Abstract
Plant mitochondria maintain metabolic communication with the cytosol through a family of carrier proteins. In Arabidopsis, a subset of 45 putative genes encoding members of this family have been identified based on generalized mitochondrial carrier features. No gene clusters are apparent and few of the predicted protein products have mitochondrial targeting sequences recognized by bioinformatic predictors. Only nine genes are currently represented by more than 10 expressed sequence tags at The Institute for Genomic Research. Analyses of public microarray experiments reveal differential expression profiles of the more highly expressed members of this gene family in different plant organs and in response to plant hormone application and environmental stresses. A comparison of this Arabidopsis carrier subset (45) to the yeast gene family (35) reveals 10 orthologous groups between the two species. Recent surveys of the Arabidopsis mitochondrial proteome by two-dimensional gel separations have not identified any of these carrier proteins, presumably because of their hydrophobicity and basicity. Isolating integral membrane proteins from Arabidopsis mitochondria, using one-dimensional electrophoresis for protein separation and tandem mass spectrometry-based sequencing of doubly charged peptides, we have unequivocally identified specific carrier gene products located in mitochondria. This approach has identified six of the nine carriers represented highly in expressed sequence tag databases: adenine nucleotide translocator (At3g8580 and At5g13490), dicarboxylate/tricarboxylate carrier (At5g19760), phosphate carrier (At5g14040), uncoupling protein (At3g54110), and a carrier gene of unknown function (At4g01100). Overall, the combined transcript and protein expression data indicates that only a small subset of the carrier family of genes provide the majority of carrier proteins of Arabidopsis mitochondria.
The transport of metabolites and other solutes across the mitochondrial inner membrane is catalyzed by a series of specific carriers that operate as exchangers/cotransporters. A superfamily of related transporters has been described in eukaryotes that contain a tripartite structure of 100 amino acid segments each consisting of two membrane-spanning α-helices separated by an extra-membrane hydrophilic loop (Walker and Runswick, 1993). These transporters operate as homodimers with a 12 transmembrane domain structure (Saraste and Walker, 1982) and are responsible for the transport of a wide variety of metabolites between mitochondria and the cytosol. Known examples include the adenine nucleotide transporter (ANT), the oxoglutarate-malate transporter (OMT), the uncoupling protein (UCP), the phosphate transporter (PiC), the dicarboxylate transporter, and the tricarboxylate transporter (Palmieri et al., 1996). Evidence from yeast also indicates that a variety of amino acid transporters and cofactor transporters belong to this superfamily (Crabeel et al., 1996; Liu and Dunlap, 1996; Palmieri et al., 1996). Studies have considered the structure, function, and import of these carriers into mammalian and yeast mitochondria (Palmieri et al., 1996). Some of these transporters have more recently been implicated as components of the mitochondrial transition pore that opens during the early events of cytochrome c-dependent programmed cell death pathway (Mig-notte and Vayssiere, 1998). Since the completion of the yeast genome, it is now apparent that 35 genes encode members of this protein family. Fewer than one-half the members of this family have been assigned a function (Belenkiy et al., 2000; Palmieri et al., 2001a, 2001c; Prohl et al., 2001); whereas expression profiling has revealed that members of unknown function are differentially expressed (Belenkiy et al., 2000). In addition, yeast two-hybrid studies suggest that some members of this family interact with noncarrier proteins including a ubiquitin-conjugating protein, a recA-like DNA repair protein, and a cellular chaperonin (Belenkiy et al., 2000).
Biochemical characterization of plant mitochondrial carrier function over the last 30 years has revealed the operation of carriers for phosphate, adenine nucleotides, mono-, di-, and tri-carboxylates, amino acids, and cofactors such as NAD+ and coenzyme A (Wiskich, 1977; Day and Wiskich, 1984; Douce et al., 1997). However, little molecular research on the genes encoding these carriers has been undertaken (Laloi, 1999). Our analysis of the Arabidopsis genome reveals a set of 45 putative mitochondrial carrier proteins based on homology with yeast and animal counterparts, but it is unclear which genes are expressed and which gene products are targeted to mitochondria. We (Millar et al., 2001) and others (Kruft et al., 2001) have recently presented proteomes of Arabidopsis mitochondria based on two-dimensional gel separations, but mitochondrial carrier proteins were not identified in either of these studies. Here, we have analyzed available evidence determining paralogous groups in the Arabidopsis carrier gene family and linking them to their yeast orthologs. We have assessed the relative expression of the gene family using publicly available expressed sequence tag (EST) and microarray data. We have then determined the actual presence of carrier proteins in Arabidopsis mitochondria using a modified proteomic approach based on isolation of integral membrane proteins, standard one-dimensional SDS-PAGE, and tandem mass spectrometry (MS/MS)-based sequencing of tryptic peptides.
RESULTS AND DISCUSSION
Identification of Mitochondrial Carrier Proteins in the Arabidopsis Genome
Many of the various substrate carriers in the inner membrane of yeast and mammalian mitochondria contain up to three copies of a 10-amino acid sequence motif known as the mitochondrial energy transfer signature (METS; Prosite PDOC00189): P-x-[DE]-x-[LIVAT]-[RK]-x-[LRH]-[LIVMFY]-[QGAIVM]. Analysis of the complete Arabidopsis genome with this METS homology motif reveals a set of 80 genes that encode putative proteins containing at least one copy of this motif. In this set of 80, a total of 45 protein products contained some common attributes of the mitochondrial carrier protein family. These genes encode 300 to 450 (350 ± 35) amino acid proteins with nine to 10 (9.8 ± 0.3) basic pIs and six putative membrane-spanning domains. Each member of this set of 45 genes shared a higher degree of sequence identity with at least one other member of the carrier family than to any other Arabidopsis gene based on BLAST analysis. This subset of carrier genes likely excludes some related members that would be included if the selection criteria were changed in some manner. For example, by widening the amino acid residue window a putative peroxisomal carrier (At5g61810) would be included. However, we consider that this set of 45 carrier genes represents a generalized mitochondrial carrier structure with a low probability of excluding such carriers and a low probability of arbitrary inclusion of non-mitochondrial carrier genes. Clearly putative carriers that do not contain the METS motif and that are longer than 450 amino acids have not been included in this analysis. For this work, the above set of 45 proteins are designated as set A. Chromosome number and gene number in Table I form the unique identifier of Arabidopsis genes, the Arabidopsis Genome Initiative gene code. Gene 08580 in Table I has an Arabidopsis Genome Initiative gene code of At3g08580 representing a physical gene locus on chromosome 3, at position 858. This gene would be physically flanked by 857 (At3g08570) and 859 (At3g08590). Using this information, the genes in set A appear evenly distributed across the five chromosomes of the Arabidopsis genome (7, 7, 8, 8, and 15). No physical clusters of two or more genes at one physical locus are evident in set A. The closest carrier gene pairs are found on chromosome 5 and are 16 genes apart. Most members of set A are separated by hundreds of genes (Table I). This means that there is no evidence of the very recent duplication of genes leading to physical clusters often observed in other Arabidopsis large gene families (Arabidopsis Genome Initiative, 2000). Only two genes are found to be duplicates residing in recent segment duplications between chromosomes 1 and 2 (At1g07030 and At2g30160) and chromosomes 4 and 5 (At2g47490 and At3g62650) based on the annotation of chromosome duplication segments (http://mips.gsf.de/proj/thal/db/gv/rv/rv_frame.html). In the latter pair, the second gene (At3g62650) is not included in set A (Table I) because it only represents the first exon of a carrier protein encoding a 152-amino acid protein and does not contain a METS.
