Abstract
Two cDNA libraries were prepared, one from leaves of a field-grown aspen (Populus tremula) tree, harvested just before any visible sign of leaf senescence in the autumn, and one from young but fully expanded leaves of greenhouse-grown aspen (Populus tremula × tremuloides). Expressed sequence tags (ESTs; 5,128 and 4,841, respectively) were obtained from the two libraries. A semiautomatic method of annotation and functional classification of the ESTs, according to a modified Munich Institute of Protein Sequences classification scheme, was developed, utilizing information from three different databases. The patterns of gene expression in the two libraries were strikingly different. In the autumn leaf library, ESTs encoding metallothionein, early light-inducible proteins, and cysteine proteases were most abundant. Clones encoding other proteases and proteins involved in respiration and breakdown of lipids and pigments, as well as stress-related genes, were also well represented. We identified homologs to many known senescence-associated genes, as well as seven different genes encoding cysteine proteases, two encoding aspartic proteases, five encoding metallothioneins, and 35 additional genes that were up-regulated in autumn leaves. We also indirectly estimated the rate of plastid protein synthesis in the autumn leaves to be less that 10% of that in young leaves.
Leaf senescence is the final stage in leaf development, and understanding senescence is important not only for purely scientific reasons, but also for practical purposes. Premature senescence leads, for example, to decreased photosynthetic capacity, and consequently lower yield. Senescence is not simply the passive death of a leaf because of aging, but is a tightly controlled process during which cell components are degraded in a coordinated fashion and, when nutrients have been relocated to other parts of the plant body, the cell finally dies (Gan and Amasino, 1997; Nooden et al., 1997). Despite the resemblance with apoptosis of animal cells (Yen and Yang, 1998), a form of programmed cell death, only a few orthologs of genes regulating apoptosis have been found in plants, indicating that there are significant differences between the processes (Koonin and Aravind, 2002). Plant cells respond to some animal apoptosis regulators (e.g. Danon et al., 2000), so there must be common elements between the processes. However, it seems as if plants have developed a unique mode of cell death (Beers, 1997) that, if understood, may give insight into processes that are important for cell integrity and viability. However, very little is known about the details of plant leaf senescence.
During the last decade, studies of leaf senescence, focusing especially on Arabidopsis, and other annual species to a lesser extent, have identified a number of senescence-associated genes (SAGs) and cellular mechanisms of senescence have begun to be elucidated, as reviewed by various authors (Buchanan-Wollaston, 1997; Nam, 1997; Quirino et al., 2000). The most obvious visual phenotype of senescence, the color changes from green to yellow, red, or orange, is the result of chlorophyll degradation, often combined with anthocyanin accumulation (Hoch et al., 2001). During senescence, photosynthesis declines and the leaf changes its metabolism from anabolism to catabolism and the chloroplasts turn into gerontoplasts. This initial decay is limited to the photosynthetic mesophyll cells, whereas epidermal cells, including stomata and cells in the phloem, stay intact and functional (Feller and Fischer, 1994). Even for the photosynthetic cells the degradation is initially only partial, and compartmentation is maintained with intact mitochondria, peroxisomes, and vacuoles. Respiration continues after photosynthesis starts to decline and mitochondria remain intact (Collier and Thibodeau, 1995). It has been suggested that mitochondria may play an important role in the process both for ATP production and for the metabolic events leading to recapture of nutrients (Feller and Fischer, 1994; Smart, 1994; Collier and Thibodeau, 1995). However, very little experimental evidence has been published to support this hypothesis. Surprisingly, leaf senescence in perennial species, especially trees (which give the most conspicuous and esthetically pleasing display of leaf senescence, at least in the temperate regions of the world) has not been studied with modern molecular genetic tools. Autumnal leaf senescence is an attractive system in which to study leaf senescence because the senescence process can be induced, at least in most trees, simply by shortening the photoperiod, as shown, for example, by Olsen et al. (1997). In contrast, senescence in annual plants tends to be induced either by some type of stress or, in monocarpic plants like soybean (Glycine max) and cereals, by the development of the seeds. There are no reasons to believe that autumnal senescence should be completely different from other types of leaf senescence but because the trigger is different, regulatory differences must exist.
We have initiated a project to understand the genetic basis of autumn senescence and describe here the first steps in this initiative: large-scale sequencing and analysis of aspen (Populus tremula) expressed sequence tags (ESTs) to identify candidate genes for regulating and mediating the process. In addition to gene identification, EST sequencing can also be used to obtain estimates of relative expression levels. Provided that no subtractive methods have been applied during library construction, relative EST abundance provides an approximate indication of the level of each transcript in the mRNA pool. If genes are grouped into broad categories (for example, according to function), the mean numbers of ESTs give a fairly good estimate of the gene transcription in each category and the EST frequency of several sets of Arabidopsis and rice (Oryza sativa) genes have been shown to roughly correspond to relative protein stoichiometries (Mekhedov et al., 2000; Ohlrogge and Benning, 2000).
Here, we present an analysis of two different sets of aspen leaf ESTs, and use the data to draw conclusions regarding gene expression during autumnal leaf senescence in aspen.
RESULTS
RNA Sampling and Preparation
From a free-growing aspen on the Umea University campus, leaf samples were harvested twice a week from August 17 until October 1, 1999, flash frozen in liquid nitrogen, and stored at −80°C until RNA preparation. The leaves showed no visible signs of autumn senescence until September 14 but then, within a week, the leaves turned yellow (Fig. 1) and started to fall. By the beginning of October, virtually all leaves had fallen, so sampling was stopped.
Figure 1.
Autumn senescence in free-growing aspen. The pictures were taken at the time of leaf sampling (11.00) during the autumn of 1999.
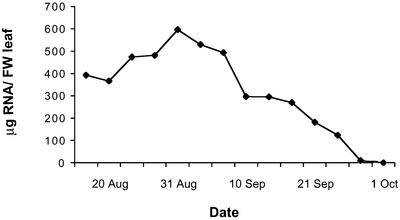
We measured the amount of extractable RNA from three independent RNA extractions of leaves from each sampling date to get a crude estimate of the kinetics of RNA disappearance as the leaf senesced. We noted an increase in the amount of extractable RNA in the first half of September, but during the latter half of the month the RNA gradually decreased in abundance and finally disappeared (Fig. 2). The amount of extractable RNA does not necessarily correspond precisely to the amount of RNA present in the sample or to the level of protein synthesis. Nevertheless, these changes indicate that chloroplast degradation may be preceded by an increase in protein synthesis, and that protein synthesis activity probably continued for about 2 weeks after the initiation of massive chlorophyll degradation.
Figure 2.
Amount of extractable RNA from aspen leaves at different dates during the autumn of 1999. Values are means of three different preparations.
