Abstract
The cultivated potato (Solanum tuberosum) shares similar biology with other members of the Solanaceae, yet has features unique within the family, such as modified stems (stolons) that develop into edible tubers. To better understand potato biology, we have undertaken a survey of the potato transcriptome using expressed sequence tags (ESTs) from diverse tissues. A total of 61,940 ESTs were generated from aerial tissues, below-ground tissues, and tissues challenged with the late-blight pathogen (Phytophthora infestans). Clustering and assembly of these ESTs resulted in a total of 19,892 unique sequences with 8,741 tentative consensus sequences and 11,151 singleton ESTs. We were able to identify a putative function for 43.7% of these sequences. A number of sequences (48) were expressed throughout the libraries sampled, representing constitutively expressed sequences. Other sequences (13,068, 21%) were uniquely expressed and were detected only in a single library. Using hierarchal and k means clustering of the EST sequences, we were able to correlate changes in gene expression with major physiological events in potato biology. Using pair-wise comparisons of tuber-related tissues, we were able to associate genes with tuber initiation, dormancy, and sprouting. We also were able to identify a number of characterized as well as novel sequences that were unique to the incompatible interaction of late-blight pathogen, thereby providing a foundation for further understanding the mechanism of resistance.
The Solanaceae family contains several species of agronomic importance such as tomato (Lycopersicon esculentum), potato (Solanum tuberosum), pepper (Capsicum annuum), eggplant (Solanum melongena), petunia (Petunia × hybrida), and tobacco (Nicotiana tabacum). Species within the Solanaceae are highly related as evidenced by conserved sequence identity at the gene level and synteny among the homologous chromosomes (Bonierbale et al., 1988; Tanksley et al., 1992; Livingstone et al., 1999). Although members of the Solanaceae family share a number of features at the genome level, potato has a number of features that makes it unique among the Solanaceae. The most important physiological feature is the development of an edible tuber from stolons and consequently, on a global scale, potato is the fourth largest crop species grown as a food source with 300 million metric tons grown annually (http://www.cipotato.org/potato/potato.htm). However, despite its significance as a major food source, the process of tuber development is not well understood at the molecular level. In addition, potato is susceptible to the late-blight pathogen (Phytophthora infestans), which is not only a historically significant disease that resulted in the deaths of millions of people (for review, see Schumann, 1991), but it also has recently reemerged as a significant pathogen on potato (Fry and Goodwin, 1997).
The development of high-throughput sequencing technology has provided a mechanism to gain insight into genomes at the DNA and the RNA level. For assessment of gene expression, multiple methodologies such as large-scale single pass sequencing of cDNA clones to generate expressed sequence tags (ESTs; Adams et al., 1993) can be utilized. This method provides a quantitative method to measure specific transcripts within a cDNA library. By increasing the depth of sequencing within a library or by broadening the diversity of tissues from which the libraries are constructed, the rate of gene discovery can be increased. For example, by sampling multiple DNA libraries that represent diverse tissues, a high rate of gene discovery can be obtained, whereas deep sampling of a single cDNA library will yield weakly expressed and rare transcripts. One issue with EST sequencing is that highly expressed transcripts are sequenced multiple times. To address this, cDNA libraries can be normalized to reduce the frequency of highly expressed genes and increase the rate of gene discovery (Soares et al., 1994). However, normalization eliminates the ability to electronically assess transcript frequency within the tissue from which the cDNA library was constructed. Other approaches to assess the coding fraction of a genome include low-pass sequencing of either the whole genome (Jander et al., 2002) or bacterial artificial chromosome clones (Barry, 2001) and sequencing fractions of the genome that are enriched in single-copy sequences through techniques such as methyl filtration and cot selection (Rabinowicz et al., 1999; Peterson et al., 2002). Although these genome-based approaches have greater power in that they identify genes without any dependence on expression levels, they require a greater economic investment than EST sequencing.
In the fall of 1999, approximately 800 sequences for potato were present in GenBank and in 2001, approximately 6,000 ESTs derived from mature potato tubers were reported by Crookshanks et al. (2001). To gain further insight into the potato genome and potato transcriptome, we performed single-pass sequencing of the 5′ ends of cDNA clones to generate approximately 62,000 ESTs for potato. The cDNA libraries sampled in this study represent major stages in the development and physiology of potato, including tissues challenged with the late-blight pathogen. Using these potato EST sequences, we were able to generate a nonredundant set of sequences and identify sequences specific to single potato tissue as well as widely expressed sequences. Using two clustering approaches, we were able to associate sequences with major physiological and developmental processes in potato growth and development. We were also able to identify sequences associated with four stages in the tuber life cycle and sequences unique to the incompatible interaction with the late-blight pathogen.
RESULTS
Sequencing Quality
All sequences were screened for quality using base-quality scores and low-quality sequences were eliminated from our analyses. A total of 83,322 sequencing reactions were performed and after quality assessment, a total of 61,940 good sequences with an average edited length of 525 bases were generated. With the exception of the sprouting eye I library, all libraries (Table I) sequenced well, with an average sequencing success of 75% with a range of 60.5% to 84.4% good sequences. The sprouting eye I library had an unsatisfactory sequencing success rate and was not deeply sequenced; a second sprouting eye library (II) was constructed and sequenced. Sequences were generated on either ABI 377 or ABI 3700 sequencing machines (Applied Biosystems, Foster City, CA) and higher edited lengths were obtained with sequences generated on the ABI 3700 (average edited length of 574) in comparison with the ABI 377-derived sequences (average edited length of 449). All good sequences were deposited in the dbEST division of GenBank.