Table I.
Members of the mitochondrial carrier superfamily from Arabidopsis
| Chr | Gene | Annotation | Accession No. | ESTs TIGR | TC TIGR | METS | Acids | MM | pI | Extension | PR | TP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 07030 | Carrier | NP172184 | 7 | TC150083 | 2 | 326 | 35,703 | 8.15 | 24 | N | 1 |
| 1 | 14140 | Putative UCP | NP172866 | 7 | TC153580 | 3 | 305 | 33,400 | 9.82 | 12 | N | M4 |
| 1 | 14560 | Carrier | NP172908 | 3 | TC166650 | 1 | 331 | 36,200 | 9.69 | 7 | N | S4 |
| 1 | 25380 | Carrier | AAG50815 | 2 | TC155478 | 1 | 311 | 33,700 | 9.42 | 10 | N | 2 |
| 1 | 72820 | Carrier | AAM19953 | 8 | TC164605 | 1 | 349 | 37,129 | 9.35 | 15 | N | 2 |
| 1 | 74240 | Carrier | NP177564 | 2 | TC169692 | 3 | 367 | 40,600 | 9.53 | 12 | N | 3 |
| 1 | 79900 | Car/acylcar carrier | AAL38806 | 7 | TC163745 | 2 | 296 | 32,252 | 9.69 | 2 | N | 1 |
| 2 | 17270 | Putative PiC | NP179319 | 3 | TC157216 | 1 | 309 | 34,200 | 9.62 | 5 | N | 2 |
| 2 | 22500 | Putative DC | AAD22351 | 32 | TC161429 | 2 | 313 | 33,357 | 9.74 | 30 | N | 4 |
| 2 | 26360 | Carrier | AAC14487 | 1 | TC158497 | 2 | 303 | 32,977 | 9.26 | 3 | N | S5 |
| 2 | 30160 | Carrier | AAC16956 | 4 | TC153500 | 1 | 331 | 35,951 | 8.82 | 12 | N | 1 |
| 2 | 37890 | Carrier | NP181325 | 2 | TC157347 | 2 | 348 | 37,911 | 9.60 | 52 | N | 2 |
| 2 | 46320 | Carrier | AAM15049 | 1 | TC169193 | 2 | 358 | 38,788 | 9.37 | 19 | N | 2 |
| 2 | 47490 | Carrier | T00435 | 6 | TC153054 | 1 | 447 | 49,784 | 10.1 | 112 | P | M3 |
| 3 | 08580 | ANT | AAL06907 | 366 | TC149364 | 2 | 381 | 41,475 | 9.84 | 65 | N | 5 |
| 3 | 20240 | Carrier | NP188659 | 2 | TC157463 | 2 | 348 | 37,903 | 9.44 | 20 | N | 3 |
| 3 | 21390 | Carrier | AY080592 | 4 | TC165144 | 2 | 327 | 35,556 | 9.47 | 27 | N | 2 |
| 3 | 48850 | Putative PiC | NP190454 | 0 | – | 1 | 336 | 39,024 | 9.36 | 55 | N | C2 |
| 3 | 51870 | Carrier | NP190755 | 6 | TC165909 | 2 | 381 | 41,817 | 9.75 | 59 | N | C5 |
| 3 | 53940 | Carrier | NP190962 | 3 | TC166581 | 1 | 358 | 38,836 | 9.62 | 67 | P | 4 |
| 3 | 54110 | UCP1 | AAM14124 | 30 | TC161370 | 3 | 306 | 32,662 | 9.62 | 4 | N | S4 |
| 3 | 55640 | Ca2+-dependent carrier | AAL34246 | 3 | TC166235 | 1 | 332 | 36,238 | 9.74 | 37 | N | C5 |
| 4 | 01100 | Carrier | CAB80919 | 27 | TC150222 | 1 | 352 | 38,325 | 9.56 | 50 | N | 2 |
| 4 | 03115 | Putative UCP | NP680566 | 0 | – | 2 | 300 | 32,450 | 10.3 | 4 | P | 2 |
| 4 | 24570 | Putative UCP (DC*) | CAB79367 | 33 | TC161368 | 2 | 313 | 32,873 | 9.62 | 32 | N | 4 |
| 4 | 26180 | Carrier | CAB79473 | 2 | TC157290 | 1 | 325 | 36,552 | 9.63 | 12 | N | 2 |
| 4 | 27940 | Carrier | CAB79596 | 4 | TC154380 | 2 | 378 | 41,492 | 9.60 | 58 | N | 2 |
| 4 | 28390 | Putative ANT | CAB79641 | 5 | TC153190 | 2 | 379 | 40,718 | 9.78 | 65 | N | 5 |
| 4 | 32400 | Putative ANT | CAB79957 | 9 | TC163368 | 2 | 392 | 42,571 | 9.42 | 95 | N | 4 |
| 4 | 39460 | Carrier | CAB44681 | 15 | TC162444 | 3 | 330 | 35,663 | 9.64 | 37 | P | C4 |
| 5 | 01340 | Carrier (DC*) | AAK96620 | 6 | TC164377 | 3 | 309 | 33,991 | 9.93 | 3 | N | 4 |
| 5 | 01500 | Carrier | AAL67106 | 6 | TC153709 | 2 | 415 | 45,090 | 9.82 | 90 | N | C3 |
| 5 | 09470 | Carrier (DC*) | NP196509 | 0 | – | 2 | 337 | 36,096 | 9.71 | 48 | N | 3 |
| 5 | 13490 | ANT | AAL85138 | 17 | TC149366 | 2 | 385 | 41,746 | 9.79 | 50 | P | 5 |
| 5 | 14040 | PiC | AAL24236 | 100 | TC149514 | 1 | 375 | 40,089 | 9.29 | 67 | N | C1 |
| 5 | 15640 | Carrier | T51393 | 9 | TC151798 | 1 | 353 | 38,062 | 9.79 | 20 | P | 2 |
| 5 | 17400 | Putative ANT | T51577 | 2 | TC167129 | 2 | 327 | 36,010 | 9.73 | 10 | N | S5 |
| 5 | 19760 | OMT (DTC*) | NP197477 | 40 | TC149811 | 1 | 298 | 31,912 | 9.35 | 3 | N | 3 |
| 5 | 26200 | Carrier | AAL24263 | 1 | TC157414 | 1 | 342 | 37,028 | 9.95 | 25 | M | 2 |
| 5 | 42130 | Carrier | NP199028 | 3 | TC168364 | 1 | 412 | 44,361 | 9.37 | 102 | P | 5 |
| 5 | 46800 | Car/acylcar carrier | BAB08924 | 9 | TC152505 | 1 | 294 | 30,454 | 9.56 | 4 | N | 2 |
| 5 | 48970 | Carrier | NP199708 | 5 | TC167019 | 2 | 339 | 37,493 | 9.55 | 28 | P | 2 |
| 5 | 56450 | Putative ANT | NP200456 | 5 | TC154893 | 2 | 330 | 36,931 | 9.72 | 15 | N | 1 |
| 5 | 58970 | UCP2 | NP568894 | 3 | TC166916 | 3 | 295 | 32,165 | 8.97 | 3 | N | S2 |
| 5 | 64970 | Carrier | NP201302 | 3 | TC161592 | 1 | 428 | 46,818 | 8.69 | 22 | N | 5 |
The chromosome (Chr) and gene location number (Gene) are indicated along with the number of ESTs (ESTs TIGR) that compose the homologous TC sequence (TC TIGR) obtained from TIGR Arabidopsis Gene Indices. The annotation of each sequence is indicated along with suggested changes to annotation (*) based on our analysis and recent literature. car/acylcar, Carnitine/acylcarnitine carrier; carrier, unknown function carrier. Accession no. refers to a database entry for this protein at National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov). The no. of mitochondrial energy transfer signatures (METS) in each sequence, predicted physical properties of molecular mass (MM), pI, and the number of amino acids at the N terminus before the first predicted transmembrane sequence (Extension). PR, Predicted localization of sequence by Predotar; M, mitochondrial targeting; P, plastidic targeting; N, other targeting. TP, Predicted localization of sequence by TargetP; M1–5, mitochondrial; C1–5. chloroplast; S1–5, secretory pathway; 1–5, other (1, high probability; 5, low probability).