EST Sequencing, Bioinformatics, and Database Construction
Two cDNA libraries were constructed and analyzed using a high-throughput DNA sequencing setup. From the autumn leaf library (harvested on September 14 as described above), 5,258 EST sequences were obtained. To get a reference data set for comparison, we sequenced 4,923 clones from a cDNA library prepared from young, but fully expanded, leaves of an aspen hybrid grown in a greenhouse (Larsson et al., 1997). The reasons for choosing this library for comparison are presented in “Discussion.” The ESTs had average (trimmed) read lengths of 349 and 355 bp in the autumn leaf and young leaf library, respectively. All clones found to contain rRNA, mitochondrial DNA, and chloroplast DNA were excluded from the analysis. EST libraries, especially from free-growing aspen, could be contaminated by sequences from fungal pathogens. Therefore, all clones that showed highest homology to a fungal sequence in SWISS-PROT/TrEMBL were manually checked and removed from the analysis if they were likely to be of fungal origin. After this curation procedure, 5,128 and 4,841 ESTs remained from the autumn leaf and young leaf libraries, respectively. To annotate the ESTs, we designed a semiautomatic system for clustering and annotation (see “Materials and Methods”) and included a quality assessment with three grades (valid, perhaps valid, and probably invalid) in the annotations. Quality was scored by dividing the BLASTX score by the self-blast score to calculate the relative confidence value (RCV; Lonsdale and Arnold, 1999) to normalize the BLASTX scores with respect to sequence length because long contigs produce higher BLASTX scores than shorter ones with the same percent similarity. It is, in our experience, easier to define a sound threshold value for “valid” annotation using RCVs than the more commonly used BLASTX scores or E values. We manually examined a large number of sequence alignments and found that RCVs of >0.35 corresponded to what we regarded as “valid” annotation. For a 500-bp sequence, this corresponds to a BLASTX score of about 300. For very long contigs, significantly higher BLASTX scores could still give a RCV of <0.35, so all BLASTX scores >300 were also regarded as “valid.” At the lower end of the quality scale, the RCV is not as good a measurement of quality. Instead, a BLASTX score of <100 is more consistent with a nonsignificant hit (as judged by manual inspection) than a low RCV or E value. Therefore, sequences with BLASTX scores of <100 were scored as “probably invalid” and the remaining annotations were denoted “perhaps valid.”
The number of genes represented in a cDNA library and the redundancy of specific genes can be estimated by clustering the EST sequences according to sequence similarity. There are several pitfalls in this procedure. ESTs obtained from the 5′ end of the clone might not overlap although they derive from the same transcript. This is because of the fact that many clones (especially those originating form large transcripts) are not full length. In addition, for highly expressed genes represented by many copies among the ESTs, clustering programs like Phrap and the TIGR assembler have a strong tendency to split them into several contigs (Liang et al., 2000). These problems could result in a significant overestimation of the number of genes represented in an EST collection. To reduce this problem, we used the Mendel database. This high-quality database organizes all plant sequences into gene family numbers (GFNs). All contigs that produced a valid or probably valid hit to an entry in the Mendel database were assigned to the corresponding GFN (together with a quality measurement, identical to that of the annotation). One GFN in our database could then consist of several contigs/singlets originating from either the same or very similar genes. The contigs/singlets that did not produce a valid hit to a Mendel entry were assigned an aspen GFN (PGFN) as a unique denominator. In the following text, we will refer (for simplicity) to each GFN/PGFN as a single gene but we are aware that although many of these contigs/singlets may originate from different parts of the same transcript, several highly homologous genes are sometimes grouped into the same GFN.
We also identified, for each sequence, the protein in the Munich Institute of Protein Sequences (MIPS) Arabidopsis database (MATDB) that gave the highest BLASTX score. Many annotations in MATDB are automatically performed and are, in our experience, less reliable than those in the other databases. However, because every gene in MATDB has been assigned to a functional class in the MIPS classification system, identification of the closest Arabidopsis homologue provided a rapid way to obtain a preliminary functional classification of our sequences, which was later subjected to extensive manual curation. All annotations were entered into a FileMaker Pro-database using appropriate scripts.
A total of 4,512 ESTs (44%) could be assigned to a Mendel gene family. In the autumn leaf library, 380 different Mendel GFNs (see “Materials and Methods”) were represented, and the young leaf library contained 460 different GFNs. Of these, 155 were shared between the two libraries. The remaining 5,669 ESTs fell into 3,717 homology groups and were given PGFNs. Only 207 PGFNs were shared between the libraries: The autumn leaf library had 2,027 unique PGFNs and the young leaf library had 1,483. Thus, the genes expressed in young leaves corresponded more often to previously characterized proteins or genes than those expressed in autumn leaves, and the pattern of gene expression in the two types of leaf differed markedly.
Different Genes Were Most Abundant in the Two Libraries
To analyze more carefully the differences in gene expression we compared, from the curated lists of annotated clusters, the most abundant ESTs in the two libraries. Genes encoding “standard” photosynthetic proteins were scarce in the autumn leaf library (Table I). Rubisco ESTs, for example, were found at a frequency corresponding to 4% of that in the young leaf library (see below), and similar frequencies were found for other genes encoding proteins involved in the photosynthetic light and dark reactions (see below). However, ESTs encoding early light-inducible proteins (ELIPs), which accumulate in the thylakoid membrane during stress, were 13 times more abundant in the autumn leaf library. Several other stress-related proteins, for example metallothionein, blight-associated protein P12 (which has homology to expansin), pollen coat protein (which is related to dehydrins), and proteases, were also frequently found among the autumn leaf ESTs.
Table I.
The 20 most abundant types of ESTs in autumn leaves
| Gene | ESTs in Autumn Leaf Library | ESTs in Young Leaf Library | Enrichment in Autumn Leaves | Significance |
|---|---|---|---|---|
| Metallothionein | 321 | 25 | 13 | >0.999 |
| Cys proteinase | 109 | 9 | 12 | >0.999 |
| ELIP | 105 | 8 | 13 | >0.999 |
| Ubiquitin | 89 | 34 | 3 | >0.999 |
| Thioredoxin | 84 | 21 | 4 | >0.999 |
| Endomembrane-associated protein | 61 | 2 | 31 | >0.999 |
| Protein translation factor SUI1 | 47 | – | – | >0.999 |
| 1-Aminocyclopropane-1-carboxylic acid oxidase | 42 | 15 | 3 | >0.999 |
| At4g22920 | 30 | – | – | >0.999 |
| Blight-associated protein P12 | 28 | 5 | 6 | >0.999 |
| (Arabidopsis 16.1-kD protein) | 25 | – | – | >0.999 |
| RbcS | 25 | 679 | 0.03 | No |
| Ubiquitin-conjugating protein | 24 | 1 | 24 | >0.999 |
| Protease FtsH2 | 22 | 2 | 12 | >0.999 |
| Pollen coat protein | 20 | 8 | 3 | >0.95 |
| Salt stress-induced hydrophobic peptide | 20 | 1 | 20 | >0.999 |
| Unknown EST | 19 | – | – | >0.999 |
| Unknown EST | 18 | – | – | >0.999 |
| Macrophage migration inhibitory factor | 18 | 2 | 9 | >0.999 |
| Peptidyl-prolyl cis-trans isomerase | 17 | 8 | 2 | >0.8 |
The nos. of ESTs in autumn and young leaf libraries, the enrichment factor in the autumn leaf library (% in autumn leaves/% in young leaves), and the significance level for differential expression are indicated.
As expected, in the young leaf library most genes with a high abundance of ESTs encoded proteins of the photosynthetic apparatus. In fact, of the 20 most abundant genes (Table II), 14 were related to photosynthesis: 679 clones (14%) represented RbcS, encoding the small subunit of Rubisco, and 219 (4.5%) represented Lhcb1, encoding the major LHC II protein. No other protein was represented by more than 2% of the clones. Other proteins related to photosynthesis included seven light-harvesting chlorophyll a/b-binding proteins, two other PS I proteins, two other PS II proteins, and three soluble proteins. One additional gene encoded a chloroplast-located protein (a thiazole biosynthetic enzyme). Among the most frequent sequences that were not related to photosynthesis were two that encoded cytosolic proteins (ubiquitin and metallothionein) and two encoding proteins that appeared to be sorted through the secretory pathway: a cell wall protein (Pro-rich protein) and a germin-like protein. It was apparent that the genes represented in the autumn leaf library were generally less well characterized than those in the young leaf library: Almost all of the genes in Table II have a very well-defined function, whereas this is only true for a minority of the genes in Table I.
