Abstract
The capsaicin (vanilloid) receptor, VR1, is a sensory neuron-specific ion channel that serves as a polymodal detector of pain-producing chemical and physical stimuli. The response of VR1 to capsaicin or noxious heat is dynamically potentiated by extracellular protons within a pH range encountered during tissue acidosis, such as that associated with arthritis, infarction, tumor growth, and other forms of injury. A molecular determinant for this important physiological activity was localized to an extracellular Glu residue (E600) in the region linking the fifth transmembrane domain with the putative pore-forming region of the channel. We suggest that this residue serves as a key regulatory site of the receptor by setting sensitivity to other noxious stimuli in response to changes in extracellular proton concentration. We also demonstrate that protons, vanilloids, and heat promote channel opening through distinct pathways, because mutations at a second site (E648) selectively abrogate proton-evoked channel activation without diminishing responses to other noxious stimuli. Our findings provide molecular evidence for stimulus-specific steps in VR1 activation and offer strategies for the development of novel analgesic agents.
Pain is initiated when noxious thermal, mechanical, or chemical stimuli excite the peripheral terminals of specialized primary afferent neurons called nociceptors (1). Tissue damage, such as that associated with infection, inflammation, or ischemia, produces an array of chemical mediators that activate or sensitize nociceptor terminals to elicit pain and promote tenderness at the site of injury. An important component of this proalgesic response is local acidosis, namely, a reduction in extracellular pH to levels below the physiological norm of ≈7.6 (2). Protons are capable of modulating the activity of a number of receptors and ion channels expressed by primary afferent nociceptors, including acid-sensitive channels of the degenerin family (3), ATP-gated channels (4, 5), and vanilloid receptors (6, 7). Analyses of native and cloned vanilloid receptors suggest that they play a role in mediating sustained proton responses in vivo (8–12). This is further supported by the profoundly reduced responses to acid in cultured dorsal root ganglion neurons and isolated skin nerve preparations of mice with a targeted disruption of the gene encoding for the cloned capsaicin (vanilloid) receptor VR1 (13).
Vanilloid receptors are nociceptor-specific cation channels that serve as the molecular target for capsaicin, the main pungent ingredient in “hot” chili peppers. VR1 is homologous to members of the transient receptor potential family of ion channels first identified in the Drosophila phototransduction pathway (6, 14). We have shown that, when expressed in heterologous systems, VR1 is also activated by noxious heat (6, 7) with a threshold (>43°C) resembling that of native heat-activated currents in cultured sensory neurons (15). Acidification of the extracellular milieu has two primary effects on VR1 function. First, extracellular protons increase the potency of heat or capsaicin as VR1 agonists, in part, by lowering the threshold for channel activation by either stimulus. For example, a decrease in extracellular pH to 6.4 induces channel activity at body temperature. Second, extracellular protons can, themselves, be viewed as VR1 agonists because further acidification (to pH < 6.0) leads to channel opening at room temperature (7).
Protons have been shown to modulate ion channels through effects on ligand binding, channel gating, or ion conduction (16–21). In the case of the capsaicin receptor, extracellular protons are believed to act primarily by increasing the probability of channel opening (7, 22), rather than by altering unitary conductance or interacting directly with a vanilloid-binding site (which may be intracellular) (6, 23). Whereas it is clear that protons serve as both modulators and activators of VR1, little is known about the mechanism(s) underlying these phenomena. Here, we identify a site (E600) within a putative extracellular domain of the VR1 protein at which mutations alter channel sensitivity to capsaicin and heat. We propose that E600 serves as an important regulatory site for proton potentiation of vanilloid receptor activity over a physiologically relevant range (pH 6–8). We also demonstrate that mutations at a second extracellular site (E648) significantly reduce proton-activated currents without altering heat- or capsaicin-evoked responses, or without eliminating the ability of protons to potentiate responses to these stimuli. These data provide molecular evidence for the existence of stimulus-specific steps in the VR1 activation process.
Materials and Methods
Molecular Biology.
Point mutations were introduced by using oligonucleotide-directed mutagenesis (20, 24). All constructs were verified by DNA sequencing. cDNAs were cloned into pCDNA3 (Invitrogen) or in the combined oocyte/mammalian expression vector pFROG3 containing 5′- and 3′-untranslated regions of Xenopus β-globin (25).
Cell Death Assay.
Dishes (35 mm) containing 60–80% confluent HEK293 cells were cotransfected with 1 μg of the indicated construct and 0.2 μg of pEGFP-C1 plasmid (CLONTECH) by using Fugene 6 reagent (Boehringer Mannheim). After 36 h, the number of dead cells was quantified by staining in 4 μM ethidium homodimer-1 (Molecular Probes). Cells expressing wild-type VR1 were incubated in medium containing 2 μM capsaicin for 6 h before cell death analysis.
Mammalian Cell Culture and Electrophysiology.