Similarity and Paralog/Ortholog Studies
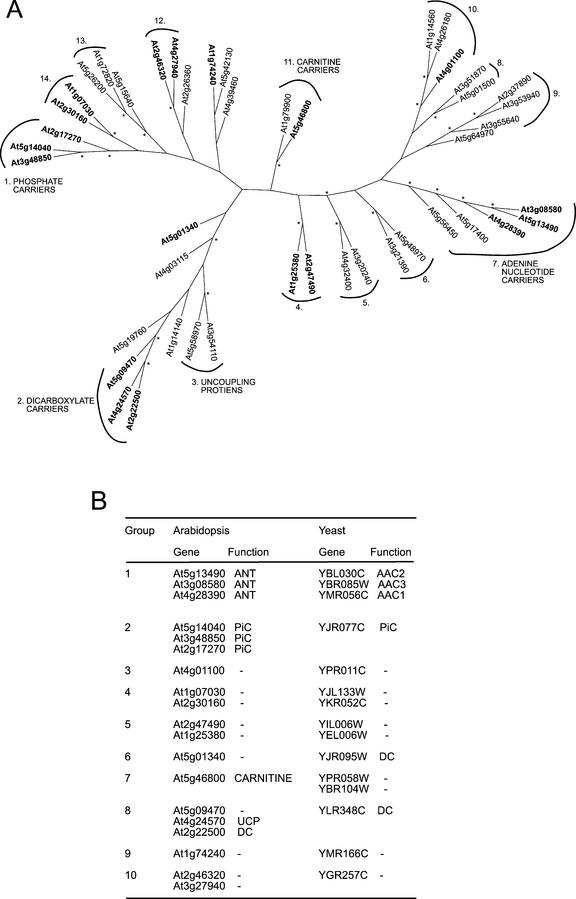
Annotation of the set A protein sequences reported by The Institute for Genomic Research (TIGR) and Munich Information Centre for Protein Sequences (MIPS) revealed a series of subfamilies predicted, on the basis of sequence similarity, to transport adenine nucleotides (6), inorganic phosphate (3), protons/fatty acids (4), carnitine/acylcarnitine (2), and dicarboxylates (2). One of the dicarboxylate carrier (DC) family members currently annotated as an OMT has recently been experimentally identified as a dicarboxylate/tricarboxylate carrier (DTC) of broad substrate specificity (Picault et al., 2002). A new annotation of this carrier as DTC has been added to Table I. A total of 28 genes of set A were annotated in TIGR and MIPS simply as carriers without any known or putative function. No link between physical chromosome location and annotated function was apparent (Table I). To further assess the relationships and the possible function of these proteins, an unrooted similarity tree of the set A sequences was constructed using ClustalX and PHYLIP (Fig. 1). To further extend similarity analysis, cross species orthologs and in-species paralogs were predicted between the Arabidopsis gene family (45 members) and the yeast carrier protein family (35 members) using INPARANOID (Remm et al., 2001). Orthologs are defined as genes in different species that originate from a single gene in the last common ancestor of both species, whereas paralogs are genes in a single species that originated from a single gene in an ancestral species.
Figure 1.
Similarity of mitochondrial carrier protein sequences. A, Un-rooted similarity tree generated using the neighbor-joining method and 1,000 bootstrap reiterations of 45 Arabidopsis carrier family sequences revealing grouping of annotated carrier functions and a range of carrier clusters of unknown function. *, Branchpoints with bootstrap values of greater than 70%. On the basis of this bootstrap cutoff, 14 groups are annotated. B, Ortholog/paralog analysis of carriers between yeast (35) and Arabidopsis (45) revealing 10 orthologous groupings. Arabidopsis genes with a yeast ortholog are bold in A. AAC, ADP/ATP carrier; carnitine, carnitine/acylcarnitine carrier; –, function unknown.
The similarity analysis revealed clustering of annotated subfamilies for ANT (7), PiC (1), carnitine/acylcarnitine (11), a group of two UCPs (3), and a group containing two DCs and a putative UCP (2). Although it is tempting to speculate on the apparent overlap between subgroups 2 and 3, they are annotated as distinct groups for historical reasons based on differences in putative function. A further nine subgroups (4, 5, 6, 8, 9, 10, 12, 13, and 14) were apparent for which no function is currently known (Fig. 1A). A total of eight genes from set A fell outside these distinct groups (At5g01340, At1g74240, At4g39460, At5g42130, At2g26360, At4g03115, At1g14140, and At5g64970). The INPARANOID analysis revealed 10 orthologous groups between Arabidopsis and yeast, containing approximately one-half the carrier genes in each species family (Fig. 1B). The three most related Arabidopsis ANTs were in a single grouping with ATP/ADP carriers from yeast (amino acid C1–3). The three Arabidopsis PiCs were grouped with the single-yeast PiC (YJR077C). Five orthologous groups were identified for which no function is known in either yeast or Arabidopsis. Interestingly, three Arabidopsis genes currently annotated as a putative UCP, a putative DC, and an unknown carrier, all grouped with the DC from yeast (YLR348C). As noted, these three genes also clustered on the similarity tree of Arabidopsis sequences (Fig. 1A). A new annotation of all three genes as putative DC has been added to Table I. The gene At5g01340, of unknown function, grouped with the yeast YJR095W succinate/fumarate carrier and is not closely associated with the above set of YLR348C-like proteins on the unrooted tree (Fig. 1A). The known DTC from Arabidopsis (Picault et al., 2002) does tentatively cluster with the YLR348C-like proteins. This suggests a broad family of putative dicarboxylate carrier proteins in Arabidopsis that may differentially function in tricarboxylic acid cycle intermediate transport both into mitochondria and out of mitochondria for the aneropleurotic functions known in plant mitochondria (Douce et al., 1997). Interestingly, known specific transporters for oxaloacetate, flavin, and tricarboxylates in yeast (Belenkiy et al., 2000) do not have clear orthologs in Arabidopsis.