Table II.
The 20 most abundant transcript types in young leaves
| Gene | ESTs in Young Leaf Library | ESTs in Autumn Leaf Library | Enrichment in Young Leaves |
|---|---|---|---|
| RbcS | 679 | 25 | 27 |
| Lhcb1 | 224 | 1 | 224 |
| Lhcb2 | 99 | 5 | 20 |
| PsbR | 96 | 8 | 12 |
| Germin-like protein | 54 | – | – |
| PsbO | 38 | 2 | 19 |
| Lhcb4 | 37 | 2 | 19 |
| Ubiquitin | 34 | 89 | 0.4 |
| Fru-bisphosphate aldolase | 33 | 6 | 6 |
| Lhcb5 | 31 | – | – |
| Lhca3 | 26 | 7 | 4 |
| Metallothionein | 25 | 321 | 0.1 |
| Thiazole biosynthetic enzyme | 23 | 2 | 12 |
| Lhcb6 | 23 | 2 | 12 |
| Unknown EST | 23 | – | – |
| Pro-rich protein | 22 | – | – |
| Carbonic anhydrase chloroplast | 19 | 1 | 19 |
| PsaF | 19 | – | – |
| Lhcb3 | 18 | – | – |
| PsaG | 18 | – | – |
The nos. of ESTs in young and autumn leaf libraries and the enrichment factor in the young leaf library (% in young leaves/% in autumn leaves) are indicated.
Five Metallothionein Genes Were Highly Expressed in Autumn Leaves
Genes encoding metallothionein, the most abundant type of EST in the autumn leaf library, have previously been shown to be senescence induced (Kagi, 1991). Metallothioneins are divided into several classes, and we found genes encoding both type 2 and 3 metallothioneins to be very abundant in the autumn library. Manual inspection of the clones showed that the transcripts apparently originated from six different genes, which we name PMt1 through 6 (Table III). Four of these had a significantly higher EST frequency in the autumn leaf library: One (PMt1) was present at higher frequency, but at a low significance level (80% probability) and one (PMt4) was not enriched in the autumn leaf library at all. For the genes PMt3, PMt5, and PMt6, we identified clones that were aberrant: Two of the 32 PMt3 clones had an 83-bp insertion, one of the 39 PMt5 clones had a 48-bp insertion, and 19 of the 231 PMt6 clones had a 65-bp insertion. We assume that these aberrants represent unspliced clones or perhaps splice variants, but we cannot rule out the possibility of the existence of almost identical genes with insertions in the exons. However, the latter possibility seems less likely because two of the three insertions result in frame shifts, so these mRNAs do not encode a protein with high homology to known metallothioneins.
Table III.
Metallothionein genes expressed in aspen leaves
| Gene | Class | Autumn | Young |
|---|---|---|---|
| PMt1 | I? | 3 | 0 |
| PMt2 | I? | 15 | 0 |
| PMt3 | II | 32 | 2 |
| PMt4 | II | 1 | 2 |
| PMt5 | III | 39 | 0 |
| PMt6 | III | 231 | 21 |
The name and class of each metallothionein gene is given, as well as the no. of ESTs from each gene found in the autumn leaf and young leaf library.
Several Cys and Aspartic Protease ESTs Were Abundant in the Autumn Leaf Library
Proteases have important roles in the senescence process (Ye and Varner, 1996; Beers, 1997; Ueda et al., 2000), and were also abundant among the clones in the autumn leaf library. However, not all types of protease were well represented among the autumn leaf ESTs. Although ESTs encoding Cys and aspartic proteases and the components of the ubiquitin system were significantly enriched, the components of the proteasome system, which is involved in the degradation of ubiquitinated proteins, were not (Table IV), indicating that this stage of leaf senescence does not involve an increase in proteasome activity. Because Cys proteases were particularly abundant and many known SAGs encode Cys proteases (Noh and Amasino, 1999a; Solomon et al., 1999), we paid particular attention to these genes. All clones encoding Cys proteases were manually checked and assigned to individual genes. In this way, we identified 12 genes coding for Cys proteases (which we named Pcyprot1–12; Table V); Pcyprot1 through 7 are quite similar (denoted Cys proteases in Table I and GFN 1,134 in the Mendel database). ESTs from five of these proteases (Pcyprot1–5, including one most similar to SAG12) were significantly enriched in the autumn leaf library but one (Pcyprot7) appeared more frequently in the young leaf library. Thus, this class of vacuolar-located proteases appears to have a role in this stage of autumn senescence, but not other Cys proteases (apparent orthologs of the Arabidopsis proteins At4g01610, At5g60360, and At4g11320). Clones encoding aspartic proteases were also checked in the same way, and we found that two rather similar genes (with 70% homology at the DNA level) encoded aspartic proteases that seemed to have higher expression in autumn leaves.
Table IV.
Proteases in young and autumn leaf libraries
| Protease Type | Autumn | Young |
|---|---|---|
| Cys proteases | 113 | 13 |
| Aspartic proteinases | 8 | 1 |
| Ubiquitin system | 119 | 56 |
| Proteasome components | 14 | 15 |
| Clp proteases | 9 | 18 |
| Other proteases | 32 | 31 |
| Total | 291 | 130 |
The nos. of ESTs found in the autumn and young leaf libraries encoding various types of proteases are indicated.
Table V.
Cys protease genes expressed in aspen leaves
| Gene | Arabidopsis Ortholog | Annotation | Predicted Sorting | Autumn | Young |
|---|---|---|---|---|---|
| Pcyprot1 | At4g16190 | Cys protease-like protein | Sec | 37 | 3 |
| Pcyprot2 | At1g47128 | Protease RD21A | Sec | 28 | 0 |
| Pcyprot3 | At5g43060 | Cys protease | Sec | 18 | 0 |
| Pcyprot4 | At5g45890 | SAG12 | Sec | 14 | 0 |
| Pcyprot5 | At4g39090 | Protease RD19A | Sec | 8 | 2 |
| Pcyprot6 | At4g39090 | Protease RD19A | Sec | 3 | 0 |
| Pcyprot7 | At4g39090 | Protease RD19A | Sec | 1 | 4 |
| Pcyprot8 | At3g45310 | Oryzain | Sec | 1 | 0 |
| Pcyprot9 | At1g02300 | Cathepsin B-like Cys proteinase | Sec | 1 | 0 |
| Pcyprot10 | At4g01610 | Cathepsin B-like Cys proteinase | Sec | 0 | 1 |
| Pcyprot11 | At5g60360 | AALP | Sec | 0 | 1 |
| Pcyprot12 | At4g11320 | Thiol protease | Sec | 0 | 2 |
Sec, Secretory pathway.
ESTs from 35 Additional Genes Were Significantly Enriched in Autumn Leaves
We have used the annotation procedure to analyze about 27,000 ESTs from five other aspen cDNA libraries (Sterky et al., 1998; R. Bhalerao, S. Jansson et al., unpublished data). All unique ESTs in the autumn leaf library may correspond to senescence-associated or stress-induced genes, but those represented by just one or a few clones could, of course, have been found there fortuitously. According to the equations of Audic and Claverie (1997), genes that are represented by four or five ESTs in 5,200 clones from one library, but do not appear among 5,000 clones in another, are significantly enriched (with 90% and 95% confidence, respectively). In the autumn leaf library, we identified 35 genes that were represented by at least four copies in the autumn leaf library, but not any other, and named these genes Paul (Populus autumn leaf) 1 through 35 (Table VI). Some of these were very abundant among the ESTs. Paul1 and Paul2 were represented by 30 and 17 ESTs, respectively, and represent aspen homologs of two Arabidopsis genes that have not been assigned any function. Six of the Pauls showed no significant homology to any gene in public databases, nine were homologous to Arabidopsis genes of unknown function, and for several others the most similar Arabidopsis protein had only a poorly defined function. At least nine of the encoded proteins seemed to be plastid located: for eight of the Arabidopsis orthologs, chloroplast-sorting signals were found and Paul5 encodes DegP1 protease, which is known to be chloroplast located and involved in the degradation of the PS II reaction center polypeptide D1. Five of the proteins encoded by the Paul genes are predicted to be sorted through the secretory pathway, two to the mitochondria and two to the nucleus, based on their respective annotations, whereas for 11 of the proteins, no sorting is predicted. For the Paul genes where we could not identify any Arabidopsis ortholog, no specific location was predicted.