HEK293 cells were maintained in DMEM (supplemented with 10% FBS, penicillin, streptomycin, and l-glutamine) and transfected with 1 μg of plasmid DNA by using lipofectamine (GIBCO/BRL). Whole-cell patch-clamp recordings were performed at 22°C 2 days after transfection. Standard bath solution contained 140 mM NaCl/5 mM KCl/2 mM MgCl2/5 mM EGTA/10 mM Hepes/10 mM glucose (pH 7.4 adjusted with NaOH). Bath solution was buffered to different pH values with either 10 mM Hepes (6.9 and 6.4) or 10 mM Mes (5.9, 5.4, 4.9, 4.4, 3.9, and 3.4). Pipette solution contained 140 mM CsCl/5 mM EGTA/10 mM Hepes (pH 7.4 adjusted with CsOH).
Oocyte Expression and Electrophysiology.
Templates were linearized with MluI and transcribed with T7 polymerase (AmpliScribe-Kit, Epicentre Technologies, Madison, WI). Oocytes were injected with 5–10 ng of VR1 or mutant cRNA. Two-electrode voltage-clamp analysis (Eh = −40 mV) was performed 4–9 days postinjection. All solutions were adjusted to constant cation concentrations: 96 mM Na+/2 mM K+/1 mM Mg2+/0.1 mM Ba2+, with chloride and buffer as counterions. Recordings of proton-activated currents in Xenopus oocytes were performed at room temperature. Solutions were buffered with 5 mM Na-Hepes (pH 7.5), 5 mM Na-Mes (pH 6.0–5.5), or 5 mM Na3-citrate (pH 5.25–3.5),and titrated with HCl. Recordings of capsaicin-activated currents were performed at room temperature in solutions buffered with 5 mM Na-Hepes (pH 7.6). Recordings of heat-evoked currents were performed as described (7, 26) using solutions buffered with a combination of 5 mM Tris/5 mM Na-Mes, titrated with HCl. All procedures involving the care and use of frogs were carried out in accordance with institutional guidelines.
Results
Initial Screen for VR1 Mutants with Altered Proton Sensitivity.
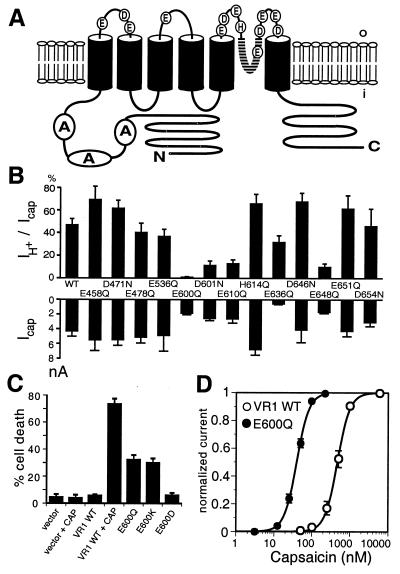
We have shown previously that acidic bath solutions evoke ionic currents when applied to outside-out, but not inside-out membrane patches excised from VR1-expressing HEK293 cells (7). Our search for sites involved in proton-dependent channel modulation or activation therefore focused on amino acids within putative extracellular loops of the VR1 protein that have side chain pKa values in a physiologically relevant range (i.e., His, Glu, and Asp residues) (Fig. 1A). We generated a series of point mutants in which these amino acids were individually replaced by residues having a similar side chain length, but lacking a protonatable moiety. The resulting VR1 mutants were transiently expressed in HEK293 cells and examined for relative responsiveness to saturating doses of capsaicin (1 μM) or protons (pH 4.4) (Fig. 1B). Mutations within the first two extracellular loops had no significant effect on the magnitude of capsaicin- or proton-evoked inward currents (proton-evoked responses were typically 30–50% the size of capsaicin responses). In contrast, several mutations within the putative third extracellular loop (E600Q, D601N, E610Q, and E648Q) showed a substantial reduction in one or both of these responses.
Figure 1.
Initial screen for VR1 mutants with altered proton sensitivity. (A) Positions of substituted acidic residues in the VR1 channel are shown in the context of a putative transmembrane topology model based on studies of the related hTRP3 channel (45). (B) Analysis of proton- and capsaicin-activated currents in transfected HEK293 cells expressing VR1 mutants. Upper bars show the mean ratios of proton (pH 4.4): capsaicin (1 μM)-activated currents. Lower bars show average absolute amplitudes of capsaicin-activated current in transfected HEK293 cells expressing wild-type or mutant VR1 receptors. Data represent means ± SEM from 4–16 experiments. (C) Higher frequency of death among HEK293 cells expressing E600 mutant channels. Percentage of green fluorescent protein-positive (transfected) HEK293 cells that stained brightly with ethidium homodimer-1 (dead cells) is shown. Data represent means ± SEM of triplicate determinations from two independent experiments. Expression of E600Q and E600K mutants increases cell death. (D) Tenfold increase in capsaicin sensitivity in E600Q mutant channels. Capsaicin dose-response curves for VR1 wild-type and E600Q mutant channels. Oocytes were perfused for 1 min with solutions of increasing capsaicin concentrations, with an intermittent wash in capsaicin-free solution for 3 min. Baseline-subtracted steady-state currents were normalized to currents maximally activated by 200 nM capsaicin for E600Q and by 5 μM capsaicin for VR1 wild type. Averaged data were fitted with the Hill equation with average parameters obtained from fits to individual cells. For VR1, EC50 = 520 ± 60 nM (n = 5), nH = 2.5 ± 0.2. For E600Q, EC50 = 40 ± 3 nM (n = 4), nH = 2.7 ± 0.2. Maximal amplitudes were 3.0 ± 0.2 μA for wild type VR1 (5 μM capsaicin) or 3.2 ± 0.2 μA for E600Q (200 nM capsaicin).