Gene Family Expression Analysis
The number of ESTs representing each of the carrier proteins was calculated using EST numbers that compose tentative consensus (TC) sequences at the TIGR Arabidopsis Gene Index database. Numbers obtained from this database will differ over time because of their use of different EST libraries as sources and the frequency with which they update their information. Numbers of ESTs in nonredundant sets have been used in a variety of organisms as crude estimates of transcript abundance, and thus they provide a baseline for assessment of which genes in a set predominate in the mRNA pool (Rafalski et al., 1998; Andrews et al., 2000). Of the set A predicted products, 42 are presently represented by ESTs, and of this set, only nine are represented by more than 10 ESTs. This set of nine, now referred to as set B, contained two different ANTs, two UCPs, a PiC, a DC, the DTC, and two carriers of unknown function. From EST data alone it could be predicted that this subset of genes represents the majority of the mitochondrial carrier capacity. Interestingly one ANT (At3g08580) is represented by more than 300 ESTs, more than three times the number of the second most abundant member of set A (At5g14040).
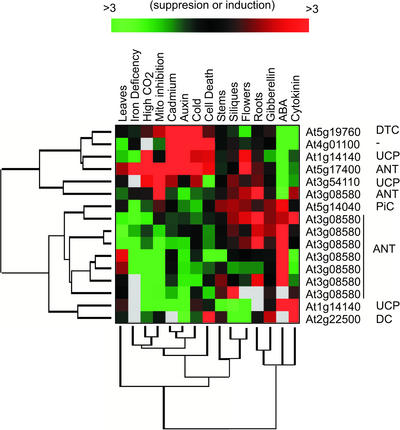
Assessment of the available data at the Stanford Microarray database revealed that only 16 of the set A carrier genes were represented on current collections of microarray slides. Of this 16, a total of eight were from our set B, those with highly abundant transcripts numbers from EST data (Table I). Only eight of the 16 carrier genes significantly changed in abundance during any of the microarray slides available and were consistently present on arrays for data analysis. This set of eight contained seven of the set B carriers along with At5g17400 for which only three ESTs were found (Table I). To assess the expression pattern of this set of eight, a series of 16 experiments based on specific treatments of wild-type plants were selected for further analysis (Fig. 2). This selection included treatments with plant hormones compared with control plants (auxin, gibberellin, cytokinin, and abscisic acid [ABA]), comparison of different plant organs compared against a reference mix of whole-plant mRNA (leaves, roots, flowers, and siliques) and a series of stress treatments compared with controls (iron deficiency, cadmium addition, cold treatment, induction of cell death, growth at high CO2, and antimycin A addition).
Figure 2.
Expression profile of Arabidopsis mitochondrial carrier family genes between tissue types and in response to hormonal signals and environmental stresses. Public information at the Stanford Microarray database was analyzed for data on expression of carrier genes in Arabidopsis. Intensities were reported as red (induced) or green (suppressed) compared with control samples. Averaged differences were subjected to log2 transformation, processed, and clustered by both genes and experiments using Cluster/Treeview (Eisen et al., 1998).
The carrier expression pattern clustered into two broad groups. Group 1 contained DTC, UCP, a carrier of unknown function, and the low abundance putative ANT (At5g17400). The expression of this group was increased in stress conditions, such as application of cadmium or auxin, exposure to cold, and induction of cell death. ABA application, on the other hand, decreased expression of this group. Expression was also down-regulated to some extent in leaves, but no trend was obvious in the other plant organs relative to whole-plant mRNA. Genes in group 2 contained PiC and the eight clones representing the abundant ANT (At3g08580). This group was decreased by the stress treatments that induced group 1, increased by ABA treatment and somewhat up-regulated in flowers, siliques, and roots compared with whole-plant mRNA. Two clones did not fit well into either grouping, DC At2g22500 and UCP At1g14140.
The co-expression of PiC and ANT in group 2 is interesting given the closely tied function of these proteins in phosphate transport and ATP/ADP transport, both functions essential for the synthesis and export of ATP for the mitochondrial matrix. The biochemical link between group 1 members remains more elusive, exacerbated by the unknown function of At4g01100 and the unclear role of the low abundance ANT. However, co-expression patterns may reflect similarity in promoter regions between carriers that need to be further investigated. Belenkiy et al. (2000) have already highlighted the value of transcript analysis for the elucidation of yeast carrier functions by correlating expression profiles with those of other known proteins to predict substrates likely to need transport under different circumstances.
Targeting Prediction of Gene Products
This family of carrier proteins are all presumed to be targeted to the mitochondrial inner membrane. In yeast, this is achieved without a cleavable presequence via a specialized import pathway (Truscott and Pfanner, 1999). Cleavable presequences do occur in some known mammalian and plant carrier proteins (e.g. ANT, PiC) but are clearly absent in others (e.g. UCP, OMT; Winning et al., 1992; Zara et al., 1992; Laloi, 1999; Truscott and Pfanner, 1999). The exact function of these cleavable regions in targeting is uncertain, but a role in import efficiency (Zara et al., 1992) or sorting between organelles (Mozo et al., 1995) has been proposed. In general, the presequences in these cases are 50 to 70 amino acids in length (Winning et al., 1992; Zara et al., 1992). Analysis of the hydrophobicity plots of set A predicted proteins, reveals that 14 have extended N-terminal regions greater than approximately 50 amino acid before the first transmembrane domain (Table I). These extensions could possibly function as targeting presequences. Similarity studies reveal At4g32400 is a homolog of the maize adenylate translocator, BRITTLE-1, which is targeted to amyloplasts not mitochondria in this monocot (Sullivan et al., 1991). The protein encoded by At3g55640 is a homolog of the known rabbit Ca2+-dependent carrier that is a member of the mitochondrial carrier superfamily but resides predominantly in the peroxisomal membrane and only at low levels in the mitochondrial membrane (Weber et al., 1997). Targeting prediction program based on neural networks to identify mitochondrial proteins in plants were used to further assess the likely intracellular location of these proteins in cells. Neither TargetP nor Predotar were able to offer complementary predictions of protein targeting for the set of carrier proteins. The divergent import pathway used by mitochondrial carriers and the possibility that N-terminal extensions have roles in import distinct from targeting may be the reason for this lack of predictability (Koehler et al., 1998; Truscott and Pfanner, 1999). Further, several N-terminal extensions on yeast carriers were recently postulated and/or shown to be non-cleaved functional additions involved in Ca2+ binding and nucleic acid metabolism (Belenkiy et al., 2000; Palmieri et al., 2001b), suggesting that not all such N-terminal extensions need be evidence of targeting or import functions at all.
