and Conformational Analysis. New York: Wiley-VCH, Inc.; 2000. The heteronuclear NOE; p. 57. [Google Scholar]
- Nikolaev BP, Provorov NA, Simarov BV, Shlyakov AM. Transformation of phosphates by free-living and symbiotic root-nodule bacteria Rhizobium meliloti followed by 31P NMR spectroscopy. Microbiology. 1994;63:16–20. [Google Scholar]
- Petritis K, Chaimbault P, Elfakir C, Dreux M. Parameter optimization for the analysis of underivatized protein amino acids by liquid chromatography and ion spray tandem mass spectrometry. J Chromatogr A. 2000;896:253–263. doi: 10.1016/s0021-9673(00)00582-3. [DOI] [PubMed] [Google Scholar]
- Poole P, Allaway D. Carbon and nitrogen metabolism in Rhizobium. Adv Microb Physiol. 2000;43:117–163. doi: 10.1016/s0065-2911(00)43004-3. [DOI] [PubMed] [Google Scholar]
- Robinson SA, Slade AP, Fox GG, Phillips R, Ratcliffe RG, Stewart GR. The role of glutamate dehydrogenase in plant nitrogen metabolism. Plant Physiol. 1991;95:509–516. doi: 10.1104/pp.95.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roby C, Martin JB, Bligny R, Douce R. Biochemical changes during sucrose deprivation in higher plant cells. Phosphorus-31 nuclear magnetic resonance studies. J Biol Chem. 1987;262:5000–5007. [PubMed] [Google Scholar]
- Rolin DB, Boswell RT, Sloger C, Tu SI, Pfeffer PE. In vivo 31P NMR spectroscopic studies of soybean Bradyrhizobium symbiosis: I. Optimization of parameters. Plant Physiol. 1989a;89:1238–1246. doi: 10.1104/pp.89.4.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolin DB, Pfeffer PE, Boswell RT, Schmidt JH, Tu SI. In vivo 31P NMR spectroscopic studies of soybean Bradyrhizobium symbiosis. Compartmentation and distribution of P metabolites. FEBS Lett. 1989b;254:203–206. [Google Scholar]
- Rosendahl L, Jakobsen I. Rhizobium strain effects on pea: the relation between nitrogen accumulation, phosphoenolpyrovate carboxylase activity in nodules and asparagine in root bleeding sap. Physiol Plant. 1987;71:281–286. [Google Scholar]
- Rosendahl L, Jakobsen I. Effects of age, supra-ambient oxygen and repeated assays on acetylene reduction and root respiration in pea. Physiol Plant. 1988;74:77–82. [Google Scholar]
- Saint-Ges V, Roby C, Bligny R, Pradet A, Douce R. Kinetic studies of the variations of cytoplasmic pH, nucleotide triphosphates (31P-NMR) and lactate during normoxic and anoxic transitions in maize root tips. Eur J Biochem. 1991;200:477–482. doi: 10.1111/j.1432-1033.1991.tb16207.x. [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Bown AW, Mclean MD. Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 1999;4:446–452. doi: 10.1016/s1360-1385(99)01486-7. [DOI] [PubMed] [Google Scholar]
- Snapp SS, Vance CP. Asparagine biosynthesis in alfalfa (Medicago sativa L.) root nodules. Plant Physiol. 1986;82:390–395. doi: 10.1104/pp.82.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spickett CM, Smirnoff N, Ratcliffe RG. An in vivo nuclear magnetic resonance investigation of ion transport in maize (Zea mays) and Spartina anglica roots during exposure to high salt concentrations. Plant Physiol. 1993;102:629–638. doi: 10.1104/pp.102.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent JI. Prolonged reduction of acetylene by detached soybean nodules. Planta. 1969;88:372–375. doi: 10.1007/BF00387466. [DOI] [PubMed] [Google Scholar]
- Streeter JG. Carbohydrate, organic acid, and amino acid composition of bacteroids and cytosol from soybean nodules. Plant Physiol. 1987;85:768–773. doi: 10.1104/pp.85.3.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter JG. Estimation of ammonium concentration in the cytosol of soybean nodules. Plant Physiol. 1989;90:779–782. doi: 10.1104/pp.90.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JT, Patrick HN, Lowther WL, Scott DB, Ronson CW. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc Natl Acad Sci USA. 1995;92:8985–8989. doi: 10.1073/pnas.92.19.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta TC, Faris MA, Macdowall FDH. Pathways of nitrogen metabolism in nodules of alfalfa (Medicago sativa L.) Plant Physiol. 1986;80:1002–1005. doi: 10.1104/pp.80.4.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesch M, de Graaf AA, Sahm H. In vivo fluxes in the ammonium-assimilatory pathways in Corynebacterium glutamicum studied by N-15 magnetic nuclear magnetic resonance. Appl Environ Microbiol. 1999;65:1099–1109. doi: 10.1128/aem.65.3.1099-1109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe TA, Bagh K, Cutler AJ, Dunstan DI, Mcintyre DD, Vogel HJ. A 14N and 15N nuclear magnetic resonance study of nitrogen metabolism in shoot-forming cultures of white spruce (Picea glauca) buds. Plant Physiol. 1989;91:193–202. doi: 10.1104/pp.91.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JK, Hughes BL, Purcell LC, Gerhardt KO, Mawhinney TP, Emerich DW. Alanine, not ammonia, is excreted from N-2-fixing soybean nodule bacteroids. Proc Natl Acad Sci USA. 1998;95:12038–12042. doi: 10.1073/pnas.95.20.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]