Table I.
Libraries and tissue sources for sequences described in this study
| cDNA Library Name | The Institute for Genomic Research (TIGR) Library ID | Tissue/Phase |
|---|---|---|
| Stolon | T1722 | Developing axillary buds of potato nodal stem cuttings culturedin vitro to induce stolon and tuber formation |
| Leaf | T1723 | Leaflets and petioles from 8-week-old greenhouse-grown plants |
| Sprouting eye I | T10212 | Sprouting eyes (2 to 15 mm) from 12–14-week postharvest tubers |
| Sprouting eye II | T10389 | Sprouting eyes (2 to 15 mm) from 12–14-week postharvest tubers |
| Dormant tuber | T10501 | Dormant tuber sections without the buds and epidermis. Tubers were stored for 1 month postharvest at 4°C prior to sampling. |
| Microtuber | T10502 | Small microtubers developed from axillary buds attached to stem explants at 7, 8, and 10 d in vitro tuberization |
| Root | T1726 | Roots from in vitro-grown stem cuttings isolated 2 weeks afterplacing the stem cuttings on the medium |
| Compatible leaf | T10427 | Leaf tissue from plants challenged with compatible P. infestansisolate US 940480. Leaf tissue was collected at 3, 6, 9, 12, 24, 48, and 72 h after inoculation. |
| Incompatible leaf | T1727 | Leaf tissue from plants challenged with P. infestans US-1. |
| Leaf tissue was collected at 1, 2, 5, 12, and 24 h post-challenge. |
Analysis of the Potato Gene Index
The ESTs were cleaned to remove low-quality regions and were then assembled and clustered to generate a gene index (Quackenbush et al., 2000, 2001). We were able to reduce the total number of ESTs to 19,892 unique sequences, with 50,789 ESTs clustering into 8,741 tentative consensus (TC) sequences leaving 11,151 sequences as singleton ESTs. With respect to annotation of the sequences generated in this study, we were able to assign a putative identification for 8,701 of the 19,892 unique sequences (43.7%) generated from the gene index build. In a search against known transposable elements from Arabidopsis, potato, and tomato, 38, 28, and 89 TC or singleton sequences, respectively, were detected within the 19,892 unique sequences (E ≤ −5), suggesting limited expression of transposable elements in potato.
The rate of gene discovery was assessed for each library by calculating the fraction of each library composed of unique sequences (no. of unique TCs plus singleton ESTs) within each library. With the exception of the sprouting eye I library, which was not deeply sampled, the proportion of sequences that were unique within libraries were similar, ranging from 51% to 64% (Table II). The number of ESTs that could be clustered into TCs was similar across all libraries (75%–88%), suggesting that transcripts were distributed approximately the same within each library.
Table II.
Statistics on clustering and redundancy within cDNA libraries and within the entire potato EST database
| Library | No. of ESTs | No. ESTs within TCs (%) | No. of TCs | No. of Singleton ESTs (%) | Unique Sequencesa (%) |
|---|---|---|---|---|---|
| Stolon | 10,286 | 8,610 (84) | 3,570 | 1,676 (16) | 5,246 (51) |
| Leaf | 10,361 | 8,476 (82) | 3,559 | 1,885 (18) | 5,444 (53) |
| Sprouting eye I | 902 | 739 (82) | 543 | 163 (18) | 706 (78) |
| Sprouting eye II | 9,230 | 7,363 (80) | 3,453 | 1,867 (20) | 5,320 (58) |
| Dormant tuber | 5,049 | 4,313 (85) | 2,184 | 736 (15) | 2,920 (58) |
| Microtuber | 5,426 | 4,785 (88) | 2,101 | 641 (12) | 2,742 (51) |
| Root | 10,190 | 8,360 (82) | 3,625 | 1,830 (18) | 5,455 (54) |
| Compatible leaf | 5,062 | 3,809 (75) | 1,881 | 1,253 (25) | 3,134 (62) |
| Incompatible leaf | 5,434 | 4,334 (80) | 2,365 | 1,100 (20) | 3,465 (64) |
| Total (all nine libraries) | 61,940 | 50,789 (82) | 8,741 | 11,151 (18) | 19,892 |
Calculated as the sum of the no. of TCs and the no. of singleton ESTs within a library.
A major advantage of EST sequencing from multiple libraries is the ability to identify genes that are putatively transcribed specifically within a certain tissue or during a particular developmental phase. Our analysis revealed that 13,068 sequences (21%) were unique to one of the nine cDNA libraries sampled (Table III). There was a 2-fold range in library specific sequences, with 14% in the microtuber library to 27% in the compatible leaf library.
Table III.
Identification of sequences specific to a single cDNA library
| Library | Library-Specific Sequences (%) |
|---|---|
| Stolon | 2,021 (20) |
| Leaf | 2,273 (22) |
| Sprouting eye I | 176 (20) |
| Sprouting eye II | 2,045 (22) |
| Dormant tuber | 851 (17) |
| Microtuber | 775 (14) |
| Root | 2,356 (23) |
| Compatible leaf | 1,371 (27) |
| Incompatible leaf | 1,200 (22) |
| Total | 13,068 (21) |
The no. of library-specific sequences was determined by adding the TC and the singleton ESTs that were detected only in a single cDNA library. The no. in parentheses are the percent of library-specific sequences within all the sequences determined from that library.
