Abstract
Tobacco (Nicotiana tabacum) genes regulated during the early stage of responses to wounding were screened by a modified fluorescence differential display method. Among 28 genes initially identified, a particular clone designated NtC7 was subjected to further analysis. Its transcripts were found to accumulate rapidly and transiently within 1 h upon treatments with not only wounding but also salt and osmotic stresses. However, jasmonic and abscisic acids and ethylene did not effectively induce NtC7 transcripts. Amino acid sequence analysis suggested NtC7 to be a new type of transmembrane protein that belongs to the receptor-like protein family, and a membrane location was confirmed in onion (Allium cepa) epidermis cells transiently expressing an NtC7-green fluorescent protein fusion protein. Seeds of transgenic tobacco overexpressing NtC7 normally germinated and grew in the presence of 500 mm mannitol, but not in the presence of 220 mm sodium chloride or 60 mm lithium chloride. Cuttings of mature transgenic leaf exhibited a marked tolerance upon treatment with 500 mm mannitol for 12 h, at which concentration wild-type counterparts were seriously damaged. These results suggested that NtC7 predominantly functions in maintenance of osmotic adjustment independently of ion homeostasis.
Plants are continuously exposed to biotic and abiotic stresses that endanger their survival. Among abiotic stresses, water stress is one of the most severe, caused by drought, salt loading, and chilling. To cope with these stresses, plants have developed various systems such as production of osmolites for osmotic adjustment, synthesis of Na+/H+ antiporters for ion sequestration, and many others (Bohnert et al., 1995). The operation of these systems usually requires three steps: osmotic stress recognition, signal transduction, and production of components for the physiological response. Knowledge on the first and second steps in plants remains relatively limited, and is mostly available from experiments with bacteria and yeast (Saccharomyces cerevisiae).
The first step is mainly mediated through the osmosensor, which recognizes change in osmotic pressure. In Escherichia coli and yeast, osmotic stress is detected by the osmosensors EnvZ and SLN1, respectively (Maeda et al., 1994; Mizuno, 1998). A similar protein, AtHK1, has been found in Arabidopsis (Urao et al., 1999), although its function in planta awaits determination. All have been identified as transmembrane two-component His kinases. In yeast, another type of sensor, SHO1, has also been detected, which is a transmembrane protein equipped with an SH3 domain (Maeda et al., 1995). Some of these sensor proteins form homodimers, the conformation easily changing upon mechanical stimuli to the membrane (Yaku and Mizuno, 1997; Tao et al., 2002). Such conformational alteration is considered to relay the signal into the cell interior (Posas et al., 1996; Lohrmann and Harter, 2002).
The second step identified so far is the mitogen-activated protein (MAP) kinase phosphorylation cascade (Wurgler-Murphy and Saito, 1997). In yeast, osmotic signals perceived by the two osmosensors, SLN1 and SHO1, are transduced to an MAP kinase (HOG1) through MAP kinase kinase (PBS2; Reiser et al., 2000). HOG1 ultimately activates the synthesis of glycerol to serve as the compatible solute (Albertyn et al., 1994). Whether or not a similar phosphorylation cascade functions in the osmosignaling pathway in plants is currently not clear. In contrast, the third step has been relatively well studied in plants, and a number of genes have been identified and characterized as osmotic stress regulated (Bohnert et al., 1995). The late embryogenesis abundant proteins are examples, being known to respond to and reduce the effects of osmotic and cold stresses (Thomashow, 1998). Many other genes encoding proteins involved in osmolite biosynthesis, transporters, and regulatory functions have also been isolated (Zhu et al., 1997).
In the present study, we initially screened genes involved in very early stage responses to wounding, and identified a particular gene encoding a membrane-located receptor-like protein, NtC7. Here, we report that NtC7 plays important roles in the early response to osmotic stress in tobacco (Nicotiana tabacum) plants.
