Abstract
The classical genetic map of Arabidopsis contains 462 genes with mutant phenotypes. Chromosomal locations of these genes have been determined over the past 25 years based on recombination frequencies with visible and molecular markers. The most recent update of the classical map was published in a special genome issue of Science that dealt with Arabidopsis (D.W. Meinke, J.M. Cherry, C. Dean, S.D. Rounsley, M. Koornneef [1998] Science 282: 662–682). We present here a comprehensive list and sequence-based map of 620 cloned genes with mutant phenotypes. This map documents for the first time the exact locations of large numbers of Arabidopsis genes that give a phenotype when disrupted by mutation. Such a community-based physical map should have broad applications in Arabidopsis research and should serve as a replacement for the classical genetic map in the future. Assembling a comprehensive list of genes with a loss-of-function phenotype will also focus attention on essential genes that are not functionally redundant and ultimately contribute to the identification of the minimal gene set required to make a flowering plant.
Before the advent of modern genomics, the existence of a gene was often first revealed when a mutant with a visible phenotype was recovered. From pea (Pisum sativum) plants with wrinkled seeds to fruitflies (Drosophila melanogaster) with altered eye pigmentation, mutants have long played a central role in genetic analysis. Recent advances in molecular biology have made it possible to identify large numbers of genes with mutant phenotypes in model organisms and to move toward a synthesis of classical genetics and structural genomics. This report provides one such synthesis for a model plant. The sequence-based map of genes with mutant phenotypes described here should provide a foundation for the long-term goal of determining which genes in Arabidopsis give a phenotype when disrupted by mutation. This information is needed to address from a genetic perspective the question of functional redundancy in Arabidopsis and to identify those genes capable of generating phenotypic diversity.
The first comprehensive map of Arabidopsis genes with mutant phenotypes was published 20 years ago (Koornneef et al., 1983). Included on that map were 76 genes with phenotypes ranging from altered trichome morphology and seed coat pigmentation to reduced surface waxes and increased hypocotyl length. Most of the genes were assigned map locations based on analysis of F2 plants produced following self-pollination of heterozygotes. Backcrosses of heterozygotes to parental homozygotes were avoided to minimize the total number of crosses performed. Precise gene orders were often not resolved because two-point crosses were involved, which meant that genes were placed on the map by comparing recombination frequencies obtained with different pairs of linked markers.
Large numbers of mutants were added to the classical map over the next 15 years, including embryo defectives with a seed phenotype that enabled heterozygous F2 plants to be identified without progeny testing. This feature reduced the number of plants required to obtain accurate mapping data and resulted in further map enhancements (Patton et al., 1991). The most extensive study presented recombination data for 169 embryo-defective mutants and estimated locations of 110 EMB genes (Franzmann et al., 1995). Recombination percentages were converted into centiMorgans using the Kosambi (1944) mapping function. Maps with the most consistent gene order were constructed by determining the minimal chi-square for all recombination data combined (Jensen and Jorgensen, 1975). Several computer programs were used to facilitate map construction and integration (Patton et al., 1991; Stam, 1993). Despite these important advances, inconsistencies in recombination data soon made it necessary to place new genes on the map by hand. This approach was used to construct the map of 284 loci published by Koornneef (1994).
Genetic maps of molecular markers were also being constructed during this time. The types of markers involved quickly expanded to include restriction fragment length polymorphisms (Chang et al., 1988; Nam et al., 1989; Fabri and Schaffner, 1994), random-amplified polymorphic DNAs (Reiter et al., 1992), cleaved-amplified polymorphic sequences (Konieczny and Ausubel, 1993), simple sequence length polymorphisms (Bell and Ecker, 1994), and amplified fragment-length polymorphisms (Alonso-Blanco et al., 1998). A recombinant inbred map developed by crossing Columbia and Landsberg ecotypes (Lister and Dean, 1993) soon became established as the standard for genetic placement of molecular markers. Mutant genes could be placed on this map by determining recombination percentages with linked molecular markers. This resulted in the establishment of two parallel types of genetic maps with mutant genes: the classical map and the recombinant inbred map. Integration of these maps proved difficult because chromosome lengths and recombination estimates were not identical. Because many people were interested in gene isolation through map-based cloning, genes with mutant phenotypes were often mapped only in relation to linked molecular markers. Furthermore, tagged mutants identified from T-DNA insertion lines did not require mapping to clone the disrupted gene and therefore often did not contribute to map enhancements. As a result, the number of mutant genes added to the classical map began to diminish.