As expected, several transcripts were widely expressed in all cDNA libraries sampled. Forty-eight different TCs were identified that contained ESTs derived from all nine libraries, and, thus, most likely represent typical “housekeeping” genes (Tables IV and V). The most abundant transcript detected was putatively identified as heat shock cognate protein 80 (360 ESTs; TC 65), followed by catalase (250 ESTs; TC 8) and elongation factor 1-alpha (233 ESTs; TC 19; Table V). If the sprouting eye I library was excluded, a total of 111 TCs were identified that contained ESTs from all eight cDNA libraries. In addition to the 48 TCs detected in all nine cDNA libraries, 6,776 additional TCs could be detected in at least two different cDNA libraries (Table IV), whereas 1,917 TCs were unique to a single cDNA library.
Table IV.
Distribution of sequences in multiple cDNA libraries
| Libraries Represented | No. of TCs |
|---|---|
| 9 | 48 |
| 8 | 105 |
| 7 | 172 |
| 6 | 298 |
| 5 | 482 |
| 4 | 853 |
| 3 | 1,546 |
| 2 | 3,320 |
| 1 | 1,917 |
| Total | 8,741 |
TCs were examined for this distribution among all nine cDNA libraries and the total no. of TCs that were distributed in multiple cDNA libraries was determined.
Table V.
Putative function of the most abundant sequences present in all nine cDNA libraries
| TC | No. of ESTs in TC | Tentative Annotation |
|---|---|---|
| TC65 | 360 | Heat shock cognate protein 80 |
| TC8 | 250 | Catalase (CAT1) |
| TC19 | 233 | Elongation factor 1-alpha (EF-1-alpha) |
| TC91 | 199 | Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) |
| TC33 | 181 | Elongation factor 1-alpha (EF-1-alpha) |
| TC4447 | 176 | L3 ribosomal protein |
| TC4413 | 151 | DnaJ protein |
| TC31 | 119 | S-adenosylmethionine decarboxylase (SAMDC) |
| TC4439 | 107 | Alpha-tubulin |
| TC4459 | 104 | Translationally controlled tumor protein (TCTP; P23) |
| TC4461 | 95 | Chaperonin-60 beta chain precursor, nuclear gene encoding chloroplast protein |
| TC4451 | 85 | Phosphoglycerate kinase (cytosolic isoenzyme) |
| TC4490 | 80 | Annexin p34 |
| TC149 | 76 | Cys protease |
| TC4492 | 76 | Mitochondrial formate dehydrogenase precursor |
| TC150 | 75 | Phospho-2-dehydro-3-deoxyheptonate aldolase 1 chloroplast precursor (DAHP synthetase 1) |
| TC4414 | 63 | DnaJ-like protein |
| TC4488 | 63 | Elongation factor 2 |
| TC165 | 61 | (pSTH-2 protein) Pathogenesis-related genes |
| TC167 | 61 | UDP-Glc:protein transglucosylase |
| TC4507 | 61 | AT5g28840/F7P1_20 |
Analysis of Sequence Origin in the Late-Blight Pathogen-Challenged Libraries
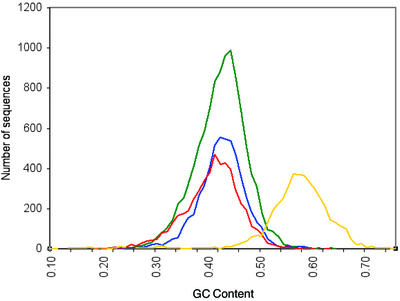
RNA from the late-blight pathogen-challenged libraries was isolated from leaf tissue challenged with high concentrations of the late-blight pathogen. Because the pathogen is invasive and establishes hyphae within the leaf tissue, we were unable to separate pathogen tissue from host tissue. As a consequence, it is possible that a portion of the sequences derived from late-blight pathogen-challenged tissues is of pathogen, not host, origin. We utilized two independent methods, GC content and sequence similarity, to assess the frequency of late-blight pathogen sequences within the two late-blight pathogen-challenged libraries. The average GC content for potato is 42.7% GC as determined from 51,444 ESTs generated in this study from non-pathogen-challenged tissues. In contrast, the average GC content of late-blight pathogen is 56.6% GC as determined from a total of 4,314 publicly available late-blight pathogen ESTs (GenBank dbEST release 128, February 2002). Analyses of the GC content profiles of ESTs from the healthy leaf and the two late-blight pathogen-challenged leaf libraries (incompatible and compatible) are similar, suggesting that there is not a substantial number of late-blight pathogen sequences present in either of the two late-blight pathogen-challenged libraries (Fig. 1). The GC content analysis was consistent with results based on sequence similarity. We searched the ESTs from the two late-blight pathogen libraries against 4,314 publicly available late-blight pathogen ESTs. Using a high stringency cutoff (95% identity over 100 bases) to identify sequences of late-blight pathogen origin, a total of six ESTs from the late-blight pathogen-challenged incompatible leaf library and a total of six ESTs from the late-blight pathogen-challenged compatible leaf library, were similar to publicly available late-blight pathogen ESTs. The late-blight pathogen sequences had similarity to typically highly abundant sequences including ribosomal proteins, elongation factors, and glyceraldehye-3-phosphate dehydrogenase (data not shown). In a more expansive search of a private collection of 77,916 late-blight pathogen ESTs using BLASTN, a total of 440 ESTs from the compatible library and 248 ESTs from the incompatible library matched a late-blight pathogen sequence using a cutoff of 1e-30 (T. Randall, personal communication). The GC content of these contaminating late-blight pathogen ESTs ranged from 27% to 59% GC, falling within the GC content distribution profile of ESTs from the healthy leaf library. These data suggest that a small fraction of the sequences obtained from the late-blight pathogen-challenged leaf libraries were derived from the oomycete pathogen.