Cell Death Induced by Neutralization of a Putative Proton-Sensing Site.
HEK293 cells expressing wild-type VR1 channels can be rapidly and efficiently killed by addition of capsaicin to the growth medium (6) (Fig. 1C). Similarly, constitutive activation of the VR1-related transient receptor potential channel leads to photoreceptor cell degeneration in the fly eye (27). We were therefore intrigued by the fact that HEK293 cells expressing E600Q mutant VR1 channels showed markedly reduced viability and reasoned that poor viability of E600Q-transfected cells might be caused by heightened activity of the mutant channel under normal culture conditions. Indeed, replacement of this Glu residue with Gln (E600Q) or a positively charged amino acid (E600K) resulted in significant cell death, whereas substitution with an acidic residue (E600D) was not deleterious (Fig. 1C). This observation suggested that a decrease in negative charge at the E600 site favors channel activation.
Xenopus oocytes expressing E600K mutant channels also showed reduced viability (≈80% cell death within 4 days of cRNA injection) which was prevented by including the VR1 antagonist ruthenium red (1 μM) in the incubation medium. Unlike HEK293 cells, oocytes expressing other E600 mutants survived reasonably well, presumably because the relatively low incubation temperature (16°C) for oocytes spared them from premature death (see below). This enabled us to conduct more detailed functional studies using the oocyte system and to ask whether mutations at the E600 site alter the sensitivity of VR1 to capsaicin or heat.
Capsaicin- and Heat-Sensitivities Are Dramatically Potentiated by Mutations at E600.
For wild-type channels, acidification of the extracellular bath solution (from pH 7.6 to 6.3) increases the size of capsaicin-evoked currents by increasing agonist potency (6, 7). If mutations in E600 affect this potentiation process, then we would expect to observe a difference in capsaicin sensitivity of these channels. Strikingly, we found that replacement of Glu with Gln (E600Q) produced a greater than 10-fold leftward shift in the capsaicin dose-response curve, with EC50 values of 520 and 40 nM for wild-type and mutant receptors, respectively (Fig. 1D). Moreover, channels bearing a Lys at this position (E600K) had even higher agonist sensitivity, showing saturated current responses at 50 nM capsaicin (not shown). Thus, E600Q mutants resemble wild-type channels operating under acidic conditions.
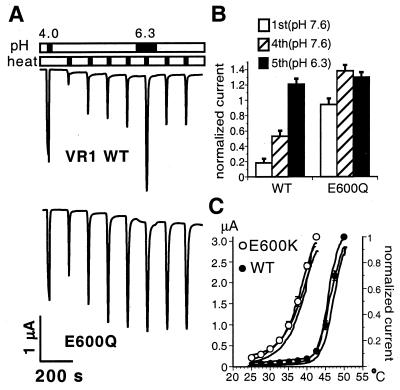
A similar effect was observed for thermal activation. When consecutive heat pulses are applied to cells expressing wild-type VR1 channels, peak current values typically increase, reaching a plateau after the third or fourth stimulus. Bath acidification to pH 6.3 potentiates this stabilized current in a reversible manner (ref. 7; Fig. 2A Upper). The E600Q mutant differed from the wild-type receptor in two significant ways. First, the initial heat stimulus produced a relatively large peak current response that was much closer to the final plateau value than that typically observed with wild-type channels (Fig. 2A Lower). Second, bath acidification failed to potentiate peak currents beyond this steady-state value (Fig. 2 A Lower and B). Similar results were obtained with oocytes expressing E600A and E600S mutants (not shown). Once again, the E600K mutant showed the most dramatic sensitization phenotype (Fig. 2C). These channels were already activated at temperature thresholds (30–32°C) well below normal (43°C), resembling the heat sensitivity of wild-type channels at pH 6.3 (7). The decrease in thermal threshold caused by introduction of a positive charge at this position could readily account for the poor viability of cells expressing this mutant, especially in transfected mammalian cells, where an incubation temperature of 37°C should favor channel activation.
Figure 2.