Table VI.
The most strongly expressed genes unique for the aspen autumn leaf library
| Gene | Annotation (Closest Arabidopsis Relative) | Targeting | No. of Clones | BLAST Score |
|---|---|---|---|---|
| Paul1 | At4g22920a | Chlpb | 30 | 460 |
| Paul2 | At3g02040 | Chlp | 17 | 445 |
| Paul3 | – | – | 11 | – |
| Paul4 | NAD-dependent formate dehydrogenase (At5g14780) | Mit | 10 | 1,145 |
| Paul5 | DegP1 protease precursor (At3g27925) | (Chlp) | 9 | 563 |
| Paul6 | At2g20890 | Chlp | 9 | 388 |
| Paul7 | At4g13250 | – | 7 | 285 |
| Paul8 | NTPRP27 (At2g15220) | Sec | 6 | 865 |
| Paul9 | ABC like (At3g07700) | – | 6 | 227 |
| Paul10 | At4g27360 | – | 6 | 340 |
| Paul11 | Arg-Ser-rich protein (At1g16610) | Chlp | 6 | 110 |
| Paul12 | At3g26580 | Chlp | 5 | 622 |
| Paul13 | At5g13800 | Chlp | 5 | 504 |
| Paul14 | – | – | 5 | – |
| Paul15 | At5g18130 | – | 5 | 125 |
| Paul16 | – | – | 5 | – |
| Paul17 | – | – | 5 | – |
| Paul18 | Disease resistance protein | Sec | 5 | 268 |
| Paul19 | Beta amylase (At4g17090) | Chlp | 5 | – |
| Paul20 | Nt-sube80 protein | Sec | 4 | 564 |
| Paul21 | Purple acid phosphatase (At3g17790) | Sec | 4 | 857 |
| Paul22 | – | – | 4 | – |
| Paul23 | Purple acid phosphatase | Sec | 4 | 129 |
| Paul24 | – | – | 4 | – |
| Paul25 | Myrosinase-binding proteins (At1g52000) | – | 4 | 187 |
| Paul26 | Pathogenesis-related protein YPR10 (At1g24020) | – | 4 | 270 |
| Paul27 | Receptor-like kinase (At1g16130) | – | 4 | 208 |
| Paul28 | Accelerated cell death 2 (At4g37000) | Chlp | 4 | 186 |
| Paul29 | – | – | 4 | 187 |
| Paul30 | Myb-like transcription factor (At1g01060) | (Nu) | 4 | 265 |
| Paul31 | Mitogen-activated protein kinase (At5g58350) | – | 4 | 230 |
| Paul32 | Oxidoreductase (At3g12790) | – | 4 | 650 |
| Paul33 | At5g60800 | – | 4 | 113 |
| Paul34 | RNA-binding protein (At1g60650) | Mit | 4 | 166 |
| Paul35 | Zinc finger protein (At3g21890) | (Nu) | 4 | 235 |
Localization is based on the predicted sorting signals of the Arabidopsis orthologs or (within parentheses) by function.
MATDB protein code.
chlp, Chloroplast; mit, mitochondria; sec, secretory; nu, nuclear.
Although none of the Paul genes, to our knowledge, have homologs previously claimed to be directly involved in leaf senescence, several have functions or expression patterns that relate to stress or senescence. Paul20 encodes a protein homologous to At3g54040 (Nt-sube80 protein), which is known to be induced by elicitors and photoassimilate accumulation (Herbers et al., 1995). Paul4 (NAD-dependent formate dehydrogenase) has been reported to be present mainly in mitochondria of non-photosynthetic tissues, but it was strongly induced by a number of stress treatments in a study by Desfrancssmall et al. (1993). Paul28 showed similarity to Acd (Accelerated cell death) 2, encoding a red chlorophyll catabolite reductase, which may protect cells from death by catabolizing chlorophyll breakdown products that could cause photooxidative stress if not degraded (Mach et al., 2001). Several Paul genes encoded putative regulatory proteins (transcription factors and kinases) and several showed homology to proteins induced by biotic stress (e.g. Paul8 and Paul26).
Representation of Known SAGs
Previous studies of other types of senescing leaves have identified many genes induced during senescence. To see whether some of these were expressed at higher levels in autumn leaves than in young leaves, we compared EST frequencies for all genes listed as senescence associated by Buchanan-Wollaston (1997 and refs. therein; Table VII) that have not already been discussed above (Cys and aspartic proteases, ubiquitin, ubiquitin-conjugating protein, metallothionein, and 1-aminocyclopropane-1-carboxylic acid oxidase). This list is certainly not covering all genes shown to be senescence induced, but could be regarded as a representative collection. Cytochrome P450 and MIP proteins were excluded because of problems with identifying the aspen orthologs to the senescence-induced members of these large multigene families.
Table VII.
EST frequencies for known SAGs in the autumn and young leaf libraries
| Gene or gene product | Autumn | Young |
|---|---|---|
| Glyoxysomal malate dehydrogenase | 3 | 2 |
| Fructose bisphosphate aldolase | 3 | 13 |
| GapC | 4 | 5 |
| Beta galactosidase | 0 | 1 |
| Phospholipase D | 2 | 0 |
| Cytosolic glutamine synthetase | 5 | 7 |
| Asp synthetase | 1 | 0 |
| Ferritin | 9 | 0 |
| ATP sulfurylase | 2 | 0 |
| Glutathione-S-transferase | 8 | 8 |
| Catalase | 1 | 5 |
| PR1 | 13 | 1 |
| Chitinase | 3 | 4 |
| NADH:ubiquinone oxidoreductase | 2 | 0 |
Known SAGs that were significantly (95% confidence level) enriched in the autumn leaf library were ferritin and the pathogenesis-related protein PR1. Phospholipase D, Asn synthetase, ATP sulfulyase, chitinases class III, and NADH-ubiquinone oxidoreductase subunit K also had higher clone frequencies in the autumn leaf library, but these enrichments were not statistically significant.
A number of genes reported to be senescence associated were not enriched in the autumn leaf library, including beta-galactosidase, glutathione S-transferase, catalase, chitinase class I and the glyoxisomal forms of NAD+ malate dehydrogenase, Fru-bisphosphate aldolase, cytosolic Gln synthetase, and glyceraldehyde-3-phosphate dehydrogenase. However, we cannot exclude the possibility that in some of these cases there could be problems involved in the identification of the true aspen ortholog to a senescence-specific form of the protein. Several known SAGs were not found in either library (although some were found in other libraries, not derived from leaf tissue). These included ribonuclease RNS2, malate synthase, isocitrate lyase, phosphoenolpyruvate carboxykinase, and pyruvate orthophosphate dikinase.
Of the 26 analyzed genes previously found to be senescence induced, we found expression patterns consistent with such induction for 13 (eight of which were statistically significant), eight appeared not to be up-regulated in this stage of senescence as compared with young leaves, and for five genes, we obtained no data on their expression patterns.