RESULTS
Identification of NtC7
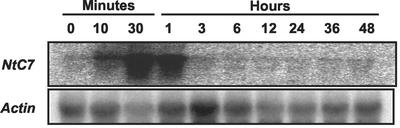
Screening for wound-responsive genes by fluorescence differential display (FDD), we initially identified 28 cDNA fragments that were found to change their levels within 3 h after wound treatment (data not shown; Hara et al., 2000). Among them, a particular clone whose transcripts were rapidly induced after wounding was subjected to a preliminary northern analysis. Total RNA samples were isolated from leaf discs 0, 15, 45, 90, and 180 min after wound treatment, and hybridization was performed using a 337-bp fragment amplified with PCR as the probe. The transcripts of this clone were found to begin to accumulate as early as within 10 min, reaching a maximum level at 1 h, and then to decline to the initial level after 3 h (Fig. 1). Because of such an early transient response, the clone, designated as NtC7 (tobacco C7), was further characterized in the present study.
Figure 1.
Accumulation of NtC7 transcripts upon wounding. Healthy leaves were detached and wounded by cutting into pieces with a pair of scissors and floated on water. The wounded leaves were harvested at the indicated time points. Blots containing 35 μg of RNA per lane were successively subjected to hybridization with the NtC7 fragment obtained by FDD and actin cDNA.
Sequence Properties and Genomic Organization
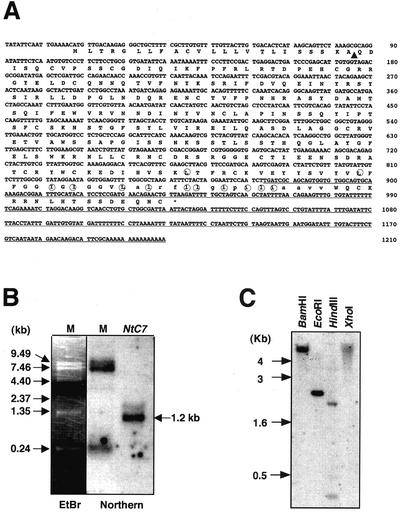
A 1,210-bp cDNA of NtC7 was isolated from a cDNA library constructed from mRNAs isolated from wound-stressed leaves (accession no. AB087235; Fig. 2A). Northern hybridization using this fragment as the probe showed the size of the corresponding transcript to be approximately 1.2 kb, indicating that the cDNA obtained was nearly full length (Fig. 2B). Southern hybridization analysis indicated NtC7 to hybridize to a discrete single fragment after digestion of genomic DNA with various restriction enzymes (Fig. 2C). Because tobacco used in this assay is amphidiploid, the results suggest that a single copy of NtC7 originated from one of the ancestral parents, either Nicotiana sylvestris or Nicotiana tomentosiformis.
Figure 2.
Analyses of the NtC7. A, The nucleic acids are presented on the top line and the derived one-letter amino acid sequence is shown below. The stop codon is indicated by an asterisk. The original NtC7 clone obtained by FDD screening is underlined. The position of cleaving site of the predicted signal peptide is indicated by a black triangle. The predicted transmembrane region is indicted by small letters. Leu and Ile near the transmembrane domain are marked with circles. The accession number of the cDNA is AB087235 under the name of NtC7. B, Northern-blot hybridization. Total RNA was probed with the full-length NtC7 cDNA, and transcript size was estimated from the migration position of marker RNAs (M) stained with ethidium bromide (EtBr) and signals because of cross hybridization. C, Southern-blot analysis. Genomic DNA (20 μg per lane) was digested with the indicated restriction endonuclease, separated on an agarose gel, blotted, and probed with a radioactively labeled NtC7 fragment.