The most recent update to the classical genetic map contains 462 loci distributed over five chromosomes and 469 total centiMorgans (Meinke et al., 1998). This map includes 131 genes with a seed phenotype and 110 genes initially placed on the recombinant inbred map and then transferred to the classical map after adjusting for differences in estimated chromosome lengths. Problems with this combined map soon became apparent as more genes were cloned and their relative locations on the physical map contradicted their estimated locations on the genetic map. Resolving these inconsistencies was difficult and was not given a high priority, despite the importance of genetic maps in model systems. A fresh approach was therefore needed to update and correct the classical map.
We decided to address this problem by focusing on mutants disrupted in known genes that could be assigned a physical location on the sequenced chromosomes. Our goal was to construct a sequence-based map of genes with mutant phenotypes to replace the classical genetic map and to provide a new standard for dealing with the chromosomal locations of mutant genes. We started with an initial list of 115 genes already noted as cloned on the genetic map (Meinke et al., 1998) and supplemented this with information from The Arabidopsis Information Resource (TAIR; http://www.Arabidopsis.org), extensive literature searches of publications listed in PubMed (http://www.ncbi.nlm.nih.gov), examination of abstracts from posters at recent Arabidopsis meetings, and requests for community additions and corrections to draft spreadsheets. The initial results of this effort are described here with the first sequence-based map of 620 mutant genes of Arabidopsis. Additional details collected during construction of this map document the methods used for gene isolation, the general phenotype of mutant alleles, the predicted function of protein products, and the dramatic increase in the number of genes identified in recent years. Although the total number of genes that can mutate to give a phenotype remains to be determined, the results presented here make it possible to begin comparisons with other model systems and to assess from a genetic perspective the extent of functional redundancy in the Arabidopsis genome.
RESULTS AND DISCUSSION
Criteria for Including Genes on the Map
Three questions had to be resolved before determining which genes should be included on a sequence-based map: (a) what constitutes a mutant phenotype; (b) should the map be limited to loss-of-function mutants; and (c) what level of confidence of gene identification should be required? We decided to include any gene with a dominant or recessive mutant phenotype that could be detected through visual inspection, cellular characterization, or biochemical analysis under standard greenhouse or specialized laboratory conditions. In this way, it was possible to include a broad range of mutants and to facilitate comparisons between the classical genetic and sequence-based maps. Dominant mutants for which a loss-of-function phenotype remains to be identified will need to be removed from future lists that are limited to genes with essential functions. We did not consider a change in gene expression pattern or metabolic profile alone sufficient to constitute a phenotype because such alterations may turn out to be characteristic of most gene disruptions in Arabidopsis. Including these genes might therefore overshadow more established mutants and result in a map filled with genes with subtle loss-of-function defects. Also excluded were genes that gave a phenotype only when inactivated by antisense or gene silencing or when overexpressed through activation tagging or introduction of a cloned wild-type allele. The reason was once again to focus on loci defined by mutation and not by experimental gene manipulation in order to present an updated map that was similar in scope to the classical genetic map but consistent with the genome sequence.
Suppressor and enhancer mutations were more problematic because it was difficult to establish consistent guidelines for what to exclude. We decided not to include cases where a double knockout in redundant genes was required to give a phenotype because these alterations could not be attributed to a single locus. But genes with mutant phenotypes detected only in specific ecotypes or genotypes were included provided the genetic lines involved did not appear to have a mutation in a redundant gene. Enhancers and suppressors that gave no phenotype by themselves but modified the phenotype of a nonredundant gene knockout were therefore included. Because we did not require that gene identities be confirmed through molecular complementation or sequencing of duplicate mutant alleles, the possibility exists that some genes listed here may later need to be removed. Genetic maps have also required consolidation when two mapped loci were later found to be allelic. We discovered at least nine such cases in the course of updating the classical map (CBB3 and DWF3; DET2 and DWF6; FUS4 and FUS8; DOC1 and TIR3; PAS3 and GK; ELL and FK; AGR and EIR; RPP11 and RPP13; RPP4 and RPP5).