Figure 1.
GC content of leaf, late-blight pathogen-challenged leaf libraries, and late-blight pathogen ESTs. A total of 10,361 ESTs were used for the healthy leaf analysis (green), 5,434 ESTs for the incompatible leaf library (blue), 5,062 ESTs for the compatible leaf library (red), and 4,314 ESTs for the late-blight pathogen sequences (yellow).
EST Frequency Clustering Analyses to Identify Broad Patterns of Gene Expression
As described above, at least 5,000 ESTs were produced from non-normalized cDNA libraries that were derived from tissue sources chosen to represent biological processes specific to potato. Because of this relatively deep sampling approach, the frequency of ESTs in a given library can be used to infer the transcriptome in the cells from which the library was derived. This approach has advantages in comparison with hybridization-based approaches in that the deep sampling approach is a more quantitative method and able to distinguish closely related paralogous genes (for review, see Ohlrogge and Benning, 2000).
As a first step, we performed a clustering analysis to assess the relatedness of each library based on EST abundance (Ewing et al., 1999). First, we compiled 8,188 TCs into a “matrix file” containing the frequency of ESTs corresponding to each TC in each of seven libraries that represent leaf challenge with late-blight pathogen and the four stages of tuber development (stolon, microtuber, dormant tuber, and sprouting eyes). We did not include the sprouting eye I library because of the low number of sequences. The R statistic described by Stekel et al. (2000) was used to identify the most highly significant differences in EST abundance for each TC among the libraries. To limit our analyses to those genes that were the most differently expressed within the tissues, only TCs with an R > 12 (254 in total), which represent a mixture of moderate to highly expressed genes, were used for the hierarchical clustering analysis. This value provides a 99.9% “true positive” rate (Stekel et al., 2000). From the hierarchical clustering analysis of this set, it is evident that the three leaf libraries form a well-supported branch (100%) distinct from the group composed of the libraries associated with tuber development: stolon, microtuber, dormant tuber, and sprouting eye II (Fig. 2). This result was not unexpected because photosynthesis-related cDNAs are more abundant in the three leaf libraries relative to the four tuber-associated libraries (Fig. 2). We also observed clusters of TCs that were more abundant in tuber-associated tissues. For example, TC 4,437 (Suc synthase), TC 4,503 (UTP Glc-1-phosphate uridyltransferase; both involved in starch synthesis), and TC39 (polyubiquitin 5) are highly enriched in the four libraries derived from tuber-related tissues, whereas TCs corresponding to class I patatin and proteinase inhibitors were more abundant in both tuber libraries and the sprouting eye II library but not the stolon library (Fig. 2). Although there is a considerable overlap in the frequency of ESTs in these two main clusters of libraries, several strongly supported subclusters such as compatible leaves with incompatible leaves and stolons with microtubers and sprouting eye II were also observed (Fig. 2). We obtained the same clustering of libraries when lower cutoffs (more genes) were used (data not shown).
Figure 2.
Hierarchical clustering analysis of differentially expressed transcripts. A set of 254 differentially expressed TCs (R >12) and seven libraries were clustered as described in “Materials and Methods.” The frequencies of a given TC from each library are indicated by increasing intensities of red. A frequency of zero is denoted with black. Bootstrapping (1,000 replicates) was used to determine the level of support for each branch. Black branches are 100% supported and yellow branches have 60% to 70% support. The libraries are: dormant tuber (1), sprouting eye II (2), microtuber (3), stolon (4), healthy leaf (5), late-blight pathogen-challenged compatible leaf library (6), and late-blight pathogen-challenged incompatible leaf library (7).
As an alternative to the hierarchical clustering analysis described above we also employed k means clustering (for review, see Quackenbush, 2001) to identify biologically relevant clusters of genes. In this analysis, we used a dataset including more genes (1,159 TCs) having a minimum of six ESTs comprising the TC and using R > 5. First, figures of merit (Yeung et al., 2001) were calculated. From this analysis, 27 clusters were found to be optimally predictive for the k means clustering algorithm and were consistent with the results obtained through hierarchal clustering (Figs. 3 and supplemental data Fig. 1; all supplemental data are available at www.plantphysiol.org). In the k means-derived cluster shown in Figure 3A, similar levels of expression of photosynthesis-related genes were apparent in the three leaf libraries, including multiple subunits and isozymes of Rubisco (TC4395, TC4392, TC4393, and TC4381) and photosystem components (TC4517, TC4509, TC283, and TC242). Within this cluster, a subcluster of genes with higher expression levels in the two late-blight pathogen-challenged leaf libraries was evident. Sequences present in both late-blight pathogen-challenged libraries include multiple defense related genes including 1,3-beta glucanase (TC749), endochitinase (TC4544), peroxidase (TC4609 and TC494), pathogenesis-related protein PR-1 (TC4643, TC302, and TC4642), and pathogenesis-related protein P2 (TC4485).
Figure 3.
Nonhierarchical (k means clustering analysis) of transcripts. A, Cluster of genes associated with leaf tissues; B, cluster of genes associated with stolon, microtuber, dormant tuber, and sprouting eyes. The frequencies of a given TC from each library are indicated by increasing intensities of red. A frequency of zero is denoted by black. The libraries are: dormant tuber (1), sprouting eye II (2), microtuber (3), stolon (4), healthy leaf (5), late-blight pathogen-challenged compatible leaf library (6), and late-blight pathogen-challenged incompatible leaf library (7).