Characterization of the NtC7 Protein
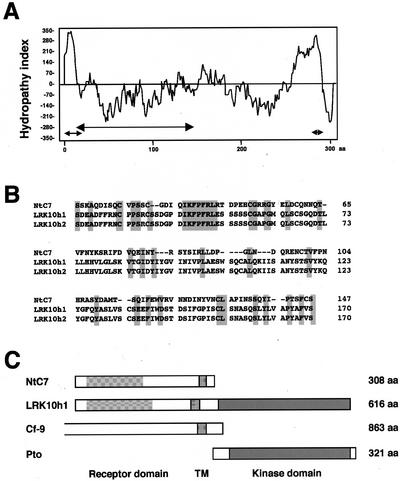
The protein encoded by NtC7 cDNA was predicted to consist of 308 amino acid residues with a relative molecular mass of about 33.9 kD (Fig. 2A). A hydropathy plot (Kyte and Doolittle, 1982) indicated the NtC7 polypeptide to possess hydrophobic regions at both N-terminal (amino acids 1 to 23) and C-terminal (amino acids 275 to 291) ends (Fig. 3A). Analysis using PSORT, a computer program for the prediction of protein localization sites in cells (Nakai and Horton, 1999), indicated that the N-terminal region is likely to serve as a signal peptide, cleaved at amino acid positions between 23 and 24 (A/Q; Fig. 2A). The C-terminal region was predicted to function as a transmembrane domain (Fig. 2A). A BLAST search (Altschul et al., 1990) showed the NtC7 protein to have similarities with rice LRK10 homologs (Feuillet and Keller, 1999) in the N-terminal region (amino acids 20 to 147; Fig. 3, A and B). LRK10 is a product of leaf rust disease resistance genes originally found in wheat (Feuillet et al., 1997). A distinct feature of NtC7, however, is the lack of a kinase domain, which is present in LRK10 homologs (Fig. 3, B and C). In this context, NtC7 rather structurally resembled tomato Cf-9, a receptor for avr-9 of C. fulvum (Jones et al., 1994; Fig. 3C).
Figure 3.
Properties of NtC7. A, Hydropathy plot of the NtC7 polypeptide. Hydropathy analysis was performed using a window of nine amino acids. A putative signal peptide, a region homologous with LRK10, and a putative transmembrane domain are indicated with arrows. B, Alignment of NtC7 with LRK10 homolog 1 (accession no. AAC27489; LRK10h1) and LRK10 homolog 2 (accession no. AAC02535; LRK10h2) from rice (Oryza sativa) was performed with the ClustalW program. Identical residues shared among the three are shaded. C, Pattern diagrams of plant receptor-like proteins. The molecular size of each is shown in numbers of amino acids (aa) on the right side. LRK10h1 is a rice homolog of wheat (Triticum aestivum) LRK10 (accession no. T06793), Cf-9 is a tomato (Lycopersicon esculentum) receptor for avr-9 of Cladosporium fulvum (accession no. AAA65235). For reference, Pto, a protein kinase proposed to interact with Cf-like proteins, is also shown (accession no. A49332). The homologous region between NtC7 and LRK10 homolog 1 (LRK10h1) in the receptor domain, a putative transmembrane domain (TM), and a kinase domain are indicated by hatched, shaded, and black boxes, respectively.
Cellular Localization
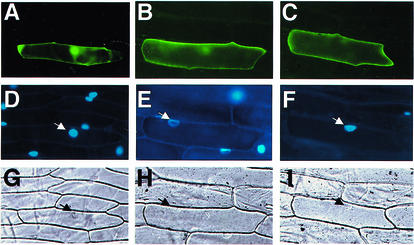
To identify the cellular localization, a reporter gene encoding GFP was fused to NtC7, and subjected to transient assay using onion (Allium cepa) epidermis cells (Fig. 4). After biolistic bombardment, individual cells were observed for localization of NtC7 by GFP fluorescence (Fig. 4, A–C), using DAPI staining for nuclei (Fig. 4, D–F), and interference contrast images for whole-cell structures (Fig. 4, G–I). Cauliflower mosaic virus (CaMV) 35S::GFP control construct [psGFP(S65T)] showed GFP signals in both the cytoplasm and the nucleus (Fig. 4A). The CaMV 35S::NtC7-GFP (pNtC7-GFP) showed GFP signals predominantly at the membrane (Fig. 4B). This pattern was identical with that of a positive control, CaMV 35S::inflorescence meristem receptor-like kinase 3 (IMK3)-GFP (pIMK3-GFP), a plasmid containing a cDNA for IMK3 of Arabidopsis (Takemura et al., 2000), showing GFP signal at the membrane (Fig. 4C). The results suggested NtC7 to be a membrane-located protein.