Isolation of Mitochondrial Integral Membrane Proteins
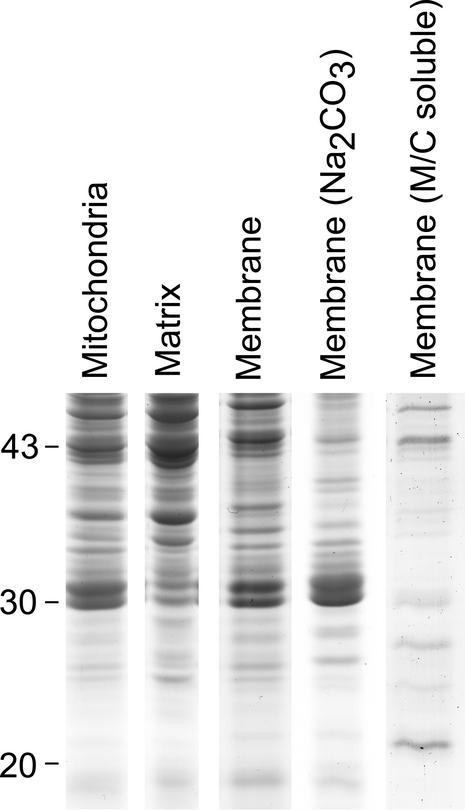
To clarify which carrier proteins are targeted and accumulate in plant mitochondria, and to possibly define an essential complement of carriers that might be required for mitochondrial operation in plants, we used a proteomic approach to determine which gene products are detectable in purified Arabidopsis mitochondria. Previous attempts to define the mitochondrial proteome in Arabidopsis by ourselves and others (Kruft et al., 2001; Millar et al., 2001) failed to identify any members of the carrier protein family. This was most likely attributable to the hydrophobicity of these carriers, which makes them intractable for isoelectric focusing (IEF)/SDS-PAGE-based separation. Here, we used a protocol developed for microsomes that used a 100 mm Na2CO3, pH 11.5, wash to strip peripheral proteins from the membrane, leaving a subset of integral membrane proteins (Fujiki et al., 1982a, 1982b). A complementary approach using chloroform-methanol solubility for selective purification of integral membrane proteins was recently published by Ferro et al. (2000). We decided to compare these two methods for the selective solubilization of plant mitochondrial carrier proteins.
Arabidopsis cell culture mitochondria were purified by two Percoll gradients as outlined by Millar et al. (2001). A membrane fraction was isolated by repeated freeze thawing and centrifugation. Integral membrane proteins were initially separated from the bulk peripheral membrane protein by Na2CO3 treatment. Comparison of whole mitochondria, soluble matrix, membrane, and integral membrane proteins by one-dimensional SDS-PAGE revealed that a series of 30- to 38-kD protein bands were selectively retained (Fig. 3). Membrane fractions were also treated with chloroform:methanol ratios of 8:1, 7:2, 2:1, 1:1, 1:2, 2:7, and 1:8 (v/v). Insoluble proteins were removed by centrifugation and soluble proteins dried from the organic solvents by a stream of N2 and then solubilized with SDS-based sample buffer. The majority of proteins were found in the insoluble fractions. There was little difference between the different ratios of organic solvents used (data not shown). Figure 3 (lane 5) shows the protein pattern from the 1:1 ratio of chloroform:methanol. There is notably little retention of the 30- to 38-kD proteins, but some selective solubilization of two proteins at 27 and 22 kD.
Figure 3.
Arabidopsis mitochondrial proteins (20–40 kD) following fractionation by membrane association and solubility. Mitochondrial samples were fractionated into water-soluble matrix proteins (matrix) and a membrane fraction (membrane). The membrane was then further fractionated by removal of peripheral proteins using Na2CO3 or solubilization in 1:1 methanol:chloroform (M/C soluble) to provide integral membrane protein fractions. All samples were separated by SDS-PAGE and stained with Coomassie G250.
MS
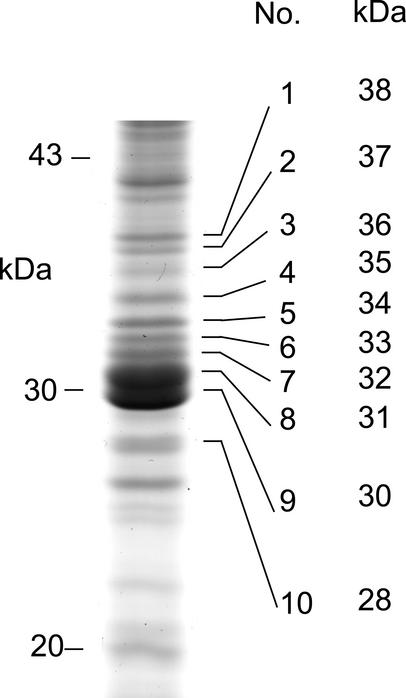
The 38- to 28-kD proteins bands present in the Na2CO3 stripped membrane fraction were excised in approximately 1-kD steps from the one-dimensional SDS-PAGE and labeled as bands 1 to 10, respectively (Fig. 4). Putative carriers greater than 38 kD were consequently excluded from this analysis. We expected each of these protein bands to contain a variety of mitochondrial proteins. Our previous experience with peptide mass fingerprinting MS in Arabidopsis (Millar et al., 2001) indicated that such data would be difficult to decipher. We chose an alternative MS/MS-based approach to identify individual tryptic peptides from each protein band and matched these data back to the Arabidopsis databases to develop a picture of the proteins present in each band. Spectra of trypsinated samples from each band were generated by electrospray ionization-TOF and a list of doubly charged peptide peaks selected from each TOF spectra. MS/MS data was then obtained from these peptides by collision induced dissociation of selected peptides using electrospray ionization-Q-TOF. A total of 189 peptides were characterized in this manner and used in searches of the Arabidopsis-predicted proteome using BLAST with the de novo sequencing data from MS/MS spectra at NCBI and direct searches using MS/MS spectra at Mascot (Table II).
Figure 4.
Ten protein bands (28–38 kD) from Na2CO3-treated mitochondrial membrane samples used for MS detection of carrier proteins. Mitochondrial proteins were separated by SDS-PAGE and stained with Coomassie G250. Visualized protein bands (1–10) were excised, destained, and in-gel digested with trypsin to completion, and peptides extracted and used for MS/MS analysis shown in Table II.
Table II.