The k means-derived cluster in Figure 3B reveals moderately expressed genes with elevated expression in the four tuber-related tissues: stolons, microtubers, dormant tubers, and sprouting eyes. This cluster overlaps with the results from the hierarchal clustering including TC 4437 (Suc synthase), TC4503 (UTP Glc-1-phosphate uridyltransferase; both involved in starch synthesis), and TC39 (polyubiquitin 5), yet provides an expanded set of genes coregulated in these four tuber-related tissues.
Genes Unique to the Incompatible Interaction with Late-Blight Pathogen
The clustering analyses described above reveal patterns of expression present in the more moderately expressed transcripts present within our gene index. However, other genes, not highly expressed in either a single or multiple tissues, are also relevant to biological processes. To identify genes important in the establishment of the resistance response, we identified sequences that are expressed exclusively in response to the incompatible interaction with late-blight pathogen. TCs (100) and annotated singleton ESTs (329) that are unique to the incompatible leaf library are listed in supplementary Table SI. Among the genes in this set are sequences with similarity to genes previously implicated in defense responses including a class IV chitinase (TC6866), class II chitinase (BQ046750), disease resistance proteins D and E from tomato (BQ047199 and BQ047222), avr9/Cf9-induced protein 111B (TC8251 and BQ046602), and genes involved in synthesis of secondary metabolites (TC 8 410, TC3979, and BQ047054). One interesting set of sequences unique to the incompatible leaf library encodes orthologs of the Hcr (homologs of Cladisporium fulvum resistance) genes that are Leu-rich repeat-encoding resistance genes (Parniske and Jones 1999). Four singleton ESTs (BG589307, BG591114, BQ047142, and BG590641) that encode orthologs of Hcr2A-0A from tomato, Hcr9–0 from tomato, HcrVf2 from Malus floribunda, and Hcr9–9E from Lycopersicon pimpinellifolium, respectively, were present in the late-blight pathogen-challenged incompatible leaf library. A singleton EST encoding an ortholog of the Cf-4A from tomato (BQ046539) was also detected in the incompatible leaf library. In addition to these defense-related and resistance genes, a number of genes implicated in signal transduction and gene regulation were unique to the late-blight pathogen-incompatible leaf library, including multiple protein kinases, calmodulin-binding proteins, and transcription factors. Other genes with functions in metabolism were unique to the late-blight pathogen-challenged incompatible interaction. However, a large percentage of TCs and singleton ESTs (69.7%) have either no match or match an unknown or hypothetical protein.
Transcripts Differentially Expressed during Tuber Development, Tuber Dormancy, and Sprouting
Tuber development, dormancy, and sprouting are processes of agronomic interest that occur below ground and are not found in other model plant species. To best study this process, an in vitro tuberization system was developed (Hendriks et al., 1991). Stolons and microtubers were derived from the axillary buds of nodal stem cuttings. Because of the similarity of the tissues employed, a comparison between the libraries should reveal genes induced during tuber initiation and outgrowth. We employed a series of pair-wise comparisons between successive developmental stages to identify genes up-regulated relative to the preceding stage. Genes up-regulated during each stage of development were identified by calculating percent frequencies for each TC in each library and then calculating the fold differences in frequencies between libraries. We also calculated a probability value for each gene in each two-library comparison (Audic and Claverie, 1997).
Supplementary Table SII lists the genes that exhibit changes in gene expression during the early stages of tuber development (stolon versus microtuber). Among the most conspicuous increases in EST abundance correspond to transcripts encoding the storage protein patatin (TC4432 and TC4433) and proteinase (TC139) and peptidase (TC215) inhibitors. This was previously observed in the in vitro system (Hendricks et al., 1991). Fructokinase (TC4493) and Suc synthase (TC4437) were also previously shown to be highly induced in the tuberization system (Appeldoorn et al., 2002). In addition to the expected differences, there are a number of genes in this set with either no match in the database or match a protein of unknown function and which are likely important for tuberization. Supplemental Table SIII lists genes up regulated during tuber dormancy (dormant tuber versus microtuber). Among the genes induced, greater than 10-fold include annexin p34 (TC4490), which is involved in membrane organization and traffic, a gene similar to probable aminopeptidase from Arabidopsis (TC296), Glc-6-phosphate 1-dehydrogenase (TC4555), and hexameric polyubiquitin (TC47). Genes differentially expressed during sprouting are shown in Supplemental Table SIV. The most obvious feature is the large number of TCs involved in translation (translation elongation factor eEF1B-alpha [TC4535] and nine TCs corresponding to ribosomal proteins). High expression of these genes is expected as the sprouting eyes are undergoing rapid cell division.
Similarity of Potato with Tomato
The generation of 62,000 potato ESTs and a gene index with 19,640 unique sequences provides a powerful resource to assess sequence conservation between potato and other Solanaceae species. Using the TIGR Tomato Gene Index (release 8.0, www.tigr.org/tdb/tgi), which is composed of 155,054 tomato ESTs and expressed transcripts, we compared the sequence identity between potato and tomato. As shown in Table VI, there is a high degree of sequence conservation between tomato and potato. At the nucleotide level using a cutoff of e-10, a total of 15,987 potato sequences (7,998 TCs and 7, 989 ESTs) had sequence similarity with a sequence in the tomato gene index. This represents 80.4% of all the unique sequences in the potato gene index. Increasing the stringency to e-25 reduced the number of potato sequences with a match to the tomato gene index to 14,165 sequences (71.2% of the total potato sequences). Similar results were observed using TBLASTX instead of TBLASTN.
Table VI.