A Sequence-Based Map of Mutant Genes
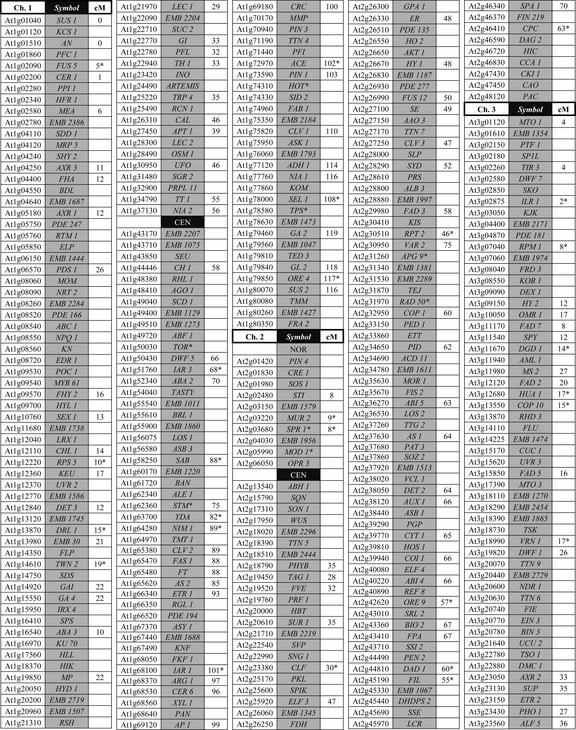
The assembled list of 620 cloned genes with mutant phenotypes is presented in Figure 1. An expanded spreadsheet with full gene names, alias symbols, gene classes, mutant phenotypes, predicted functions, and reference labs is provided in Table S-I (supplemental data can be viewed at http://www.plantphysiol.org). Genes listed in Figure 1 are arranged by locus number, a unique identifier that corresponds to the physical location of a predicted gene along the length of the chromosome (Arabidopsis Genome Initiative, 2000). The gene order presented here is therefore consistent with the published genome sequence. Because adjacent genes are often assigned locus numbers that differ in value by 10, regardless of gene size or intergenic distances, the proximity of two genes can be estimated by comparing their locus numbers. The precise number of intervening genes and base pairs can then be obtained from current annotation (http://www.Arabidopsis.org). Locations of mutant genes are displayed on a sequence-based physical map in Figure 2. Enlarged versions that include gene symbols and locus numbers are given in Figures S-1 and S-2 (supplemental data can be viewed at http://www.plantphysiol.org).
Figure 1.
An ordered list of 620 Arabidopsis genes with mutant phenotypes. Genes are arranged by locus number, starting with the top of chromosome 1. Sequences are available at TIGR (http://www.tigr.org) and TAIR (http://www.Arabidopsis.org). Gene symbols with an asterisk conflict with other registered symbols that correspond to different genes (see Table S-I). Numbers in the centiMorgan column represent estimated locations on the genetic map. Numbers with an asterisk designate genes placed on the recombinant inbred map and later transferred to the classical map. Sequence gaps are noted for the centromere (CEN) and nucleolar organizer (NOR) regions.
Figure 2.
A sequence-based map of genes with mutant phenotypes. Gene locations are marked with horizontal lines. A single line at this scale may represent two or more neighboring genes. The length of each chromosome is proportional to its sequence. Centromeric gaps are marked by short constrictions.
Chromosomal Distribution of Mutant Genes
Three approaches were used to examine the chromosomal distribution of mutant genes. The first was to compare the relative numbers of genes on each chromosome. The results as shown in Table I are similar to those found with the classical genetic map and the sequenced genome as a whole. Genes with an embryo-defective (emb) phenotype are the most common class included on both types of maps. This is consistent with the large number of genes known to have essential functions during seed development (McElver et al., 2001).
Table I.
Distribution of genes on the classical genetic and sequence-based maps of Arabidopsis
| Map Feature | Classical Genetic | Sequence-Based | Total Genomea |
|---|---|---|---|
| Total loci | 462 (100.0%) | 620 (100.0%) | 29,084 (100.0%) |
| Chromosome 1 | 122 (26.4%) | 156 (25.2%) | 7,378 (25.4%) |
| Chromosome 2 | 70 (15.2%) | 106 (17.1%) | 4,814 (16.6%) |
| Chromosome 3 | 80 (17.3%) | 102 (16.4%) | 5,987 (20.6%) |
| Chromosome 4 | 77 (16.7%) | 103 (16.6%) | 4,319 (14.8%) |
| Chromosome 5 | 113 (24.4%) | 153 (24.7%) | 6,586 (22.6%) |
| Total EMB | 131 (28.4%) | 140 (22.6%) | Est. 500 to 750b |
Based on current annotation of the Arabidopsis genome (TIGR Release 3.0; http://www.tigr.org).