Photosynthesis Was Down-Regulated But Lipid Metabolism and Respiration Were Up-Regulated
The MIPS functional classification scheme is not always appropriate for plant genes. For example, there is no single class for “photosynthesis”: Rubisco and the Calvin cycle enzymes are found in class 01.05.01.05.01 (metabolism, C compound and carbohydrate metabolism, C compound and carbohydrate utilization, autotrophic CO2-fixation, and Calvin cycle), the proteins of the PSs are found in class 02.30 and the chloroplastic ATPase is found, together with mitochondrial ATPase, in class 02.11. Therefore, we constructed a slightly modified MIPS classification scheme, differing from the original in some subclasses within class 1 (metabolism) and 2 (energy) and classified all genes according to it. The modified scheme (named UPSC-MIPS) is presented in the supplementary material.
Based on the functional classification of the clones, we compared the classes of genes that were expressed in the two libraries. We did not attempt to classify genes with BLASTX scores under 100 (“not classified” in Fig. 3), and those that were most similar to a plant gene without a known function, typically an Arabidopsis open reading frame, were put in the category “unclassified.” The functional classification for each of these genes is included in the list of clones in the supplementary material.
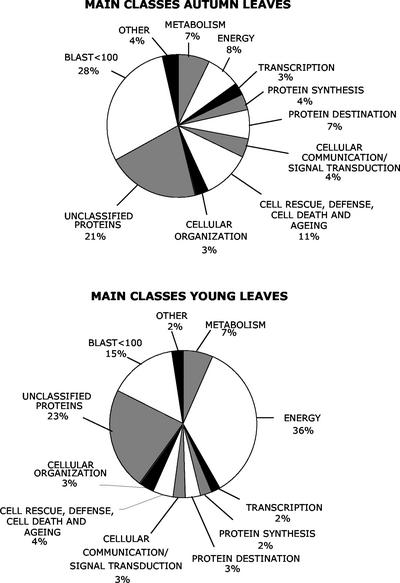
Figure 3.
Functional classification (according to the UPSC-MIPS classification scheme) of autumn leaf and young leaf ESTs.
In Figure 3, the percentage of clones found in the different main UPSC-MIPS classes is shown, and the distribution of clones in the subclasses of class 01 (metabolism) and 02 (energy) is shown in Table VIII. The full list of clones in the different classes is found in the supplementary material. The fraction of clones in class 01 (metabolism) was the same in the two libraries, but the subclasses C compound and carbohydrate metabolism, lipid, fatty acid and isoprenoid metabolism, nucleotide metabolism, and nitrogen and sulfur metabolism were more strongly represented among the clones in the autumn leaf library than in the young leaf library.
Table VIII.
Functional classification (according to Umea Plant Science Centre [UPSC]-MIPS classification scheme) of autumn leaf and young leaf ESTs belonging to the metabolism and energy classes
| Functional Classes | Autumn Leaves | Young Leaves |
|---|---|---|
| % | ||
| Metabolism | 7.0 | 6.6 |
| Amino acid metabolism | 1.2 | 1.2 |
| Nitrogen and sulfur metabolism | 0.2 | 0.1 |
| Nucleotide metabolism | 0.4 | 0.2 |
| C compound and carbohydrate metabolism | 2.8 | 2.1 |
| Lipid, fatty acid, and isoprenoid metabolism | 1.3 | 1.0 |
| Metabolism of vitamins, cofactors, and prosthetic groups | 0.3 | 0.8 |
| Secondary metabolism | 0.9 | 1.2 |
| Energy | 7.9 | 35.1 |
| Glycolysis and gluconeogenesis | 0.8 | 0.5 |
| Tricarboxylic acid pathway | 0.2 | 0.2 |
| Electron transport | 0.9 | 0.6 |
| Light reaction | 0.9 | 15.8 |
| Rubisco and Calvin cycle | 1.2 | 14.9 |
| Photorespiration | 0.6 | 1.0 |
| Pentose phosphate pathway | 0.1 | 0.1 |
| Glyoxylate cycle | 0.2 | 0.1 |
| Oxidation of fatty acids | 0.3 | 0.1 |
| Other photosynthesis activities | 2.5 | 1.7 |
| Other energy activities | 0.1 | 0.2 |
The differences between the libraries were much more pronounced in the class 02 (energy). For instance, the subclass photosynthesis contained 5.2% of the clones in the autumn leaf library, compared with 33% in the young leaf library. Major differences were also found within the photosynthesis subclass. Subclass 02.30.01.01 (photosynthetic light reaction), for example, accounted for 0.9% of the ESTs in the autumn leaf library, compared with 16% in the young leaf library, an almost 20-fold reduction. On the other hand, subclass 02.30.02.05 (photorespiration) was much less depleted (0.6% versus 1.0%).
Several authors have suggested that lipid metabolism provides energy for the senescing leaf (Gut and Matile, 1988; Wanner et al., 1991). We found enrichment in autumn leaves of ESTs in the classes 01.06 (lipid and fatty acid metabolism), 02.01 (glycolysis and gluconeogenesis), and 02.11 (electron transport and membrane-associated energy conservation).
The classes from the list of most abundant genes that seemed to be most enriched in the autumn leaf library, 06 protein destination (including 06.13, proteolysis), and 11 Cell rescue, defense, death, and aging, were much better represented in the autumn than in the young leaf library (7% versus 3% and 11% versus 4%, respectively). The fraction of ESTs without significant homology to any gene in public databases was almost twice as large in the autumn leaf library (28% versus 15%), whereas the fraction of “unclassified” clones (homologous to a gene without assigned function) was about the same in the two libraries.
Transcript Profiling
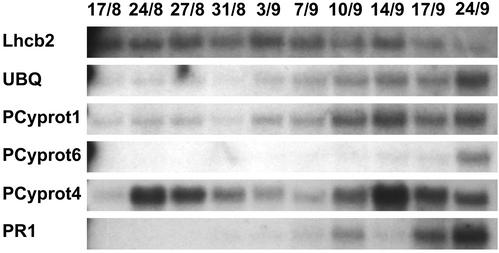
We analyzed transcript abundance for five of the genes: ubiquitin, PR1, and three Cys proteases (Pcyprot 1, 4, and 6), which we identified as putative SAGs in aspen during the autumn in leaves of a free-growing aspen tree. As a comparison, one apparently down-regulated gene (Lhcb2) was also analyzed.
The expression patterns of the two types of gene were, as expected, strikingly different: Although the Lhcb2 mRNA level decreased steadily during the autumn, all five putative SAGs showed an increase in transcript abundance (Fig. 4). However, the patterns were different. Pcyprot6 and PR1 mRNAs accumulated to high levels only in the very late stages of senescence, ubiquitin and the Pcyprot 1 showed a more gradual increase and the Pcyprot4 transcript showed biphasic behavior, with one peak at August 24 and another at September 14. This supports the hypothesis that many of the genes we identified as potential SAGs in the EST material are SAGs.
Figure 4.
RNA gel-blot analysis of mRNA transcripts during senescence. Total RNA was isolated from leaves at different dates during the autumn of 1999 and hybridized with labeled cDNA probes for Lhcb2, polyubiquitin (UBQ), three Cys proteases, and pathogenesis-related protein 1 (PR1).