Sequence conservation between potato and tomato
| Search Program/ Cutoff Criterion | TC | EST | Total |
|---|---|---|---|
| e-10 | |||
| BLASTN | 7,998 | 7,989 | 15,987 |
| TBLASTX | 8,020 | 7,874 | 15,894 |
| e-25 | |||
| BLASTN | 7,624 | 6,541 | 14,165 |
| TBLASTX | 7,598 | 6,272 | 13,870 |
A total of 19,892 unique potato sequences (TCs and singleton ESTs) were searched against a gene index of 32,317 unique tomato sequences (TCs and singleton ESTs; www.tigr.org/tdb/lgi) using either BLASTN or TBLASTX.
DISCUSSION
To gain insight into the potato transcriptome, we report in this study the generation of approximately 62,000 EST sequences from potato. Because the inherent nature of EST sequencing results in the redundant sequencing of identical transcripts, we reduced the redundancy in the EST dataset by creating a gene index. Through a series of clustering and assembly processes, we were able to reduce the approximately 62,000 ESTs into 19,892 unique sequences. Based on sequence similarity to known genes, we assigned a putative function to 8,701 (43.7%) of the unique sequences. We also identified transcripts unique to single cDNA libraries that represent differentially expressed genes and sequences broadly expressed in all sampled libraries that represent “housekeeping” genes.
The sequences reported in this study provide a significant improvement in our understanding of potato because the ESTs generated in this study were derived from biologically and agronomically relevant tissues. Aerial tissue was represented in three leaf libraries: healthy leaves and two pathogen-challenged leaf libraries. Below-ground tissue was represented by four libraries: stolon, microtuber, dormant tuber, and root. An intermediate between aerial and below-ground tissue was represented by the sprouting eye libraries. The process of tuber development is represented in four libraries: stolon, microtuber, dormant tuber, and sprouting eyes, whereas the healthy and challenged leaf libraries represent differential responses (compatible versus incompatible) to pathogen challenge. Because none of the libraries were normalized and greater than 5,000 ESTs are available from each, these libraries can be utilized not only for gene discovery but also for comparative analyses of gene expression.
Using two clustering methods, we were able to identify patterns of expression specific to the tissues of agronomic and biological interest. For aerial tissue, genes present in all three leaf libraries were identified along with a set of genes associated with late-blight pathogen infection. Although the former represents well-documented photosynthetic-related transcripts, the later represents those genes induced by pathogen infection yet present in both the compatible and incompatible interaction. The expression of defense-related genes such as chitinase, beta-glucanase, and pathogenesis-related proteins in both compatible and incompatible interactions has been documented previously (Maleck et al., 2000). Because both late-blight pathogen libraries used in this study were constructed from RNA isolated from multiple time points of the interaction with the pathogen, we are not able to assess temporal expression of these defense genes in the infection process and, as a consequence, cannot correlate a temporal gene expression pattern with the incompatible interaction. In addition to the clusters of genes involved in pathogen responses, we were able to identify clusters of genes associated with tuber development from tuber formation through the sprouting stage.
We were able to identify genes specific to the resistance response by examining sequences unique to the incompatible leaf library. In total, 1,200 sequences (100 TCs and 1,100 singleton ESTs) were specific to the incompatible interaction. As expected, a subset of these genes had similarity to genes previously implicated in defense responses that encoded antimicrobial factors. Another subset encodes protein kinases and transcription factors that may have a role in signal transduction and gene regulation events specific to the resistance response. For example, two singleton ESTs with similarity to the tomato Pti4 and Pti6 transcription factors (BQ047502 and BQ047690) were unique to the incompatible library and belong to the ethylene response family of transcription factors that bind to cis elements in the promoters of pathogenesis-related proteins, thereby activating their expression (Gu et al., 2002). Although a putative function could be assigned to 370 (41 TCs and 329 singletons) of these sequences, the remaining sequences (836) have no annotation or match unknown or hypothetical proteins and represent a new source of genes potentially involved in the resistance response. The TCs and singleton ESTs in supplemental Table SI are considered unique to the incompatible leaf library because they did not cluster with any other ESTs from the other eight libraries. However, the annotation associated with some of these sequences indicated overlap in function with sequences in other libraries sampled in this study. Whether these are the same gene or related members of a gene family is unknown at this time. With the availability of full-length sequences, deeper sequencing of these libraries, and/or expression studies, we will be able to further characterize the function of these genes in the incompatible interaction.
Potato tubers are economically important plant organs that have no counterpart among other plant models. Morphologically, the tuber is a modified stem with greatly reduced leaves, axillary buds, and internodes with a greatly expanded stem radial axis. In nature, tubers generally originate from stolon tips (modified underground stems) and are subject to control by photoperiod, light intensity, temperature, and nutrient availability (for review, see Jackson, 1999). In addition to morphological changes, tubers accumulate large amounts of starch (20% of fresh weight) and characteristic proteins such as patatin. The tuber life cycle consists of induction, initiation, enlargement, dormancy, and sprouting stages (for review, see Fernie and Willmitzer, 2001). Our libraries represent four of these stages: induction (stolon library), initiation (microtubers), dormancy (dormant tubers), and sprouting (sprouting eyes). We identified genes with a potential role in tuber development by comparison of EST frequencies between libraries representing successive stages of development. Many of the genes identified in our study were expected based on previous studies. However, many of the TCs, particularly among those unique to microtubers and tubers, show no similarity to known genes and may represent genes that may be manipulated for potato tuber improvement.