Figure 4.
Membrane localization of NtC7 in onion epidermal cells. Onion bulbs were bombarded with gold particles coated with psGFP(S65T) (A, D, and G), pNtC7::green fluorescent protein (GFP; B, E, and H), or pIRM3::GFP plasmids (C, F, and I). The proteins were transiently expressed and individual cells were observed by epifluorescence for GFP (A–C), by staining with 4′,6 diamidino-2-phenylindole (DAPI; D–F) or under interference contrast (G–I). Nuclei identified with DAPI staining and in interference contrast images are indicated by arrows (D–I).
Expression Analysis
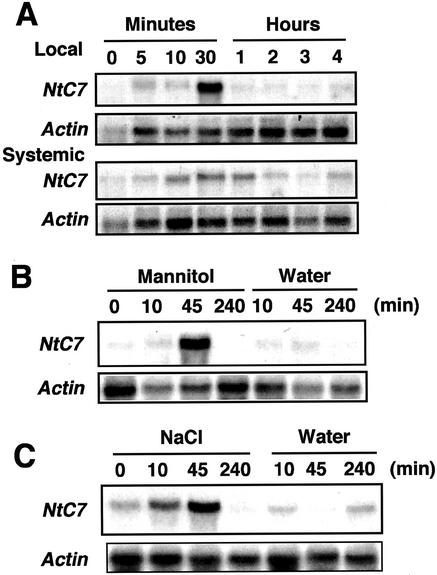
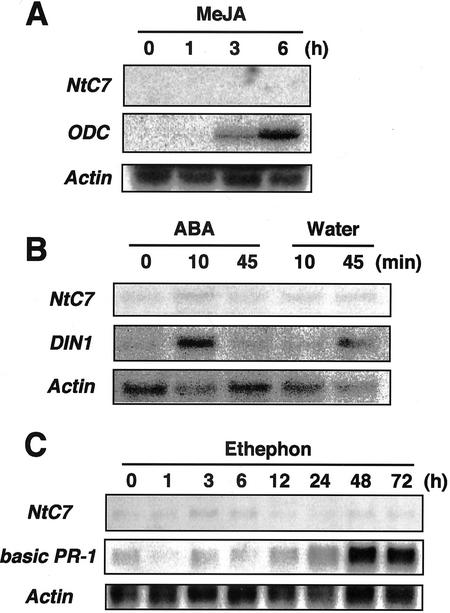
Transcript accumulation of NtC7 was analyzed in leaves subjected to abiotic stress conditions (Fig. 5). Because NtC7 was primarily identified from leaf discs floated on water, the stress was possibly from wounding, osmotic changes, or a combination of the two. To distinguish these, a healthy leaf of an intact plant was cut with a pair of scissors, and RNA was extracted from both wounded and adjacent unwounded leaves. Northern hybridization indicated simple injury to be sufficiently effective to locally and systemically induce NtC7 transcripts (Fig. 5A). Osmotic stress was achieved by 500 mm mannitol treatment to leaves that were detached and left to absorb water for 4 h before further processing to depress initial wound effects. Transcripts temporarily accumulated by 45 min after osmotic shock, but were diminished 4 h later (Fig. 5B). In control leaves, which were kept in water for the same period, transcript accumulation was not induced (Fig. 5B). Treatment with 200 mm NaCl also induced NtC7 transcripts showing a similar accumulation pattern as that for osmotic stress (Fig. 5C). Because wound and osmotic signals are often transmitted through jasmonic acid (JA), abscisic acid (ABA), and ethylene, respectively, detached leaves were also treated with these chemicals and transcript induction was estimated (Fig. 6). To confirm treatment efficacy, samples were hybridized with cDNAs for ODC (Orn decarboxylase) that responds to JA, DIN1 that responds to ABA (Hara et al., 2000), and basic PR-1 that responds to ethephon (Hiraga et al., 2000). Although all chemicals correctly induced the marker transcripts, neither of them induced NtC7 transcript accumulation (Fig. 6). These observations indicate NtC7 to respond to both wounding and osmotic stresses independently of JA, ABA, and ethylene.