Identification of carrier proteins from MS/MS data
| Spot No. | Gel MM | MS/MS Spectra | Match to At | Match Carrier | Annotation of Carrier | Chr Locus | MM | GRAVY |
|---|---|---|---|---|---|---|---|---|
| 1 | 38,000 | 15 | 9 | 4 | Putative carrier protein | At4g01100 | 38325 | 0.004 |
| 2 | 37,000 | 6 | 4 | 1 | Putative carrier protein | At4g01100 | 38325 | 0.004 |
| 3 | 36,000 | 8 | 1 | 0 | ||||
| 4 | 35,000 | 13 | 7 | 0 | ||||
| 5 | 34,000 | 20 | 16 | 7 | PiC | At5g14040 | 40089 | 0.145 |
| 6 | 33,000 | 18 | 10 | 0 | ||||
| 7 | 32,000 | 14 | 10 | 0 | ||||
| 8 | 31,000 | 37 | 22 | 8 | ANT | At3g08580 | 41475 | −0.121 |
| 3 | ANT | At5g13490 | 41746 | −0.111 | ||||
| 4 | DTC | At5g19760 | 31912 | 0.129 | ||||
| 9 | 30,000 | 45 | 35 | 5 | UCP | At3g54110 | 32662 | 0.176 |
| 10 | 28,000 | 13 | 4 | 0 |
A total of 189 MS/MS spectra derived from 10 protein bands representing 28- to 38-kD mitochondrial membrane proteins were matched to the Arabidopsis database using Mascot or derived amino acid sequence compared via near exact match BLAST at NCBI. Total spectra derived from each band (MS/MS spectra), the no. successfully matched to an Arabidopsis gene (Match to At), and the no. matched to carrier family proteins (Match carrier) are indicated. The predicted molecular mass (MM) and hydrophobicity (GRAVY) of each identified carrier are noted.
Identification of Arabidopsis Carrier Proteins
A total of nine peptides from spot 8 could be matched to two related ANT gene products (At3g08580 and At5g13490); of these, five were specific to At3g08580, one was specific to At5g13490, and two peptides were found in both sequences. This provided clear evidence that both of these gene products were present in mitochondria. Seven unique peptides matched to a PiC gene product (At5g14040), four to the DTC carrier (At5g19760), and five to a UCP (At3g54110). A total of five unique peptides that matched a putative carrier protein (At4g01100), were identified in spot 1 and 2 at 37 to 38 kD. The 27- and 22-kD proteins solubilized by chloroform-methanol in Figure 3 were also sequenced and found not to encode any carrier proteins (data not shown). Comparison of apparent molecular mass and the predicted molecular mass from the matched gene products revealed that UCP (At3g54110), DTC (At5g19760), and At4g01100 were all within 1 to 2 kD of predicted size of the precursor protein suggesting these proteins contain no or a very small cleaved targeting presequences (Table II). Only the UCP and DTC contain several amino acids before the first transmembrane domain. Interestingly, At4g01100 contains 50 amino acids at the N terminus but its apparent molecular mass clearly indicates that this is unlikely to have been cleaved on import (Table II). In contrast, both the ANTs and the PiC proteins found were 6 to 10 kD smaller than predicted. Both of these members of the carrier family have been reported to contain extended N-terminal presequences in other plant and mammalian species (Winning et al., 1992; Zara et al., 1992; Laloi, 1999). Most of the carrier proteins identified had positive grand average of hydropathicity (GRAVY) scores indicating their hydrophobicity (Table II). Very few proteins with positive GRAVY scores were identified in our previous study of the Arabidopsis mitochondrial proteome (Millar et al., 2001). This set of six identified carrier proteins located in the mitochondrial membrane, set C, were all members of set B derived from representation in EST databases and, of course, all members of set A derived from genome sequencing information.
During the sequencing of these carrier proteins, a variety of other noncarrier mitochondrial proteins were identified; these included prohibitins (At1g03860, At5g40770, and At3g27280), voltage-dependent anion carriers (At5g67500 and At3g01280), COXII (NP085487), COX6b-1 (At4g28060), AOX1a (At3g22370), NADH-UQ oxidoreductase (23 kD; At1g79010), translocase of the outer membrane TOM40 (At3g20000), malate dehydrogenase (At3g15020), γ-ATP synthase subunit (At2g33040), and FAD ATP synthase subunit (At2g21870). These mainly known membrane, hydrophobic components further confirm the carbonate extraction as a valuable purification step and also provide direct evidence of the purity of the mitochondrial membrane fractions used for the carrier analysis.
Link between Hydrophobicity and Identification
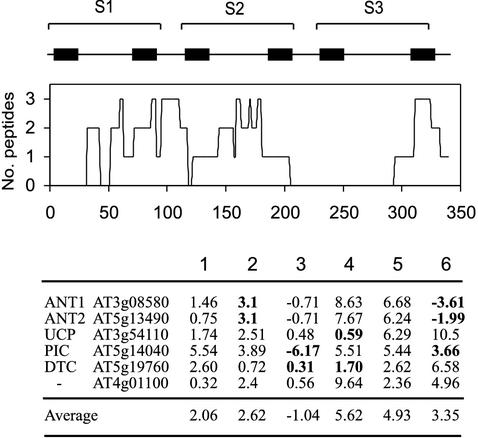
Hydrophobic proteins are rarely identified in proteomic analysis. This is potentially attributable to a number of factors including their insolubility in IEF experiments and the difficulty of extracting hydrophobic peptides from gel matrices and ionizing them by electrospray or laser desorption (Ferro et al., 2000; Molloy, 2000). Taking the carrier proteins as a model set of proteins with a very similar six-transmembrane domain structure, we investigated whether we could relate the positional information of peptide matching to the location and relative hydrophobicities of the transmembrane segments in set C proteins. These identifications were made from one-dimensional SDS-PAGE and therefore we could isolate the effect of gel matrices and MS ionization from IEF-related protocols. Figure 5 shows that peptide hits across the six transmembrane structures of the carrier proteins were not entirely random. There were no hits between amino acid positions 200 and 300, which correspond to the segments surrounding transmembrane regions 4 and 5. We used a new hydrophobicity indicator based on an experimentally determined whole-residue hydropathy scale (WW scale; Jayasinghe et al., 2001), including a constraint for the peptide backbone, to score the hydrophobicity of the six transmembrane domains of set C proteins across a 19-amino acid window. The greater the score, the greater the relative hydrophobicity. Where a peptide directly traversed this 19-amino acid window is shown in bold in Figure 5. Region 5 was consistently the most hydrophobic across the set C protein sequences, and no peptides were identified that traversed this region. Highly hydrophobic domains at position 4 were present in a number of the proteins. Two peptides were identified that traversed this domain, but these were from DTC and UCP where hydrophobicity of domain 4 was unusually low. The highest hydrophobicity of a transmembrane domain across which a peptide traversed was domain 6 of PiC. This degree of hydrophobicity may represent an approximate upper limit for extraction and ionization. Low hydrophobicity transmembrane helices in the carrier family of yeast have been proposed to form the transmembrane path in the transport of polar molecules (Belenkiy et al., 2000). The diversity of position and the number of such low hydrophobic transmembrane domains may be valuable in the future for predicting targets for mutagenesis to identify transport function and further classify functional groups within the carrier superfamily.