The genome size of a number of the major crop species (including potato) are in the range of gigabases (Arumuganathan and Earle, 1991) and, as a consequence, current and future insight into the genomes of these species will be through partial genome projects such as single pass sequencing of cDNA clones to generate ESTs. As a consequence, the approximately 62,000 ESTs generated in this study provide a major resource for studying potato biology. These sequences also provide a rich resource for comparative genomics and in a comparison of 19,892 unique potato sequences derived from 61,940 potato ESTs with 32,317 unique tomato sequences derived from 155,054 tomato ESTs, approximately 80% of the potato ESTs were similar to a tomato sequence. Through additional genomic efforts such as gene expression profiling, we will be able to further define the role the sequences identified in this study have in growth and development of potato.
MATERIALS AND METHODS
Library Construction
Libraries were generated from mRNA isolated from multiple tissues of potato (Solanum tuberosum). All libraries were constructed from potato cv Kennebec, with the exception of the stolon and microtuber libraries, which were constructed from the potato cv Bintje. All libraries were directionally cloned into pBluescript vectors (Stratagene, LaJolla, CA) and after ligation, cloned into SOLR cells (Ausubel et al., 1994). The healthy leaf, sprouting eye, stolon, root, and tuber libraries were constructed in the Steve Tanksley lab. The healthy leaf library was constructed from leaflets and petioles obtained from greenhouse-grown (8-week-old) plants. The sprouting eye libraries were constructed from 2- to 15-mm germinating eyes from Kennebec tubers. The stolon library was constructed from developing axillary buds of potato nodal stem cuttings cultured on a medium that induces tuber formation (Bachem et al., 1996). The microtuber library was constructed using in vitro-grown tubers. The dormant tuber library was constructed from internal tuber tissue (excluding epidermal and bud tissue) that had been stored for 1 month after harvest at 4 C. The root library was constructed from roots grown in vitro on CM medium.
Two libraries were constructed from late-blight pathogen (Phytophthora infestans)-challenged leaf tissue. The BLPI library was constructed in the Barbara Baker lab after challenging incompatible leaves with late-blight pathogen US-1 (US 940501; 450,000 sporangia mL−1) in the Biotron (University of Wisconsin, Madison). RNA was isolated from leaf tissue collected at 1, 2, 5, 12, and 24 h post-challenge. The PPC library was constructed in the William Fry lab after challenging leaves with the compatible late-blight pathogen isolate US 940480 (20,000 sporangia mL−1). RNA was isolated from tissue collected at 3, 6, 9, 12, 24, 48, and 72 h after inoculation.
Sequencing Methodology
Clones were grown for 18 h in yeast tryptone media (Biofluids, Rockville, MD). Templates were prepared using the Eppendorf-5 Prime Direct Bind prep kit (Eppendorf, Boulder, CO). The 5′ ends of the cDNA clones were sequenced on ABI 377 or 3700 sequencing machines using standard sequencing methods. Bases were called using either phred (Ewing and Green, 1998; Ewing et al., 1998) or the TraceTuner program (Paracel, Pasadena, CA). Vector and low-quality bases were trimmed using an in-house program.
Computational Methods
EST sequences were trimmed to eliminate vector, adaptor, and low-quality sequences. Sequences sharing greater than 94% identity over 40 or more contiguous bases with unmatched overhangs less than 30 bases in length were placed into clusters. Overlaps based exclusively on low-complexity regions were excluded. Each cluster was assembled at high stringency using the Paracel Transcript Assembler (version 2.6.2, http://www.paracel.com; Huang and Madan, 1999) to produce TC sequences. Alignments containing gaps (or inserts) longer than nine nucleotides were discarded, allowing for the segregation of possible alternative splice forms. Sequences not assembled into a TC were termed singleton ESTs. The TCs and the singleton ESTs were searched against a nonredundant protein database to provide a putative function with a minimum of 30% identity over 20% of the length of the protein required for a TC or singleton EST to be annotated (Quackenbush et al., 2000, 2001). Transposable element sequences of Arabidopsis, potato, and tomato (Lycopersicon esculentum; 93, 12, and 99 sequences, respectively) were downloaded from the GenBank as of October 31, 2002. BLASTN (WU-BLAST 2.0, http://blast.wustl.edu; Altschul et al., 1990) was used to identify the presence of transposable elements in the 19,892 TC and singleton EST sequences using a cutoff criterion of E ≤ −05. Tomato and potato EST sequences were searched using WU-BLAST 2.0 (W. Gish, unpublished data; http://blast.wustl.edu; Altschul et al., 1990). Potato EST data described in this study are available online (http://www.tigr.org/tdb/potato/plantphysiologypaper/specialgeneindex).
Digital analyses of gene expression were performed with the TIGR MultipleExperimentViewer software (version 1.1; Quackenbush, 2001) by using transcript abundance in each TC inferred from the EST frequency for that TC in all seven libraries. Only TCs that were composed of at least six ESTs were used for the cluster analyses. Hierarchical clustering (Eisen et al., 1998) with statistical support for the nodes of the trees, based on resampling the data, was performed. k Means clustering (Soukas et al., 2000) with initial calculation of the figures of merit (Yeung et al., 2001) was also performed.
Supplementary Material
ACKNOWLEDGMENTS
We wish to acknowledge the bioinformatic and information technology support from Michael Heaney, Susan Lo, Vadim Sapiro, Corey Irwin, Eddy Arnold, Rajeev Karamchedu, Jeff Shao, and Billy Lee. We also wish to thank Tamara Feldblyum and members of the TIGR sequencing core facility. The authors are grateful to the critical reading and comments of Norman Lee.