Based on relative frequency of embryo-defective mutants and duplicate alleles (Franzmann et al., 1995; McElver et al., 2001).
The second approach was to look for large gaps or clusters on the sequence-based map. As shown in Figure 2, mutant genes are distributed throughout the length of each chromosome with the notable exception of centromeric regions. We considered two alterative explanations for these gaps: (a) genes with mutant phenotypes might be preferentially excluded from centromeric regions; or (b) the absence of mutant genes in these regions might simply reflect the known scarcity of functional genes around the centromere. We attempted to distinguish between these models by looking at loci predicted to fall between the mutant genes located just above and below each centromere constriction (e.g. At1g37130 and At1g43170 for chromosome 1). The genetically defined centromere is positioned within these gaps for chromosomes 1, 2, 3, and 5 and extends somewhat north of the boundary defined by At4g04890 and At4g04770 on chromosome 4 (Copenhaver et al., 1999). Of the nearly 2,700 sequenced loci assigned to these gaps in the current genome annotation (http://www.tigr.org), approximately 50% appear to be pseudogenes, transposons, or repeat elements, 20% are annotated as encoding hypothetical proteins with no database matches, and another 20% correspond to putative proteins. Fewer than 300 loci in these combined regions appear to be promising candidates for active genes with defined functions. Based on the observed frequency of genes with mutant phenotypes elsewhere in the genome (620/29,084 = 2.1%), six of these 300 genes should have already been found to give a mutant phenotype. The failure to identify such mutants, if confirmed in future studies, could reflect an overestimation of functional genes in the centromere, inaccessibility of these regions to traditional mutagens, or increased levels of functional redundancy. For the most part, however, the distribution of genes with mutant phenotypes in Arabidopsis mirrors the distribution of functional, protein-coding genes throughout the genome.
A final approach used to look at gene distribution was to determine how often genes with mutant phenotypes were positioned next to each other on the chromosome. In 95 cases of 620 examined, two mutant genes are separated by five genes or fewer based on current annotation: 21 gene pairs are physically adjacent, 15 are separated by a single gene, 23 by two genes, 10 by either three or five genes, and 16 by four genes. We identified one adjacent pair (At4g03050 and At4g03060) that appears to represent a tandem duplication involving similar gene functions, and another cluster of three adjacent genes (At1g08540, At1g08550, and At1g08560) with different functions and mutant phenotypes. One interesting gene (SIN1/SUS1/CAF/DCL1; At1g01040) with an essential role in growth and development (Golden et al., 2002) was found at the extreme north end of chromosome 1. The presence of adjacent pairs of mutant genes is particularly intriguing in light of the frequent occurrence of tandem gene duplications and associated redundancy in Arabidopsis. Whether these clusters define small regions of the genome with unusual structural features or limited functional redundancy remains to be determined. We have nevertheless demonstrated that genes with mutant phenotypes in Arabidopsis are not always surrounded by dispensable genes with little relevance to growth and development.