Figure 5.
Physical distribution of peptides matched by MS/MS on the six-transmembrane domain structure of identified carrier proteins. Number of peptides matching at each point of the 350 amino acid sequences of all six carrier proteins are graphed. The hydrophobicity of each transmembrane domain in each carrier protein was calculated using the augmented Wimley-White (aWW) scale at http://blanco.biomol.uci.edu/mpex. Where an identified peptide directly bridges the putative transmembrane domain, the domain hydrophobicity is shown in bold.
A Complex Gene Family with a Small Number of Abundant Members
Understanding the complexity from genome sequencing and then assessing the dominant, active members of gene families for directed study is a vital activity in post-genomics research. At first glance, the array of 45 mitochondrial carrier genes identified by Arabidopsis genome sequencing suggested a very complex set of functions, many of which were unknown based on annotation. However, further analysis of the expression of this gene family through EST databases, microarrays, and proteomic approaches, coupled with ortholog/paralog studies of similarity, indicates that a small subset of these genes predominate. Of set B, a collection of nine genes that were highlighted by EST analysis, seven were present in microarray databases and another six were clearly identified by MS (set C). The putative carrier At4g01100, of unknown function, is an immediate target for future work. We show here that this carrier, which has a yeast ortholog also of unknown function, is induced by stress conditions, suppressed by plant hormones, and is present in the inner mitochondrial membrane. It should be noted that this investigation was limited to carriers expressed under cell culture conditions and further members may be present in whole plants. The systematic analysis that reduced the diversity of set A to a manageable experimental set of abundant proteins provides the basis for future analysis of knock-out and knock-down Arabidopsis lines.
MATERIALS AND METHODS
Gene Identification, Sequence Alignments, Hydropathy Plots, and Paralog/Ortholog Analysis
Putative carrier proteins were initially identified by searches for Prosite Motif (PDOC00189) at the MIPS Annotated Arabidopsis Database (http://mips.gsf.de/). Unrooted similarity trees were made using ClustalX (Thompson et al., 1997) and PHYLIP program (Phylogeny Inference Package, v3.5c, Department of Genetics, University of Washington, Seattle; Felsenstein, 1989). Orthologs between yeast and Arabidopsis carrier sequences and in-paralogs within the sequences from each species were determined using the fully automated INPARANOID program that bypasses error-prone multisequence alignment and phylogenetic roots to ortholog assignment by using two-way best pair wise matches for clustering (Remm et al., 2001). Transmembrane domains were identified in carrier proteins using sliding window hydrophobicity analysis (19 amino acid) with the augmented Wimley-White scale at http://blanco.biomol.uci.edu/mpex; for consistent identification of the six transmembrane regions, CONH was set to −0.15. The hydrophobicity values presented in Figure 5 were obtained using CONH = −0.15.
Microarray and EST Data
Estimates of the number of ESTs representing each carrier gene were made using TC sequence coverage at TIGR Gene Index for Arabidopsis v9.0, release date September 19, 2002 (http://www.tigr.org/tdb/tgi.shtml). Carrier sequences were aligned to a TC using BLAST. The Stanford Microarray database was analyzed using the advanced search function to identify clone numbers for the carrier gene family and then selecting and reporting on spot intensities in Arabidopsis microarray comparisons based on tissue-specific expression, hormone treatments, and stress response (http://genome-www5.stanford.edu/cgi-bin/SMD/mad.pl). Intensities were reported as red (induced) or green (suppressed) compared with control samples. Averaged differences were subjected to log2 transformation, processed data was subjected to a self-organizing map algorithm followed by complete linkage hierarchical clustering of both genes and experiments using Pearson correlation (non-centered) through Cluster/Treeview (Eisen et al., 1998).
Targeting Programs
Subcellular targeting of predicted protein sequences were performed with TargetP (http://www.cbs.dtu.dk/services/TargetP/) as directed by (Emanuelsson et al., 2000) and by Predotar (http://www.inra.fr/Internet/Produits/Predotar/) as directed on this Web site.
Arabidopsis Mitochondria Protein: Isolation, Fractionation, and Electrophoresis
Mitochondria were isolated from dark-grown Arabidopsis cell suspension cultures according to Millar et al. (2001) using Percoll density gradient centrifugation. Subfractionation of mitochondria was done as follows. Mitochondria were resuspended to a final protein concentration of 2 mg mL−1 in 10 mm TES-KOH (pH 7.5), freeze-thawed in liquid N2 three times, and the suspension was centrifuged for 10 min at 18,000g, yielding a supernatant containing soluble proteins and a pellet containing membrane associated and integral membrane proteins. The pellet was resuspended in 100 mm NaCO3 (pH 11.5) and then centrifuged for 10 min at 18,000g, yielding a supernatant containing membrane associated proteins and a pellet containing integral membrane proteins. One-dimensional SDS-PAGE was performed according to standard protocols using 14% polyacrylamide gels and a Tris-Gly buffer system.
Quadrupole Time-of-Flight (Q-TOF) MS and Data Analysis
Q-TOF MS/MS was performed on a Q-TOF MS (Q-STAR Pulsar, Applied Biosystems, Foster City, CA) using an IonSpray source. Proteins to be analyzed were cut from the one-dimensional PAGE gel and digested with trypsin according to Millar et al. (2001), injected in 50% methanol, 0.1% formic acid, selected doubly charged peptides fragmented by N2 collision, and analyzed by MS/MS. Mass spectra and collision MS/MS data were analyzed with Analyst QS software (Applied Biosystems). Masses of collision-induced ion fragments were searched against the equivalent theoretical masses derived from the NCBI protein database using Mascot (http://www.matrixscience.com) or used for prediction of amino acid sequence with BioAnalyst and subjected to near exact match BLASTP analysis at http://www.ncbi.nlm.nih.gov/BLAST/.
ACKNOWLEDGMENTS
We acknowledge the critical reading and helpful comments on this manuscript by Prof. David Day and Dr. Jim Whelan.
Footnotes
This work was supported by the Australian Research Council Discovery Program (grants to A.H.M., D. Day, and J. Whelan). A.H.M. was supported by an Australian Research Council Queen Elizabeth II fellowship.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.009985.