Footnotes
This work was supported by the National Science Foundation Plant Genome Research Program (grant no. DBI–9975866 to B.J.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.013581.
LITERATURE CITED
- Adams MD, Soares MB, Kerlavage AR, Fields C, Venter JC. Rapid cDNA sequencing (expressed sequence tags) from a directionally cloned human infant brain cDNA library. Nat Genet. 1993;4:373–380. doi: 10.1038/ng0893-373. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Appeldoorn NJ, Sergeeva L, Vreungdenhil D, van der Plas LH, Visser RG. In situ analysis of enzymes involved in sucrose to hexose-phosphate conversion during stolon-to-tuber transition of potato. Physiol Plant. 2002;115:303–310. doi: 10.1034/j.1399-3054.2002.1150218.x. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- Audic S, Claverie J-M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994. [Google Scholar]
- Bachem CW, van der Hoeven RS, de Bruijn SM, Vreugdenhill D, Zabeau M, Visser RG. Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J. 1996;9:745–753. doi: 10.1046/j.1365-313x.1996.9050745.x. [DOI] [PubMed] [Google Scholar]
- Barry GF. The use of the Monsanto draft rice genome sequence in research. Plant Physiol. 2001;125:1164–1165. doi: 10.1104/pp.125.3.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonierbale MR, Plaisted MRL, Tanksley SD. RFLP maps of potato and tomato based on a common set of clones reveal modes of chromosomal evolution. Genetics. 1988;120:1095–1103. doi: 10.1093/genetics/120.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crookshanks M, Emmersen J, Welinder KG, Nielsen KL. The potato tuber transcriptome:analysis of 6077 expressed sequence tags. FEBS Lett. 2001;506:123–126. doi: 10.1016/s0014-5793(01)02888-5. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred: II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred: I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Ewing RM, Ben Kahla A, Poirot O, Lopez F, Audic S, Claverie J-M. Large-scale statistical analysis of rice ESTs reveal correlated patterns of gene expression. Genome Res. 1999;9:950–959. doi: 10.1101/gr.9.10.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Willmitzer L. Molecular and biochemical triggers of potato tuber development. Plant Physiol. 2001;127:1459–1465. [PMC free article] [PubMed] [Google Scholar]
- Fry WE, Goodwin SB. Resurgence of the Irish potato famine fungus. Bioscience. 1997;4 7:363–371. [Google Scholar]
- Gu YQ, Wildermuth MC, Chakravarthy S, Loh YT, Yang C, He X, Han Y, Martin GB. Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses expressed in Arabidopsis. Plant Cell. 2002;14:817–831. doi: 10.1105/tpc.000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks T, Vreugdenhil D, Stiekema WJ. Patatin and four serine proteinase inhibitor genes are differentially expressed during tuber development. Plant Mol Biol. 1991;17:385–394. doi: 10.1007/BF00040633. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SD. Multiple signaling pathways control tuber induction in potato. Plant Physiol. 1999;119:1–8. doi: 10.1104/pp.119.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 2002;129:400–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone KD, Lackney VK, Blauth JR, van Wijk R, Jahn MK. Genome mapping in Capsicum and the evolution of genome structure in the Solanaceae. Genetics. 1999;152:1183–1202. doi: 10.1093/genetics/152.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K, Levine K, Euglem T, Schimd J, Lawton KA, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Benning C. Unraveling plant metabolism by EST analysis. Curr Opin Plant Biol. 2000;3:224–228. [PubMed] [Google Scholar]
- Parniske M, Jones J. Recombination between diverged clusters of the tomato Cf-9 plant disease resistance gene family. Proc Natl Acad Sci USA. 1999;96:5850–5855. doi: 10.1073/pnas.96.10.5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DG, Schulze SR, Sciara EB, Lee SA, Bowers JE, Nagel A, Jiang N, Tibbitts DC, Wessler SR, Paterson AH. Integration of Cot analysis, DNA cloning, and high-throughput sequencing facilitates genome characterization and gene discovery. Genome Res. 2002;12:795–807. doi: 10.1101/gr.226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J. Computational analysis of microarray data. Nat Rev. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. The TIGR gene indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 2001;29:159–164. doi: 10.1093/nar/29.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J. The TIGR gene indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 2000;28:141–145. doi: 10.1093/nar/28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz PD, Schutz K, Dedhia N, Yordan C, Parnell LD, Stein L, McCombie WR, Martienssen RA. Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat Genet. 1999;23:305–308. doi: 10.1038/15479. [DOI] [PubMed] [Google Scholar]
- Schumann GL. The Irish potato famine and the birth of plant pathology. In: Schumann GL, editor. Plant Diseases: Their Biology and Social Impact. St. Paul, MN: American Phytopathological Society; 1991. pp. 1–24. [Google Scholar]
- Soares MB, Bonaldo MF, Jelene P, Su L, Lawton L, Efstratiadis A. Construction and characterization of a normalized cDNA library. Proc Natl Acad Sci USA. 1994;91:9228–9232. doi: 10.1073/pnas.91.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas A, Cohen P, Socci ND, Friedman JM. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 2000;4:963–980. [PMC free article] [PubMed] [Google Scholar]
- Stekel DJ, Git Y, Falciani F. The comparison of gene expression from multiple cDNA libraries. Genome Res. 2000;10:2055–2061. doi: 10.1101/gr.gr-1325rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP et al. High density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung KY, Haynor DR, Ruzzo WL. Validating clustering for gene expression data. Bioinformatics. 2001;17:309–318. doi: 10.1093/bioinformatics/17.4.309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.