Prepotentiated phenotype of E600Q and E600K mutant channels. (A) Modulation of heat-activated currents by extracellular protons in cells expressing VR1 wild-type (Upper) or E600Q mutant (Lower) channels. To compare current sizes, channels were first activated for 20 s by acidic solution (pH 4.0) at room temperature. [In oocytes, E600Q channels showed robust activation at low pH (see below)]. To record heat-activated currents, bath temperature was then elevated from room temperature to 47°C within 10 s. This procedure was repeated seven times in 2-min intervals. During the fifth heat application, bath pH was decreased from 7.6 to 6.3. (B) Sensitization of heat-activated currents and potentiation by protons in VR1 wild-type (n = 6) and E600Q mutant (n = 4) channels. Heat-evoked peak currents, recorded as in A, were normalized to pH 4-activated steady-state currents at room temperature. Averaged values for peak currents at the first (pH 7.6), fourth (pH 7.6), and fifth heat stimulus (pH 6.3) are shown. Error bars represent SEM. (C) Lower threshold for heat activation of E600K mutant channels. Solid lines show representative heat-activated currents at the first heat stimulus recorded from three independent oocytes (left axis). Open (E600K) and closed (VR1) circles display mean heat-activated currents normalized to peak amplitudes at the maximal activation temperature [42.5°C for E600K (n = 7), 50°C for VR1 wild type (n = 6), right axis]. Heat activation as in A. Error bars represent SEM.
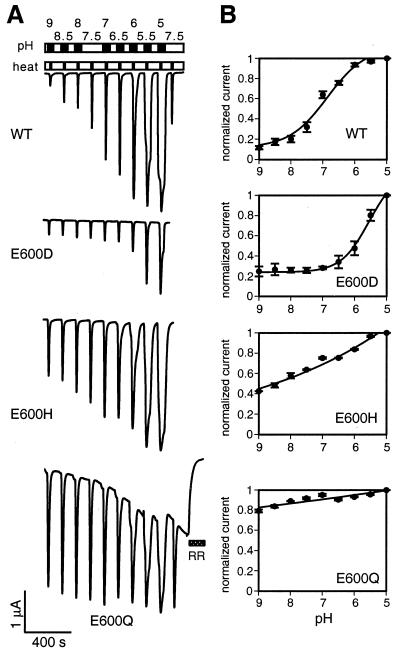
For cells expressing wild-type VR1, we found that the most dynamic increase in thermal responses occurred when levels of extracellular protons increased from normal physiological concentration (pH 7.5) to that typically associated with tissue acidosis (pH 7.0–6.0). This relationship was described by a dose-dependent response curve with half-maximal potentiation occurring at ≈pH 7.0 (Fig. 3). Peak currents tripled within this pH range and saturated at higher proton concentrations. In our analysis of E600 mutants, we found that the pH dependence of thermal activation was related to the side-chain charge of the residue at position 600. For example, heat-evoked currents in the E600D mutant were potentiated only when the bath pH dropped below 6.5. E600H mutants, on the other hand, showed continuous potentiation over the entire pH range tested (9.0 to 5.0), but the extent of potentiation was significantly less than that observed for wild-type or E600D channels. For E600Q or other mutants having nontitratable amino acids at this position (not shown), the magnitude of heat-evoked currents was largely independent of extracellular pH. We also noticed that cells expressing E600Q channels developed an increasing basal current with repetitive thermal stimulation (Figs. 2A Lower and 3A), a phenomenon that was exacerbated by acidification and eliminated by addition of ruthenium red (10 μM) (Fig. 3 Bottom). Taken together, our data show that introduction of neutral or positive residues at the E600 site potentiates responses to capsaicin or heat, whereas introduction of a residue with lower pKa (E600D) decreases channel sensitivity to these stimuli.
Figure 3.
Mutations in E600 affect proton-dependent potentiation of heat-activated currents. (A) Titration analysis of proton-dependent potentiation of heat-activated currents in VR1 wild-type or E600D, E600H, or E600Q mutant channels. Channels were first sensitized by four subsequent heat pulses to 48°C (not shown). Upper bar shows the pH application protocol. Oocytes were perfused in bath solutions with a pH indicated above the bar for 1 min each (black), interrupted by perfusion at pH 7.5 (white). The lower bar shows the heat application protocol. For each pH, bath temperature was elevated from room temperature (white) to 42°C (black) for VR1 wild-type, E600H, and E600Q, or to 48°C for E600D. Both wild-type (WT)- and the E600Q-expressing cells were again tested at pH 7.5. The E600Q-expressing cells were subsequently perfused with standard solution containing 10 μM ruthenium red (RR). (B) Dose-response analysis of proton-mediated potentiation of heat-activated currents in oocytes expressing VR1 wild-type (n = 4), E600D (n = 3), E600H (n = 3), or E600Q (n = 3) channels. Recordings as in A. For VR1 wild type, E600D and E600H, currents before application of solution of the indicated pH were subtracted from heat-evoked peak currents. For E600Q, currents after ruthenium red block were subtracted from peak currents. Current values were normalized to currents at pH 5.0. Averaged currents are shown with error bars representing SEM.
Proton Activation Range Is Also Determined at a Key Extracellular Residue on VR1.