LITERATURE CITED
- Andrews J, Bouffard G, Cheadle C, Lu J, Becker K, Oliver B. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogastertestis. Genome Res. 2000;10:2030–2043. doi: 10.1101/gr.10.12.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Belenkiy R, Haefele A, Eisen M, Wohlrab H. The yeast mitochondrial transport proteins: new sequences and consensus residues, lack of direct relation between consensus residues and transmembrane helices, expression patterns of the transport protein genes, and protein-protein interactions with other proteins. Biochim Biophys Acta. 2000;1467:207–218. doi: 10.1016/s0005-2736(00)00222-4. [DOI] [PubMed] [Google Scholar]
- Crabeel M, Soetens O, De Rijcke M, Pratiwi R, Pankiewicz R. The ARG11 gene of Saccharomyces cerevisiaeencoded a mitochondrial integral protein required for arginine biosynthesis. J Biol Chem. 1996;271:25011–25018. doi: 10.1074/jbc.271.40.25011. [DOI] [PubMed] [Google Scholar]
- Day D, Wiskich J. Transport processes of isolated plant mitochondria. Physiol Veg. 1984;22:241–261. [Google Scholar]
- Douce R, Aubert S, Neuburger M. Metabolite exchange between the mitochondrion and the cytosol. In: Dennis DT, Turpin DH, Lefebvre DD, Layzell DB, editors. Plant Metabolism. Essex, UK: Addison Wesley Longman; 1997. pp. 234–251. [Google Scholar]
- Eisen M, Spellman P, Brown P, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Ferro M, Seigneurin-Berny D, Rolland N, Chapel A, Slavi D, Garin J, Joyard J. Organic solvent extraction as a versatile procedure to identify hydrophobic chloroplast membrane proteins. Electrophoresis. 2000;21:3517–3526. doi: 10.1002/1522-2683(20001001)21:16<3517::AID-ELPS3517>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard A, Fowler S, Lazarow P. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982a;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard A, Fowler S, Lazarow P. Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J Cell Biol. 1982b;93:103–110. doi: 10.1083/jcb.93.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasinghe S, Hristova K, White S. Energetics, stability and prediction of transmembrane helices. J Mol Biol. 2001;312:927–934. doi: 10.1006/jmbi.2001.5008. [DOI] [PubMed] [Google Scholar]
- Koehler C, Jarosch E, Tokatlidis K, Schmidt K, Schweyen R, Schatz G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- Kruft V, Eubel H, Jansch L, Werhahn W, Braun HP. Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 2001;127:1694–1710. [PMC free article] [PubMed] [Google Scholar]
- Laloi M. Plant mitochondria carriers: an overview. Cell Mol Life Sci. 1999;56:918–944. doi: 10.1007/s000180050484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Dunlap J. Isolation and analysis of the arg-13 gene of Neurospora crassa. Genetics. 1996;143:1163–1174. doi: 10.1093/genetics/143.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignotte B, Vayssiere J. Mitochondria and apoptosis. Eur J Biochem. 1998;252:1–15. doi: 10.1046/j.1432-1327.1998.2520001.x. [DOI] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giege P, Leaver CJ. Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 2001;127:1711–1727. [PMC free article] [PubMed] [Google Scholar]
- Molloy M. Two-dimensional electrophoresis of membrane proteins using immobilised pH gradients. Anal Biochem. 2000;280:1–10. doi: 10.1006/abio.2000.4514. [DOI] [PubMed] [Google Scholar]
- Mozo T, Fischer K, Flugge U, Schmitz U. The N-terminal extension of the ADP/ATP translocator is not involved in targeting to plant mitochondria in vivo. Plant J. 1995;7:1015–1020. doi: 10.1046/j.1365-313x.1995.07061015.x. [DOI] [PubMed] [Google Scholar]
- Palmieri F, Bisaccia F, Capobianco L, Dolce V, Fiermonte G, Iacobazzi V, Indiveri C, Palmieri L. Mitochondrial metabolite transporters. Biochim Biophys Acta. 1996;1275:127–132. doi: 10.1016/0005-2728(96)00062-x. [DOI] [PubMed] [Google Scholar]
- Palmieri L, Agrimi G, Runswick MJ, Fearnley IM, Palmieri F, Walker JE. Identification in Saccharomyces cerevisiaeof two isoforms of a novel mitochondrial transporter for 2-oxoadipate and 2-oxoglutarate. J Biol Chem. 2001a;276:1916–1922. doi: 10.1074/jbc.M004332200. [DOI] [PubMed] [Google Scholar]
- Palmieri L, Pardo B, Lasorsa FM, del Arco A, Kobayashi K, Iijima M, Runswick MJ, Walker JE, Saheki T, Satrustegui J et al. Citrin and aralar1 are Ca(2+)-stimulated aspartate/glutamate transporters in mitochondria. EMBO J. 2001b;20:5060–5069. doi: 10.1093/emboj/20.18.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Rottensteiner H, Girzalsky W, Scarcia P, Palmieri F, Erdmann R. Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J. 2001c;20:5049–5059. doi: 10.1093/emboj/20.18.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picault N, Palmieri L, Pisano I, Hodges M, Palmieri F. Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria. J Biol Chem. 2002;277:24204–24211. doi: 10.1074/jbc.M202702200. [DOI] [PubMed] [Google Scholar]
- Prohl C, Pelzer W, Diekert K, Kmita H, Bedekovics T, Kispal G, Lill R. The yeast mitochondrial carrier Leu5p and its human homologue Graves' disease protein are required for accumulation of coenzyme A in the matrix. Mol Cell Biol. 2001;21:1089–1097. doi: 10.1128/MCB.21.4.1089-1097.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski J, Hanafey M, Miao G, Ching A, Lee J, Dolan M, Tingey S. New experimental and computational approaches to the analysis of gene expression. Acta Biochim Pol. 1998;45:929–934. [PubMed] [Google Scholar]
- Remm M, Storm C, Sonnhammer E. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Saraste M, Walker JE. Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett. 1982;144:250–254. doi: 10.1016/0014-5793(82)80648-0. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Strelow L, Illingworth C, Phillips R, Nelson O. Analysis of maize brittle-1 alleles and a defective suppressor-mutator-induced mutable allele. Plant Cell. 1991;3:1337–1348. doi: 10.1105/tpc.3.12.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott K, Pfanner N. Import of carrier proteins into mitochondria. Biol Chem. 1999;380:1151–1156. doi: 10.1515/BC.1999.146. [DOI] [PubMed] [Google Scholar]
- Walker JE, Runswick M. The mitochondrial transport protein superfamily. J Bioenerg Biomembr. 1993;25:435–446. doi: 10.1007/BF01108401. [DOI] [PubMed] [Google Scholar]
- Weber F, Minestrini G, Dyer J, Werder M, Boffelli D, Compassi S, Wehrli E, Thomas R, Schulthess G, Hauser H. Molecular cloning of a peroxidomal Ca2+dependent member of the mitochondrial carrier superfamily. Proc Natl Acad Sci USA. 1997;94:8509–8514. doi: 10.1073/pnas.94.16.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winning B, Sarah C, Purdue P, Day C, Leaver CJ. The adenine nucleotide translocator of higher plants is synthesized as a large precursor that is processed upon import into mitochondria. Plant J. 1992;2:763–773. [PubMed] [Google Scholar]
- Wiskich J. Mitochondrial metabolite transport. Annu Rev Plant Physiol. 1977;28:45–69. [Google Scholar]
- Zara V, Palmieri F, Mahlke K, Pfanner N. The cleavable presequence is not essential for import and assembly of the phosphate carrier of mammalian mitochondria but enhances the specificity and efficiency of import. J Biol Chem. 1992;267:12077–12081. [PubMed] [Google Scholar]