Time and Method of Gene Isolation
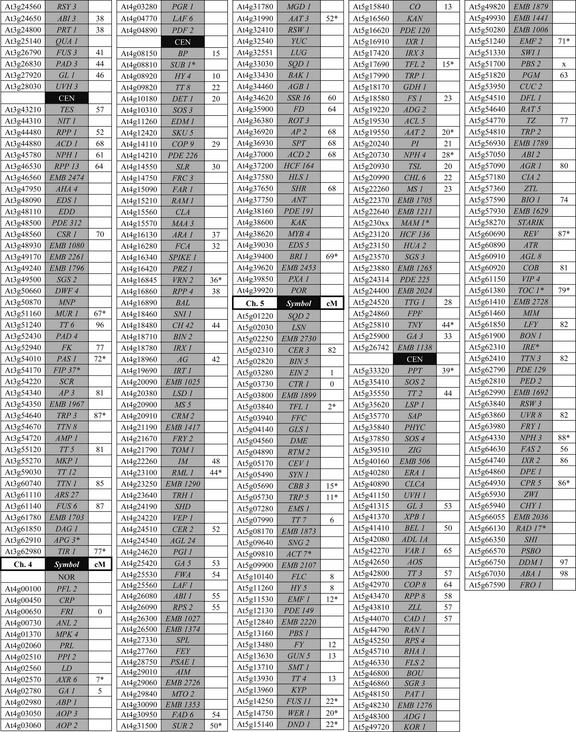
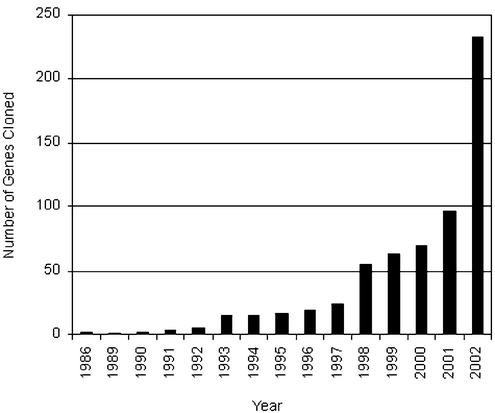
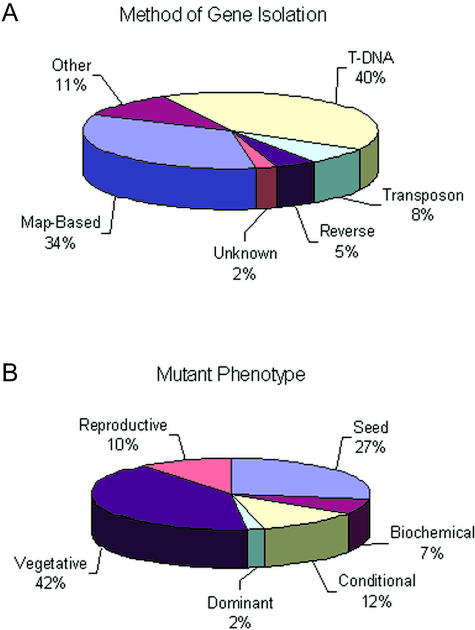
There has been a dramatic rise in recent years in the number of mutant genes cloned and characterized at a molecular level. This trend, documented in Figure 3, reflects improvements in methods for map-based cloning, widespread availability of transposon and T-DNA insertion lines, and longstanding efforts of the Arabidopsis community to characterize mutants obtained through forward genetics. At least 45 of the 76 loci (59%) found on the original classical map (Koornneef et al., 1983) and 237 of the 462 loci (51%) on the updated map (Meinke et al., 1998) have now been cloned. One-half of the 235 mapped genes that remain to be identified are embryo defectives, many of which were given a low priority for gene isolation because they were not tagged (Franzmann et al., 1995). A current version of the classical map with cloned genes highlighted is presented in Figure S-3. Methods used to identify these genes are summarized in Figure 4A. Within the next several years, the number of genes identified through reverse genetics is likely to increase sharply as more emphasis is placed on screening insertion lines for knockouts in specific genes of interest. The rate-limiting step for characterizing genes with mutant phenotypes will then shift from trying to isolate the disrupted gene to searching for a subtle or conditional phenotype.
Figure 3.
Date of initial publication or public release of gene identities associated with a mutant phenotype.
Figure 4.
Classification of mapped genes according to method of identification and phenotype of mutant alleles. A, Method used to determine the identity of each mutant gene included on the sequence-based map. “Other” includes cases where promising candidate genes with consistent functions and map locations were analyzed directly for altered nucleotide sequence or protein function. B, Phenotype class of a representative mutant allele for each gene identified. Refer to “Materials and Methods” for definitions.
Phenotypes of Disrupted Genes
Mutant phenotypes should be associated with individual alleles rather than a single gene because weak and strong alleles may produce different types of abnormalities. We nevertheless decided to place the 620 mutant genes described here into broad phenotypic classes based on known gene disruptions in order to assess the diversity of genes and mutants included. The results of this subjective but informative effort are shown in Figure 4B. The relative frequency of each phenotype class reflects not only the number of genes available to be disrupted but also the amount of attention devoted to that class of mutants by members of the community. Dominant mutants have been placed in a separate category to highlight their inclusion and to acknowledge the absence of a knockout phenotype.