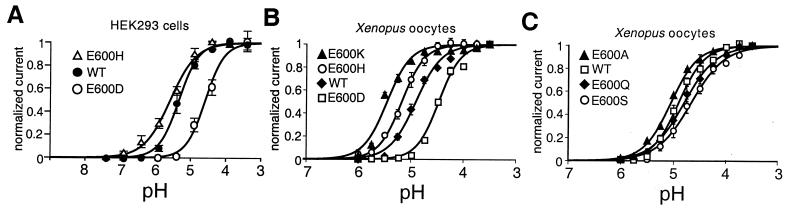
Proton-evoked activation of VR1 at room temperature is achieved under high extracellular proton concentrations (pH < 6). This response may simply be an extreme form of potentiation whereby the thermal threshold for VR1 activation is decreased to 24°C, or it may require additional temperature-independent steps that involve the protonation of sites other than E600 within the channel protein. In the latter case, mutations at E600 would be expected to set the range of proton sensitivity (in much the same way that they do for other noxious stimuli), but not to eliminate proton-evoked channel activation at room temperature. Indeed, our data support this model because dose-response curves for proton-evoked channel activation varied with the predicted pKa of the side chain at this position. In HEK293 cells or oocytes expressing E600D, wild-type, or E600H receptors, the half-maximal pH (pH0.5) for channel activation at room temperature was 4.6, 5.3, or 5.6, respectively (Fig. 4 A and B). These shifts in pH0.5 did not show a strict linear correlation with predicted pKa values, but this could reflect effects of the protein or membrane microenvironment on side-chain charge, or additional influences of side-chain shape on receptor activation. Nevertheless, the overall effect was similar to that which we observed for proton potentiation of capsaicin- or noxious heat-evoked currents.
Figure 4.
Analysis of E600 mutants suggests direct titration by protons. (A) Proton dose-response curves recorded from transiently transfected HEK293 cells show a direct relationship between agonist potency and side-chain pKa in wild-type, E600D, and E600H mutants. pH values producing half-maximal responses were 5.34 ± 0.01, 4.55 ± 0.02, and 5.58 ± 0.02, respectively. The Hill equation was used for curve fitting. Values represent the means ± SEM from 4–17 separate experiments. (B) Correlation of side-chain pKa at position 600 with proton sensitivity. Dose-response curves for proton activation of E600 mutants with protonatable side chains recorded from Xenopus oocytes. Cells were perfused for 20 s with solutions of decreasing pH, with an intermittent wash at pH 7.5 for 40 s. Baseline-subtracted currents were normalized to steady-state currents at pH 3.5. Figure shows averaged data fitted with the Hill equation with average parameters obtained from fits to individual cells. For E600K, EC50 = 5.5 ± 0.1 (n = 5), nH = 2.1 ± 0.2; E600H, EC50 = 5.2 ± 0.1 (n = 3), nH = 2.0 ± 0.2; VR1 wild-type, EC50 = 4.9 ± 0.1 (n = 9), nH = 1.9 ± 0.1; E600D, EC50 = 4.5 ± 0.1 (n = 3), nH = 2.3 ± 0.3. Error bars represent SEM. Maximally activated current amplitudes at pH 3.5 were: VR1 wild type, 2.7 ± 0.2 μA; E600K, 4.4 ± 0.2 μA; E600H, 2.7 ± 0.1 μA; E600D, 2.4 ± 0.2 μA. (C) Dose-response curves for proton activation of E600 mutants with neutral side chains recorded from Xenopus oocytes. Fits as in B. For E600A, EC50 = 5.0 ± 0.1 (n = 3), nH = 1.7 ± 0.1; E600S, EC50 = 4.7 ± 0.1 (n = 3), nH = 1.4 ± 0.1; E600Q, EC50 = 4.8 ± 0.1 (n = 4), nH = 1.5 ± 0.1. Maximally activated current amplitudes at pH 3.5 were: E600Q, 2.8 ± 0.2 μA; E600S, 2.6 ± 0.1 μA; and E600A, 2.8 ± 0.1 μA.
We also examined the effects of introducing residues with nontitratable side chains at position 600. In oocytes, these mutant channels exhibited proton dose-response curves and maximal current amplitudes resembling those of the wild-type receptor (Fig. 4C). When expressed in HEK293 cells, these same mutants showed a greater variation in the magnitude of proton-activated currents, some significantly smaller than those observed with wild-type channels. The reason for this discrepancy between the two expression systems is unclear, but it could reflect greater desensitization of proton-evoked responses in HEK293 cells or a sampling bias introduced by the higher incidence of death among HEK293 cells expressing these mutants. These observations suggest that potentiation of proton-evoked responses requires more pronounced changes in side-chain charge (e.g., E600K or E600D) than does potentiation of vanilloid- or heat-evoked responses.
Taken together, our data show that the charge at position 600 plays a critical role in determining the pH sensitivity range of channel activation by noxious stimuli, including vanilloids, protons, and heat. This potentiation may involve the direct titration of E600 by extracellular protons, or E600 may influence the protonation of a nearby site. Moreover, because nontitratable E600 mutants can still be activated by low pH solutions, we conclude that proton-evoked channel activation and proton-mediated potentiation are distinct processes.
Stimulus-Specific Activation Pathways Revealed by Proton-Insensitive Receptors.