DISCUSSION
The coloration of the leaves of deciduous trees in the fall in temperate regions is perhaps the most striking example of leaf senescence. Therefore, it is rather surprising that there are no data on gene expression during autumn leaf senescence. This type of leaf senescence is probably very similar to senescence in, for example, detached leaves, but there must also be differences. The autumnal leaf senescence program is induced in all the leaves by decreasing day length, regardless of whether they are stressed by other environmental factors. There is also a lot of natural variation in the regulation of the process. For instance, in an adaptation to the earlier autumns in the north, trees from higher latitudes start senescing earlier than trees from lower latitudes (Pauley and Perry, 1954). We have embarked on a project to elucidate the genetic basis of autumn senescence in aspen leaves, and describe here the first steps and results. We have performed large-scale EST sequencing to isolate candidate SAGs and to study the differences in overall gene expression between senescing leaves and young, but fully expanded, leaves. As a reference material, we have chosen young, greenhouse-grown leaves to maximize gene finding. This is assuming the comparison will detect not only differences in gene expression dependent on senescence, but also differences induced by various environmental stresses imposed on leaves of the free-growing aspen. Further studies will reveal which of the genes discussed in this paper that are truly senescence associated and those that are induced by various stress treatments, but even at this stage we can draw several interesting conclusions about gene expression in autumn leaves.
Estimating expression levels from EST frequencies is an indirect method and there are both technical and biological limitations to such an analysis. For example, uneven efficiency in the reverse transcription of mRNAs of different sizes, size fractionation, and the possible recalcitrance of some genes toward cloning in Escherichia coli are all problems that may affect the results. Despite these limitations, the “digital northern” approach has a major advantage over traditional northern or most DNA chip array experiments because it gives data on mRNA levels for individual genes relative to the total mRNA pool. mRNA levels do not necessarily correspond well to protein synthesis, and there are many well-documented examples of translational regulation of gene expression in plants. However, for most major enzymatic components, EST abundance seems to be a fair approximation of relative protein abundance (Jansson, 1999; Mekhedov et al., 2000; Ohlrogge and Benning, 2000). For all genes that we have tested so far, RNA blotting has given similar results to EST frequency analysis, although direct methods have to be used to obtain high-quality data for individual genes. We are now performing large-scale transcript profiling using DNA microarrays to get further information about the precise transcript abundance of the different genes.
The pattern of gene expression in the two libraries was strikingly different. Our data indicate that most of the metabolic characteristics previously reported for senescing leaves (down-regulation of photosynthesis and up-regulation of genes involved in protein, lipid, pigment degradation, and respiration, as well as stress-related genes; for review, see Smart, 1994) were also found in aspen autumn leaves. For the majority of genes previously shown to be senescence associated in annual plants, we found the same in autumn leaves. This confirms that the general pattern of metabolism is the same in autumn leaves as in senescing leaves of annual plants. Some of the genes previously reported to be SAGs in other systems were not well represented in the autumn leaf library. The transcript profiling that we performed on a limited number of genes shows that mRNA for several of them accumulates later in the process, and it is likely that the majority of the known SAGs will also prove to be SAGs in the autumn leaf system. However, we also believe that we will be able to identify genes whose expression patterns in autumn leaves are not mimicked in senescing leaves of annuals such as Arabidopsis. Because the process is triggered differently we expect there to be at least some regulatory proteins that have a specific role in inducing autumn senescence.
In the young, greenhouse-grown leaves a very large proportion of the mRNA pool (and, thus, protein synthesis) was devoted to synthesis of the photosynthetic apparatus: 33% of the ESTs encoded proteins known to be components of the various protein complexes involved in photosynthesis. In contrast, only 5% of the clones in the library from senescing leaves encoded photosynthetic proteins and one-half of those were stress-related photosynthetic proteins such as ELIPs. The average gene encoding a “standard” photosynthetic protein, directly involved in light reaction or CO2 fixation, was down-regulated about 20-fold in the autumn leaves. We expected gene expression in young leaves grown under non-stressed conditions to be heavily concentrated on photosynthesis, but we were surprised to find how little of the gene expression in autumn leaves, which still showed no visible sign of chlorophyll degradation, was dedicated to photosynthesis. Senescence is a strictly controlled developmental process and by the middle of September, photosynthetic gene expression had apparently been turned off and the leaves had prepared to break down their chloroplasts.
In addition to the confirmation that autumn senescence shares many features with senescence in leaves of annual plants, we also identified a number of genes whose orthologs in Arabidopsis are either unknown or have not been connected with senescence. By choosing leaves in the process of chlorophyll degradation as sources for the autumn leaf cDNA library, we believed that we could get a snapshot of the protein synthesis activity related to degradation of the chloroplasts, and possibly other cell constituents as well. Of our identified 35 Paul genes, nine encoded proteins Arabidopsis orthologs seem to be chloroplast located, and four of these have no assigned function. These are all good candidates for proteins involved in degradation of the chloroplast components, and we also found two known chloroplast proteases, DegP1 and FtsH2 (Adam et al., 2001), and one enzyme involved in chlorophyll degradation among the genes apparently up-regulated in the autumn leaves.
Another striking difference was the higher fraction of ESTs in the autumn leaf library that showed no significant homology to any known protein in public databases. This could simply be a consequence of the fact that young, green leaves have been very extensively studied and the proteins of such leaves are better characterized. Because gene prediction also relies on EST data, genes expressed in tissues that not have been subjected to EST sequencing are overrepresented among genes for which no orthologs have been found, and/or whose putative function remains unknown. This means that we may overestimate the fraction of “truly novel” genes in our data but, even so, there are many potentially interesting genes to be found in autumn leaves of aspen.
A prominent feature of the nuclear genome of plants is the large fraction of genes that appear to have originated from the cyanobacterial genome. It is believed that in the evolution of the green plant, a cyanobacterial progenitor of the chloroplast was engulfed by the eukaryotic host, becoming enclosed by a double membrane, and then permanently integrated into the plant cell as an organelle, the chloroplast. The ancestral chloroplast genome has been estimated to have consisted of around 3,200 genes, roughly 1,700 of which have been lost because of redundancy between the nuclear and plastid gene products, and about 1,400 genes appear to have been transferred to the nuclear genome, leaving only 87 plastid-encoded genes (Sato et al., 1999; Abdallah et al., 2000). EST sequencing does not give direct information concerning organelle mRNA content, but our data can also be used to indirectly estimate organelle protein synthesis in aspen leaves. The basis of the calculation is that nearly all of the proteins encoded in the organelle genomes form, together with nuclear-encoded subunits, multiprotein complexes in which the protein subunits and their relative stoichiometries are known in great detail. Because we can calculate the average EST frequency of all nuclear-encoded subunits of these complexes and assume that the plastid-encoded subunits are synthesized in matching amounts, we can estimate relative rates of chloroplast protein synthesis (details of these calculations are given in the supplemental material). Based on these calculations, we estimate plastid protein synthesis to account for 130/5,128 ≈ 2.5% of the cytoplasmic protein synthesis in autumn leaves and 1,144/4,842 ≈ 24% in young leaves. Although these figures are only indirect estimations and may not be quantitatively accurate, they strongly indicate that there is a massive down-regulation of plastid protein synthesis before any visible sign of autumn senescence.