Comparison of Genetic and Sequence-Based Maps
Information presented in Figure 1 makes it possible for the first time to assess the physical accuracy of the classical genetic map on a global scale. Although gene positions based on recombination percentages are for the most part consistent with physical locations confirmed through genome sequencing, precise locations and orders of closely linked genes are sometimes incorrect. Many of these inaccuracies can be attributed to the subjective process of transferring genes from the recombinant inbred map to the classical genetic map. Similar problems were encountered in the past when genetic maps constructed under different conditions were combined (Stam, 1993). The level of inaccuracy within each type of map is nevertheless about the same. In approximately 75% of the cases where a gene from either the classical genetic or recombinant inbred map has been cloned and sequenced, the adjacent cloned gene from the same map was placed in the correct position. In other words, the centiMorgan values increase or stay the same going down the chromosome in 75% of the cases examined when genes from the two maps are considered as separate groups. Some inconsistencies in gene placement are not surprising given the methods used to construct genetic maps. Other irregularities, such as the placement of CER3 at the top of chromosome 5 (GenBank accession no. 1669654) despite extensive genetic evidence documenting its location at the bottom of chromosome 5 (Koornneef, 1994), are more problematic and remain to be resolved. We chose not to perform a genome-wide comparison of genetic and physical map distances because many of the genetic locations were estimated based on recombination percentages with different sets of distant markers and were therefore not an accurate reflection of genetic distance. Crosses between mutants listed here could be performed in the future to compare recombination percentages and physical distances on a global scale.
Diversity of Genes Identified
Mutant genes identified to date encode proteins with a wide range of biological functions. Characterizing the full spectrum of cellular processes involved will require a higher degree of saturation and functional characterization of the entire proteome using a standardized gene ontology (GO) system adopted for model eukaryotes (Gene Ontology Consortium, 2001). At the present time, 35% of the 620 genes listed here have no GO assignment, 17% have a single functional assignment, and the remaining 48% have multiple GO assignments. Additional information on predicted protein functions can be found in Table S-1 and at TAIR.
Mutant genes also differ in predicted length from start to stop codons. We reasoned that existing mutant collections might be biased toward large genes because they represent bigger targets for random mutagenesis. This model is supported by the results presented in Table II. Small genes (<1 kb in length) are underrepresented in our collection (4% versus 25% of total) whereas large genes (>3 kb in length) are more common (42% versus 16%). These differences are also reflected in the average gene size: 3.2 kb for mutant genes and 1.9 kb for the entire genome. Large genes have already been shown to be preferred targets for T-DNA insertions that result in a seed phenotype (McElver et al., 2001). Results presented here document a similar trend for the entire collection of mutant genes identified through forward genetic screens. A significant challenge for the future is therefore to isolate and characterize large numbers of mutants disrupted in small genes with important biological functions.
Table II.
Predicted sizes of cloned mutant genes identified through forward genetic screensa
| Gene Class Analyzed | Number | Percentage of Genes Identified in Different Size Classesb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| <1 kb | 1+ kb | 2+ kb | 3+ kb | 4+ kb | 5+ kb | 6+ kb | >7 kb | ||
| Total genome | 27,288 | 25.3 | 37.9 | 20.9 | 8.1 | 3.9 | 1.9 | 0.9 | 1.1 |
| Total forward genetics | 590 | 4.1 | 24.7 | 28.8 | 17.8 | 12.4 | 4.9 | 3.2 | 4.1 |
| Map-based cloning | 208 | 3.4 | 23.6 | 34.1 | 18.3 | 8.6 | 6.2 | 2.4 | 3.4 |
| T-DNA tagged | 254 | 4.7 | 24.0 | 23.7 | 17.3 | 14.6 | 5.5 | 5.5 | 4.7 |
| Other forwardc | 128 | 3.9 | 28.1 | 30.5 | 18.0 | 14.1 | 1.5 | 0.0 | 3.9 |
Based on recent annotation from TIGR (http://www.tigr.org). Gene boundaries correspond to start and stop codons. Gene classes are defined by method of gene isolation.
Genes are placed in size groups: 1+ kb includes all predicted genes between 1.00 and 1.99 kb in length; 2+ kb includes those between 2.00 and 2.99 kb; etc.
Includes mutant genes identified by transposon tagging and other combinations of methods.
Estimating the Total Number of Mutant Genes
Despite the limitations of defining what constitutes a mutant phenotype, the question of how many Arabidopsis genes will be found to exhibit a phenotype when disrupted by mutation needs to be addressed to place the current study in perspective and to compare Arabidopsis with other model systems. The most definitive approach would be to perform a comprehensive phenotypic screen of a complete collection of individual knockout lines. This level of saturation has been reported for the yeast Saccharomyces cerevisiae (Giaever et al., 2002) but at present remains impractical for Arabidopsis, where efficient methods of gene replacement are not available and emphasis has been placed instead on screening for knockouts in existing collections of insertion lines, which are random, redundant, and incomplete.