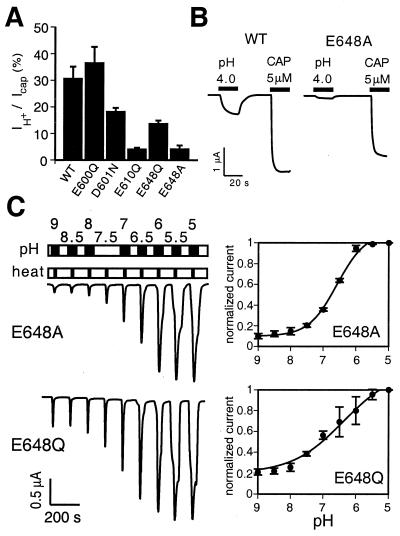
In our initial mutational screen, three other potential proton-sensitive sites were identified, and we examined these in greater detail in the oocyte system. E610Q mutants exhibited relatively small responses to all stimuli (Figs. 1B and 5A and data not shown), suggesting that this substitution had deleterious effects on channel function at large. D601N and E648Q mutants, on the other hand, exhibited a more interesting phenotype characterized by reduced proton-evoked responses with normal capsaicin sensitivity (Fig. 5A). We also constructed an E648A mutant because alanine is found at the cognate position of the rat VRL-1 channel, a heat-activated, proton-insensitive VR1 homologue (26). E648A mutants showed an even greater decrease in proton-activated current amplitudes, whereas capsaicin- or heat-evoked currents did not differ significantly from those of wild-type or E648Q channels (EC50 of E648A for capsaicin = 600 ± 140 nM versus 520 ± 60 nM for VR1 wild type). Mutants bearing neutral amino acid substitutions at this location did not appear to be constitutively active or prepotentiated, suggesting that proton-evoked channel gating does not simply involve the shielding of a negative charge at this site. Interestingly, E648A mutants seem to uniquely affect proton-evoked, but not vanilloid- or heat-evoked channel gating (Fig. 5B), arguing for the existence of stimulus-specific steps or pathways leading to VR1 activation. Moreover, E648A mutants showed normal proton potentiation of thermal responses (Fig. 5C), providing additional evidence that potentiation and proton-evoked channel activation involve the titration of different sites on the VR1 protein. That these mutations affect separate, independent processes is further supported by the phenotype exhibited by the E600Q–E648A double mutant. Like the E600Q mutant, these channels showed increased sensitivity to capsaicin and heat, and a loss of proton-mediated (pH 6.3) potentiation. However, they also had greatly reduced proton-evoked (pH 4) current responses at room temperature, reminiscent of the E648A mutant (not shown).
Figure 5.
Reduction in proton-activated currents in mutants at position 648. (A) Bar graph showing average ratios of proton- to capsaicin-activated steady-state amplitudes in oocytes expressing VR1 wild type (n = 5), or E600Q, D601N, E610Q, E648Q, or E648A (n = 4 each) channels. Cells were perfused for 20 s at pH 4.0, washed for 40 s, then perfused for 20 s with solution containing 5 μM capsaicin. Mutation of E648 to Ala drastically reduces proton-activated currents. Error bars represent SEM. (B) Representative proton- and capsaicin-activated currents in VR1 wild type (Left) and mutant E648A (Right) channels. Protocol as in Fig. 3A. (C) Dose-response analysis of heat-activated currents in E648 mutants resembles that of wild-type receptors. Performed as in Fig. 3 A and B.
Discussion
Our findings provide molecular evidence that protons modulate VR1 activity by interacting with specific amino acid residues on the extracellular surface of the channel protein. Cells expressing wild-type receptors show especially dynamic modulation of heat-evoked currents between pH 8 and 6, a sensitivity range that matches the extent of local acidosis attained during most forms of tissue injury (2, 28). VR1 mutants bearing other amino acids at the E600 site (E600D, E600H, or E600Q) do not exhibit the same robust and dynamic increase in heat-evoked currents over this pH range, highlighting the physiological significance of having Glu at this position. The E600 configuration also tunes VR1 to respond with a thermal threshold of ≈43°C under normal physiological conditions (pH 7.5), a set point that is significantly above body temperature, yet moderate enough to serve as an effective detector of potentially injurious heat. Indeed, the presence of other residues at this site might very well have adverse consequences in vivo because mutant channels containing a Lys or Gln at this position are sufficiently active at 37°C to promote the death of transfected mammalian cells in culture. Such hyperactivating mutations in the VR1 gene might result in loss of nociceptors, in much the same way that constitutively activating mutations of other ion channels lead to neurodegeneration (29–34), or systemic treatment of neonatal rats with capsaicin induces death of VR1-expressing primary afferent neurons (35). Among rare individuals who suffer from congenital insensitivity to pain, some lack a subset of unmyelinated small-diameter nociceptors as a consequence of defects in the high-affinity nerve growth factor receptor (trkA) gene (36–38). Our results suggest that a similar phenotype could result from hyperactivating mutations in the VR1 gene.