We found no evidence for a conversion of peroxisomes to glyoxysomes, like in senescing rape (Brassica napus) leaves (Vicentini and Matile, 1993) during this stage of senescence; the key enzymes of the glyoxylate cycle (malate synthase and isocitrate lyase) and of gluconeogenesis (phosphoenolpyruvate carboxykinase and pyruvate orthophosphate dikinase) were not found among the ESTs from autumn leaves. However, the changes in subclasses of “energy” indicate that respiration and mitochondrial energy conversion were important in autumn leaves and that mitochondria have already taken over the chloroplasts' role as energy-generating organelles even before massive chloroplast breakdown has occurred. Formate dehydrogenase (one of the Paul genes) was strongly induced in the autumn leaves and can be involved in energy production in mitochondria without the participation of the tricarboxylic acid cycle. Alternatively, this enzyme may be involved in metabolism of C1 compounds, although the source of these putative metabolites cannot be defined at present. The senescing leaves are still source leaves because the phloem presumably transports recycled nutrients away from the leaves into the overwintering parts of the tree. The exact nature of the compounds transported is unknown, but amino acids with high nitrogen content (e.g. Gln and Asn) are obvious candidates. An important challenge for further research will be to define the metabolic pathways involved in nutrient recapture from senescing leaves.
Our data indicate that Cys and aspartic proteases may play an important role during chloroplast degradation, whereas at least the ubiquitin system (as evident from the RNA blot data) is not up-regulated until a later stage of senescence. It has been shown in other systems that the proteasome components do not accumulate during senescence (Bahrami and Gray, 1999), but the enzymes of the ubiquitin pathway do (Belknap and Garbarino, 1996). It is possible that the proteasomes present are sufficient to degrade the ubiquitinated proteins that, presumably, accumulate during later stages of autumn senescence. The senescence-associated Cys proteases may all be located in the vacuole or endoplasmic reticulum bodies (Hayashi et al., 2001). This would be consistent with the secretory system having an important role in the senescence process. One of the Cys proteases we detected seems to be the aspen ortholog to SAG12, which has very restricted expression in Arabidopsis and Brassica napus, limited to the last stages of senescence (Noh and Amasino, 1999b). The expression pattern in aspen was different, with one peak at the beginning of September, and one at the time when chlorophyll degradation started. The first peak correlated with weather factors likely to cause photooxidative stress, which would induce genes (PsbS and ELIP) that are up-regulated during light stress in aspen leaves (K. Wissel and S. Jansson, unpublished data). Because leaf senescence is probably an oxidative process (Munne-Bosch and Alegre, 2002) unfavorable conditions may trigger degradative processes that, at least in part, may be reversed if weather conditions become more favorable again.
We believe that this work, in addition to discovering genes, provides insights into gene expression in aspen leaves at a rather early stage of autumn senescence. Moreover, it illustrates the usefulness of leaves of deciduous trees as a model system to study leaf senescence, and we believe that our ongoing transcript profiling using DNA microarrays will make it possible to pinpoint a number of candidate genes for regulators of the process. Ability to control senescence will have important biotechnological implications because trees that shed their leaves too early have lower than optimal productivity, whereas if the senescence process is initiated too late, the tree does not have sufficient time to recapture nutrients and complete the hardening procedure before the winter, and, thus, is likely to suffer from growth limitations and/or frost injuries. Therefore, this study (the first, to the best of our knowledge, in which gene expression during autumn leaf senescence has been studied) may be the first step to a deeper understanding of this biologically important process.
MATERIALS AND METHODS
Plant Material
Leaves were sampled from a free-growing aspen (Populus tremula) at the University of Umea campus. About 30 leaves, from the outer part of the crown, were sampled twice a week, at 11 am on each occasion. Leaves were flash frozen in liquid nitrogen and stored at −80°C.
RNA Preparation and Blotting
Aspen RNA was prepared according to Chang et al. (1993) with the following modifications. No spermidine was used in the extraction buffer and 2.67% (v/v) β-mercaptoethanol was used instead of 2%. One additional extraction step was performed after LiCl precipitation. RNA concentrations were determined spectrophotometrically (GeneQuant, Amersham-Pharmacia Biotech, Uppsala) and equal amounts of total RNA were separated on a 1.2% (w/v) agarose gel, transferred to a Hybond N+ (Amersham, Buckinghamshire, UK) membrane, and RNA-blot analysis was performed using standard procedures.
cDNA Library Constructions
The senescence cDNA library was constructed from RNA prepared from leaves harvested on September 14, using the SMART cDNA library construction kit system (CLONTECH Laboratories, Palo Alto, CA). The young leaf cDNA library, described by Larsson et al. (1997), was prepared from hybrid aspen (Populus tremula × tremuloides T89) plants grown in fertilized peat in a greenhouse under natural light conditions with supplementary light (18-h photoperiod).
EST Sequencing
The cDNA inserts were sequenced from the 5′ end using PCR products as templates by a Biomek robot (Beckman Instruments, Fullerton, CA) in a 96-well microtiter format. PCR amplifications were performed using general vector primers and standard PCR protocols. The size and quality of the PCR products were checked by gel electrophoresis. The samples were analyzed using a DYEnamic ET Dye Terminator Kit (Amersham-Pharmacia Biotech) and a biotinylated sequencing primer (reverse sequencing primer). The sequencing reaction products were purified on a magnetic workstation (Magnatrix 1200, Magnetic Biosolutions, Stockholm) using paramagnetic beads (Dynapure, Dynal, Oslo) before the samples were loaded onto an ABI 377 (Perkin-Elmer Applied Biosystems, Foster City, CA) or MegaBACE 1000 (Amersham-Pharmacia Biotech) DNA sequencer.
Bioinformatics
Raw sequences chromatograms were processed by the Phred program (http://www.phrap.com/phred/). Vector sequences and low-quality regions were deleted using Vectorstrip (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/). The cleaned inserts were then stored in FASTA format. The whole of the above process was performed semiautomatically with the help of Perl scripts. ESTs containing rRNA, chloroplast DNA, or mitochondrial DNA were identified by the BLASTN algorithm of NCBI-BLAST (Altschul et al., 1990) followed by comparison of homologous Arabidopsis sequences (accession nos. X52322, AP000423, and Y08501/Y08502, respectively) and excluded from further analysis. Sequences were clustered into singlets and contigs using Phrap (http://www.phrap.com/) and compared locally with SWISS-PROT/TrEMBL, MATDB (downloaded from http://www.expasy.ch and http://mips.gsf.de, respectively) and the Mendel Gene Family Database (obtained from http://www.mendel.ac.uk) using BLASTX. TBLASTX was used to calculate the BLAST score of the sequence against itself (self-blast score). A set of Perl scripts was arranged for the pipeline process of running multiple blast searches, parsing the blast result files, and retrieving the annotations of the sequencing producing the best hits.
We constructed a FileMaker Pro-database with a Web interface with options like direct BLAST searches against our own database (on a local Unix server) or against MIPS or SWISS-PROT (over the Internet). There are also direct links to the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.ad.jp/kegg/) and Enzyme (http://www.expasy.ch/enzyme/) databases for easy access to more detailed information. For contigs, there is also information about the number of clones present in them, and the libraries from which they originate.
Significance levels for differential expression were calculated by the equation of Audic and Claverie (1997).
ACKNOWLEDGMENTS
We wish to thank Baram Amini, Thomas Hiltonen, Susanne Larsson, Björn Sjöblom, and Carl Zingmark for their participation during various stages of this work.
Footnotes
This work was supported by the Knut and Alice Wallenberg Foundation, by the Foundation for Strategic Research, by the Swedish Research Council (grant to S.J.), and by the Swedish Research Council for the Environment, Agricultural Sciences, and Spatial Planning (Formas; grants to S.J., J.L., and P.G.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.012732.