An alternative approach would be to take several small regions of the genome, produce knockouts for each predicted gene, perform a comprehensive screen for phenotypes on each knockout, and extrapolate the percentage of genes found to have mutant phenotypes to the genome as a whole. Such an effort is certainly feasible, and if representative regions of the genome were chosen, could provide a direct estimate of the anticipated number of target genes. A related approach would be to take results from other model systems, compensate for differences in functional redundancy, and come up with an adjusted estimate for Arabidopsis. Recent studies have revealed that approximately 14% of the 2,400 genes on chromosome 1 of C. elegans disrupted with RNAi exhibit a phenotype (Fraser et al., 2000) and that 19% of the 5,900 knockouts of yeast analyzed in a genome-wide survey exhibit a growth defect (Giaever et al., 2002). Approximately 35% of all predicted genes in Arabidopsis appear to be unique based on sequence comparisons in which similarity is defined by BLASTP value (e <10−20) and alignment (>80% of the protein). The proportion increases to 55% for C. elegans and 71% for yeast (Arabidopsis Genome Initiative, 2000). Although disrupting a duplicated gene can still result in a phenotype, increased functional redundancy should reduce the likelihood of a phenotype overall. Given this level of redundancy and phenotype detection in different model systems, we estimate that 10% of Arabidopsis genes (about 3,000 total) should give a loss-of-function phenotype that can be identified using current screening methods. This estimate may increase considerably in the future as visual and biochemical analyses of knockout mutants become more robust.
Information collected from large-scale screens of existing T-DNA collections should also provide insights into the level of saturation achieved. This requires estimating the average number of inserts per line, the percentage of inserts that fall within a single gene, the frequency of mutant phenotypes observed in the entire collection, and the percentage of mutant phenotypes associated with T-DNA insertion. Much of this information has already been obtained, particularly in relation to embryo defectives. There are about 1.5 inserts per line on average (Feldmann, 1991; Krysan et al., 1999; McElver et al., 2001), 30% of the phenotypes observed result from stable T-DNA integration (Castle et al., 1993; McElver et al., 2001), and 35% of T-DNA insertions appear to fall within an open reading frame (Krysan et al., 2002). The percentage of T-DNA insertions that disrupt the function of a typical Arabidopsis gene, however, remains to be resolved. There are an estimated 500 to 750 EMB genes based on the frequency of duplicate mutant alleles identified to date (Franzmann et al., 1995; McElver et al., 2001). If we estimate that embryo defectives represent about 20% of all mutant phenotypes in T-DNA populations, then the total number of genes that can give a mutant phenotype is 2,500 to 3,750. The current estimated level of saturation for mapped genes with mutant phenotypes is therefore 15% to 25%. Although these numbers may need to be adjusted dramatically as additional details emerge from forward and reverse genetic screens, we believe that they represent a reasonable starting point for future experiments and an important first step in the analysis of Arabidopsis genes with mutant phenotypes.
MATERIALS AND METHODS
Establishing a List of Genes with Mutant Phenotypes
Gene identities associated with mutant phenotypes were identified in part by searching PubMed (http://www.ncbi.nlm.nih.gov) for relevant publications using different combinations of keywords (Arabidopsis, gene, mutant, mutation, and protein). Abstracts of papers describing the initial cloning of a mutant gene were saved for future reference. Publications were examined for details on gene symbols, mutant phenotypes, predicted functions, and methods of gene isolation. Reference laboratories responsible for identifying the disrupted gene were noted. Chromosome locus numbers maintained at The Institute for Genomic Research (TIGR; http://www.tigr.org) were identified using BLASTP (Altschul et al., 1997) accessed through TAIR (Huala et al., 2001) to compare published sequence information with the entire Arabidopsis proteome. Additional genes were found by scanning abstracts of recent Arabidopsis meetings in Madison, Wisconsin (June, 2001) and Seville, Spain (June, 2002). Direct requests for information were made to the Arabidopsis community through TAIR and the electronic Arabidopsis newsgroup (arab-gen@net.bio.net). Symbols of mapped and well-characterized mutants not included on initial lists of cloned genes were also used to search PubMed and GenBank. Information presented here was obtained through August 15, 2002.
Classification of Mutant Phenotypes
Six phenotype classes were used to document the diversity of genes identified: seed (embryo- or endosperm-defective or seed pigment mutant), vegetative (altered germination, seedling, root, rosette, or transition to flowering), reproductive (abnormal flower, silique, seed coat, or gamete), biochemical (altered enzyme activity, product accumulation, or cellular function without other striking defects), conditional (phenotype only in certain genetic backgrounds or in response to pathogen or unusual treatment), and dominant (phenotype observed only with dominant allele). Genes with variable mutant phenotypes were assigned to the first relevant class in the order listed above. Assignments were designed to be informative and representative but could not always be definitive given the complexity of some mutant phenotypes.