Titration of free Glu residues by protons usually occurs with a pKa ≈4.3 (39), which is well below the half-maximal proton concentration of ≈ pH 7.0 that we found to be effective in potentiating VR1 responses to heat. Nonetheless, interaction of E600 with other residues within the multimeric channel complex could dramatically change the electrostatic environment of this Glu residue. For example, amino acid carboxyl-carboxylate pairs sharing a single proton can display drastically elevated pKa values within the physiological range (40). Such a mechanism is likely to occur in cyclic nucleotide-gated channels, where Glu residues that probably form carboxyl-carboxylate pairs within the channel pore are thought to mediate the proton-dependent transitions between different conductance states in these channels with a pKa ≈7.6 (17, 41). Similar pairings of pore Glu confer proton-dependent block in L-type calcium channels with an even higher pKa of ≈8.5, nearly four pH units greater than the pKa of an individual Glu residue (42–44). It can be imagined that E600 is forming similar pairings with other proton-accepting or -donating amino acid residues across the outer surface of the VR1 receptor channel. This pH-dependent process could lead to channel opening at low pH or influence the response of the channel complex to other agonists.
Intracellular protons potentiate cyclic nucleotide channel gating by directly coordinating the interaction of the ligand with an Asp in the cytoplasmic cyclic nucleotide-binding pocket of the channel, leading to stabilization of the open state (19). In contrast, protons and vanilloids apparently exert their actions on VR1 from opposite sides of the membrane (7, 22, 23), consistent with an allosteric mechanism of proton-mediated potentiation. The profound reduction in acid-evoked responses in sensory neurons from mice lacking VR1 (13) underscores the importance of proton-modulatory sites such as E600, because they may prove to be effective molecular targets for the development of novel analgesic agents that function by reducing the effects of acidosis on nociceptor excitability.
Acknowledgments
We thank W. Neuhausser for help with cell death assays and Drs. L. Jan, R. Nicoll, and R. MacKinnon for helpful discussions. M.T. thanks M. Masu for encouragement. S.-E. J. is a fellow of the German Academy of Natural Scientists Leopoldina (BMBF-LPD 9801–3); M.T. was a Comroe Fellow of the University of California San Francisco Cardiovascular Research Institute. This work was supported by grants from the National Institutes of Health and a McKnight Foundation Investigator Award to D.J.
Abbreviation
- VR1
capsaicin (vanilloid) receptor
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100129497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100129497
References
- 1.Fields H. Pain. New York: McGraw–Hill; 1987. [Google Scholar]
- 2.Reeh P W, Steen K H. Prog Brain Res. 1996;113:143–151. doi: 10.1016/s0079-6123(08)61085-7. [DOI] [PubMed] [Google Scholar]
- 3.Waldmann R, Lazdunski M. Curr Opin Neurobiol. 1998;8:418–424. doi: 10.1016/s0959-4388(98)80070-6. [DOI] [PubMed] [Google Scholar]
- 4.King B F, Wildman S S, Ziganshina L E, Pintor J, Burnstock G. Br J Pharmacol. 1997;121:1445–1453. doi: 10.1038/sj.bjp.0701286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoop R, Surprenant A, North R A. J Neurophysiol. 1997;78:1837–1840. doi: 10.1152/jn.1997.78.4.1837. [DOI] [PubMed] [Google Scholar]
- 6.Caterina M J, Schumacher M A, Tominaga M, Rosen T A, Levine J D, Julius D. Nature (London) 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 7.Tominaga M, Caterina M J, Malmberg A B, Rosen T A, Gilbert H, Skinner K, Raumann B E, Basbaum A I, Julius D. Neuron. 1998;21:531–543. doi: 10.1016/s0896-6273(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 8.Petersen M, LaMotte R H. Pain. 1993;54:37–42. doi: 10.1016/0304-3959(93)90097-9. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Simon S A. Proc Natl Acad Sci USA. 1994;91:738–741. doi: 10.1073/pnas.91.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bevan S, Geppetti P. Trends Neurosci. 1994;17:509–512. doi: 10.1016/0166-2236(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 11.Kress M, Zeilhofer H U. Trends Pharmacol Sci. 1999;20:112–118. doi: 10.1016/s0165-6147(99)01294-8. [DOI] [PubMed] [Google Scholar]
- 12.Caterina M J, Julius D. Curr Opin Neurobiol. 1999;9:525–530. doi: 10.1016/S0959-4388(99)00009-4. [DOI] [PubMed] [Google Scholar]
- 13.Caterina M J, Leffler A, Malmberg A B, Martin W J, Trafton J, Petersen-Zeitz K R, Koltzenburg M, Basbaum A I, Julius D. Science. 2000;288:306–313. doi: 10.1126/science.288.5464.306. [DOI] [PubMed] [Google Scholar]