LITERATURE CITED
- Abdallah F, Salamini F, Leister D. A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 2000;5:141–142. doi: 10.1016/s1360-1385(00)01574-0. [DOI] [PubMed] [Google Scholar]
- Adam Z, Adamska I, Nakabayashi K, Ostersetzer O, Haussuhl K, Manuell A, Zheng B, Vallon O, Rodermel SR, Shinozaki K et al. Chloroplast and mitochondrial proteases in Arabidopsis. A proposed nomenclature. Plant Physiol. 2001;125:1912–1918. doi: 10.1104/pp.125.4.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Bahrami AR, Gray JE. Expression of a proteasome alpha-type subunit gene during tobacco development and senescence. Plant Mol Biol. 1999;39:325–333. doi: 10.1023/a:1006102110889. [DOI] [PubMed] [Google Scholar]
- Beers EP. Programmed cell death during plant growth and development. Cell Death Diff. 1997;4:649–661. doi: 10.1038/sj.cdd.4400297. [DOI] [PubMed] [Google Scholar]
- Belknap WR, Garbarino JE. The role of ubiquitin in plant senescence and stress responses. Trends Plant Sci. 1996;1:331–335. [Google Scholar]
- Buchanan-Wollaston V. The molecular biology of leaf senescence. J Exp Bot. 1997;48:181–199. [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- Collier DE, Thibodeau BA. Changes in respiration and chemical content during autumnal senescence of Populus tremuloides and Quercus rubra leaves. Tree Physiol. 1995;15:759–764. doi: 10.1093/treephys/15.11.759. [DOI] [PubMed] [Google Scholar]
- Danon A, Delorme V, Mailhac N, Gallois P. Plant programmed cell death: a common way to die. Plant Physiol Biochem. 2000;38:647–655. [Google Scholar]
- Desfrancssmall CC, Ambardbretteville F, Small ID, Remy R. Identification of a major soluble protein in mitochondria from non-photosynthetic tissues as Nad-dependent formate dehydrogenase. Plant Physiol. 1993;102:1171–1177. doi: 10.1104/pp.102.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller U, Fischer A. Nitrogen metabolism in senescing leaves. Crit Rev Plant Sci. 1994;13:241–273. [Google Scholar]
- Gan SS, Amasino RM. Making sense of senescence: molecular genetic regulation and manipulation of leaf senescence. Plant Physiol. 1997;113:313–319. doi: 10.1104/pp.113.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut H, Matile P. Apparent induction of key enzymes of the glyoxylic acid cycle in senescent barley leaves. Planta. 1988;176:548–550. doi: 10.1007/BF00397663. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa NK, Nishimura M, Hara-Nishimura I. A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 2001;42:894–899. doi: 10.1093/pcp/pce144. [DOI] [PubMed] [Google Scholar]
- Herbers K, Monke G, Badur R, Sonnewald U. A simplified procedure for the subtractive cDNA cloning of photoassimilate-responding genes: isolation of cDNAs encoding a new class of pathogenesis-related proteins. Plant Mol Biol. 1995;29:1027–1038. doi: 10.1007/BF00014975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch WA, Zeldin EL, McCown BH. Physiological significance of anthocyanins during autumnal leaf senescence. Tree Physiol. 2001;21:1–8. doi: 10.1093/treephys/21.1.1. [DOI] [PubMed] [Google Scholar]
- Jansson S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- Kagi JHR. Overview of methallothoinein. Methods Enzymol. 1991;205:613–626. doi: 10.1016/0076-6879(91)05145-l. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Diff. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- Larsson S, Bjorkbacka H, Forsman C, Samuelsson G, Olsson O. Molecular cloning and biochemical characterization of carbonic anhydrase from Populus tremula × tremuloides. Plant Mol Biol. 1997;34:583–592. doi: 10.1023/a:1005849202731. [DOI] [PubMed] [Google Scholar]
- Liang F, Holt I, Pertea G, Karamycheva S, Salzberg SL, Quackenbush J. An optimized protocol for analysis of EST sequences. Nucleic Acids Res. 2000;28:3657–3665. doi: 10.1093/nar/28.18.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale D, Arnold B. Mendel ESTS: database of plant ESTs in dbEST annotated with gene family numbers and gene family names. Plant Mol Biol Rep. 1999;17:239–247. doi: 10.1093/nar/29.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA. 2001;98:771–776. doi: 10.1073/pnas.021465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhedov S, de Ilarduya OM, Ohlrogge J. Toward a functional catalog of the plant genome. A survey of genes for lipid biosynthesis. Plant Physiol. 2000;122:389–401. doi: 10.1104/pp.122.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munne-Bosch S, Alegre L. Plant aging increases oxidative stress in chloroplasts. Planta. 2002;214:608–615. doi: 10.1007/s004250100646. [DOI] [PubMed] [Google Scholar]
- Nam HG. The molecular genetic analysis of leaf senescence. Curr Opin Biotechnol. 1997;8:200–207. doi: 10.1016/s0958-1669(97)80103-6. [DOI] [PubMed] [Google Scholar]
- Noh YS, Amasino RM. Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol Biol. 1999a;41:181–194. doi: 10.1023/a:1006342412688. [DOI] [PubMed] [Google Scholar]
- Noh YS, Amasino RM. Regulation of developmental senescence is conserved between Arabidopsis and Brassica napus. Plant Mol Biol. 1999b;41:195–206. doi: 10.1023/a:1006389803990. [DOI] [PubMed] [Google Scholar]
- Nooden LD, Guiamet JJ, John I. Senescence mechanisms. Physiol Plant. 1997;101:746–753. [Google Scholar]
- Ohlrogge J, Benning C. Unraveling plant metabolism by EST analysis. Curr Opin Plant Biol. 2000;3:224–228. [PubMed] [Google Scholar]
- Olsen JE, Junttila O, Nilsen J, Eriksson ME, Martinussen I, Olsson O, Sandberg G, Moritz T. Ectopic expression of oat phytochrome A in hybrid aspen changes critical day length for growth and prevents cold acclimatization. Plant J. 1997;12:1339–1350. [Google Scholar]
- Pauley SO, Perry TO. Ecotypic variation of the photoperiodic response in Populus. J Arnold Arbor. 1954;35:167–188. [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM. Molecular aspects of leaf senescence. Trends Plant Sci. 2000;5:278–282. doi: 10.1016/s1360-1385(00)01655-1. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6:283–290. doi: 10.1093/dnares/6.5.283. [DOI] [PubMed] [Google Scholar]
- Smart CM. Gene expression during leaf senescence. New Phytol. 1994;126:419–448. doi: 10.1111/j.1469-8137.1994.tb04243.x. [DOI] [PubMed] [Google Scholar]
- Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell. 1999;11:431–443. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky F, Regan S, Karlsson J, Hertzberg M, Rohde A, Holmberg A, Amini B, Bhalerao R, Larsson M, Villarroel R et al. Gene discovery in the wood-forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proc Natl Acad Sci USA. 1998;95:13330–13335. doi: 10.1073/pnas.95.22.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Seo S, Ohashi Y, Hashimoto J. Circadian and senescence-enhanced expression of a tobacco cysteine protease gene. Plant Mol Biol. 2000;44:649–657. doi: 10.1023/a:1026546004942. [DOI] [PubMed] [Google Scholar]
- Vicentini F, Matile P. Gerontosomes, a multifunctional type of peroxisome in senescent leaves. J Plant Physiol. 1993;142:50–56. [Google Scholar]
- Wanner L, Keller F, Matile P. Metabolism of radiolabelled galactolipids in senescent barley leaves. Plant Science. 1991;78:199–206. [Google Scholar]
- Ye ZH, Varner JE. Induction of cysteine and serine proteases during xylogenesis in Zinnia elegans. Plant Mol Biol. 1996;30:1233–1246. doi: 10.1007/BF00019555. [DOI] [PubMed] [Google Scholar]
- Yen CH, Yang CH. Evidence for programmed cell death during leaf senescence in plants. Plant Cell Physiol. 1998;39:922–927. [Google Scholar]