Drawing the Sequence-Based Maps
Maps were drawn with the “Chromosome Map Tool” available at TAIR, which queries a database with the supplied locus numbers to obtain assignment information. The locus name and coordinate information are then sent to an applet, which draws all five chromosomes on the browser screen. The user can choose a zoom level to scale the picture. Figure 2 was drawn at the 100% zoom level, where one pixel on the screen equals 50 kb. Figures S-1 and S-2 were drawn at the 600% zoom level, where one pixel equals approximately 8 kb.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tanya Berardini at TAIR for providing a draft list of putative mutants associated with gene identifiers, Brian Haas at TIGR for information on gene size distributions genome wide, members of the Meinke laboratory for helpful comments on the manuscript, and the entire Arabidopsis community for providing information on gene identities.
Footnotes
This research was supported by the National Science Foundation Developmental Mechanisms and Arabidopsis 2010 Programs.
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.014134.
LITERATURE CITED
- Alonso-Blanco C, Peeters AJ, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MT. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 1998;14:259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet. 1993;241:504–514. doi: 10.1007/BF00279892. [DOI] [PubMed] [Google Scholar]
- Chang C, Bowman JL, DeJohn AW, Lander ES, Meyerowitz EM. Restriction fragment length polymorphism linkage map for Arabidopsis thaliana. Proc Natl Acad Sci USA. 1988;85:6856–6860. doi: 10.1073/pnas.85.18.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell LD et al. Genetic definition and sequence analysis of Arabidopsis centromeres. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- Fabri CO, Schaffner AR. An Arabidopsis thaliana RFLP mapping set to localize mutations to chromosomal regions. Plant J. 1994;5:149–156. [Google Scholar]
- Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J. 1991;1:71–82. [Google Scholar]
- Franzmann LH, Yoon ES, Meinke DW. Saturating the genetic map of Arabidopsis thaliana with embryonic mutations. Plant J. 1995;7:291–300. [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome 1 by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, Meinke DW, Ray A. SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 2002;130:808–822. doi: 10.1104/pp.003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W et al. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29:102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J, Jorgensen JH. The barley chromosome 5 linkage map: I. Literature survey and map estimation procedure. Hereditas. 1975;80:5–16. [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Koornneef M. Arabidopsis genetics. In: Meyerowitz E, Somerville C, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 89–120. [Google Scholar]
- Koornneef M, van Eden J, Hanhart CJ, Stam P, Braaksma FJ, Feenstra WJ. Linkage map of Arabidopsis thaliana. J Hered. 1983;74:265–272. [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Krysan PJ, Young JC, Jester PJ, Monson S, Copenhaver G, Preuss D, Sussman MR. Characterization of T-DNA insertion sites in Arabidopsis thaliana and the implications for saturation mutagenesis. OMICS. 2002;6:163–174. doi: 10.1089/153623102760092760. [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics. 2001;159:1751–1763. doi: 10.1093/genetics/159.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M. Arabidopsis thaliana: a model plant for genome analysis. Science. 1998;282:662–682. doi: 10.1126/science.282.5389.662. [DOI] [PubMed] [Google Scholar]
- Nam HG, Giraudat J, den Boer B, Moonan F, Loos WDB, Hauge BM, Goodman HM. Restriction fragment length polymorphism linkage map of Arabidopsis thaliana. Plant Cell. 1989;1:699–705. doi: 10.1105/tpc.1.7.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton DA, Franzmann LH, Meinke DW. Mapping genes essential for embryo development in Arabidopsis thaliana. Mol Gen Genet. 1991;227:337–347. doi: 10.1007/BF00273921. [DOI] [PubMed] [Google Scholar]
- Reiter RS, Williams JGK, Feldmann KA, Rafalski JA, Tingey SV, Scolnik PA. Global and local genome mapping in Arabidopsis thaliana by using recombinant inbred lines and random amplified polymorphic DNAs. Proc Natl Acad Sci USA. 1992;89:1477–1481. doi: 10.1073/pnas.89.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam P. Construction of integrated genetic linkage maps by means of a new computer package: JOINMAP. Plant J. 1993;3:739–744. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.