- 14.Montell C, Rubin G M. Neuron. 1989;2:1313–1323. doi: 10.1016/0896-6273(89)90069-x. [DOI] [PubMed] [Google Scholar]
- 15.Cesare P, McNaughton P. Proc Natl Acad Sci USA. 1996;93:15435–15439. doi: 10.1073/pnas.93.26.15435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Ellinor P T, Sather W A, Zhang J F, Tsien R W. Nature (London) 1993;366:158–161. doi: 10.1038/366158a0. [DOI] [PubMed] [Google Scholar]
- 17.Root M J, MacKinnon R. Science. 1994;265:1852–1856. doi: 10.1126/science.7522344. [DOI] [PubMed] [Google Scholar]
- 18.Varnum M D, Black K D, Zagotta W N. Neuron. 1995;15:619–625. doi: 10.1016/0896-6273(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 19.Gordon S E, Oakley J C, Varnum M D, Zagotta W N. Biochemistry. 1996;35:3994–4001. doi: 10.1021/bi952607b. [DOI] [PubMed] [Google Scholar]
- 20.Jordt S E, Jentsch T J. EMBO J. 1997;16:1582–1592. doi: 10.1093/emboj/16.7.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baukrowitz T, Tucker S J, Schulte U, Benndorf K, Ruppersberg J P, Fakler B. EMBO J. 1999;18:847–853. doi: 10.1093/emboj/18.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumann T K, Martenson M E. J Neurosci. 2000;20:RC80–RC84. doi: 10.1523/JNEUROSCI.20-11-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung J, Hwang S W, Kwak J, Lee S Y, Kang C J, Kim W B, Kim D, Oh U. J Neurosci. 1999;19:529–538. doi: 10.1523/JNEUROSCI.19-02-00529.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunkel T A, Bebenek K, McClary J. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 25.Gunther W, Luchow A, Cluzeaud F, Vandewalle A, Jentsch T J. Proc Natl Acad Sci USA. 1998;95:8075–8080. doi: 10.1073/pnas.95.14.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caterina M J, Rosen T A, Tominaga M, Brake A J, Julius D. Nature (London) 1999;398:436–441. doi: 10.1038/18906. [DOI] [PubMed] [Google Scholar]
- 27.Yoon J, Ben-Ami H C, Hong Y S, Park S, Strong L L, Bowman J, Geng C, Baek K, Minke B, Pak W L. J Neurosci. 2000;20:649–659. doi: 10.1523/JNEUROSCI.20-02-00649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine J, Taiwo Y. In: The Textbook of Pain. Wall P D, Melzack R, editors. London: Churchill Livingston; 1994. pp. 45–56. [Google Scholar]
- 29.Driscoll M, Chalfie M. Nature (London) 1991;349:588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- 30.Hong K, Driscoll M. Nature (London) 1994;367:470–473. doi: 10.1038/367470a0. [DOI] [PubMed] [Google Scholar]
- 31.Patil N, Cox D R, Bhat D, Faham M, Myers R M, Peterson A S. Nat Genet. 1995;11:126–129. doi: 10.1038/ng1095-126. [DOI] [PubMed] [Google Scholar]
- 32.Slesinger P A, Patil N, Liao Y J, Jan Y N, Jan L Y, Cox D R. Neuron. 1996;16:321–331. doi: 10.1016/s0896-6273(00)80050-1. [DOI] [PubMed] [Google Scholar]
- 33.Kofuji P, Hofer M, Millen K J, Millonig J H, Davidson N, Lester H A, Hatten M E. Neuron. 1996;16:941–952. doi: 10.1016/s0896-6273(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 34.Navarro B, Kennedy M E, Velimirovic B, Bhat D, Peterson A S, Clapham D E. Science. 1996;272:1950–1953. doi: 10.1126/science.272.5270.1950. [DOI] [PubMed] [Google Scholar]
- 35.Jancso G, Kiraly E, Jancso-Gabor A. Nature (London) 1977;270:741–743. doi: 10.1038/270741a0. [DOI] [PubMed] [Google Scholar]
- 36.Smeyne R J, Klein R, Schnapp A, Long L K, Bryant S, Lewin A, Lira S A, Barbacid M. Nature (London) 1994;368:246–249. doi: 10.1038/368246a0. [DOI] [PubMed] [Google Scholar]
- 37.Indo Y, Tsuruta M, Hayashida Y, Karim M A, Ohta K, Kawano T, Mitsubuchi H, Tonoki H, Awaya Y, Matsuda I. Nat Genet. 1996;13:485–488. doi: 10.1038/ng0896-485. [DOI] [PubMed] [Google Scholar]
- 38.Mardy S, Miura Y, Endo F, Matsuda I, Sztriha L, Frossard P, Moosa A, Ismail E A, Macaya A, Andria G, et al. Am J Hum Genet. 1999;64:1570–1579. doi: 10.1086/302422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Creighton T E. Proteins—Structures and Molecular Properties. New York: Freeman; 1993. [Google Scholar]
- 40.Sawyer L, James M N. Nature (London) 1982;295:79–80. doi: 10.1038/295079a0. [DOI] [PubMed] [Google Scholar]
- 41.Morrill J A, MacKinnon R. J Gen Physiol. 1999;114:71–83. doi: 10.1085/jgp.114.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pietrobon D, Prod'hom B, Hess P. J Gen Physiol. 1989;94:1–21. doi: 10.1085/jgp.94.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X H, Bezprozvanny I, Tsien R W. J Gen Physiol. 1996;108:363–374. doi: 10.1085/jgp.108.5.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X H, Tsien R W. J Biol Chem. 1997;272:30002–30008. doi: 10.1074/jbc.272.48.30002. [DOI] [PubMed] [Google Scholar]
- 45.Vannier B, Zhu X, Brown D, Birnbaumer L. J Biol Chem. 1998;273:8675–8679. doi: 10.1074/jbc.273.15.8675. [DOI] [PubMed] [Google Scholar]