Figure 5.
Effects of osmotic and salt stresses on NtC7 transcript accumulation. Healthy leaves were wounded by cutting with a pair of scissors, and wounded (local) and unwounded adjacent (systemic) leaves were sampled at the indicated time points, and RNAs assayed by northern hybridization with the indicated probes (A). Healthy leaves were detached and put in buffer solution. At 4 h after the first wounding, leaves were transferred to a buffer solution with or without 500 mm mannitol (B) or 200 mm NaCl (C) for the indicated time period, and RNAs were assayed by northern hybridization with the indicated probes.
Figure 6.
Effect of phytohormones on NtC7 transcript accumulation. A leaf cutting was prepared from a healthy plant, left for 4 h for acclimatization to the initial wound stress, then exposed to 50 μm methyl ester JA (A), 100 μm ABA (B), or 100 μm ethephon (C) for the indicated time period. Total RNAs were then subjected to northern assay. Control samples for ABA treatment were treated with water (Water; B).
Physiological Assay of Transgenic Seedlings
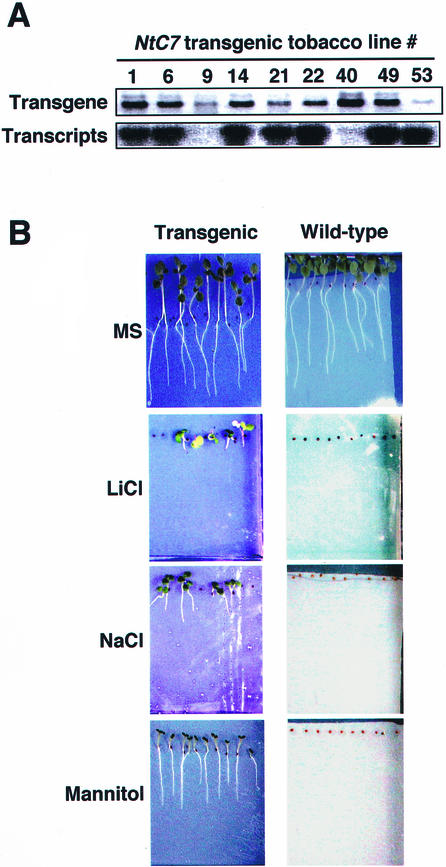
To examine the physiological function of NtC7, transgenic tobacco plants constitutively expressing NtC7 were constructed. More than 10 transgenic lines were produced, and after confirmation of integration and expression of NtC7 by PCR and northern analyses (Fig. 7A), five lines were selected for further examination. Seeds of the T3 generation of line 1 were sown on agar plates containing one-half-strength Murashige and Skoog medium supplemented with or without appropriate concentrations of mannitol, NaCl, or LiCl, and germination and growth were examined. The transgenic seedlings showed clear resistance to osmotic stress caused by mannitol at as high as 500 mm, at which concentration the growth of control wild-type seedlings was completely suppressed (Fig. 7B). In contrast, their growth was totally retarded in the presence of 220 mm sodium chloride, suggesting the plants to be susceptible to Na+ ions (Fig. 7B). This was confirmed by their sensitivity to 60 mm lithium chloride, at which concentration osmotic status of cells is not seriously affected, the toxicity of Li+ being even higher than that of Na+ ions (Fig. 7B). The same results were obtained with other transgenic lines (6, 14, 21, and 53) showing resistance to mannitol and susceptibility to salt ions (data not shown).
Figure 7.
Effects of osmotic and salt stresses on transgenic tobacco seedlings expressing NtC7. A, Integration and expression of NtC7 in transgenic lines were examined by PCR (upper; transgenes) and northern hybridization (lower; transcripts). B, Germination and growth test. A batch of 10 seeds of transgenic line 1 or wild-type plants were sown on the one-half-strength Murashige and Skoog-agar medium containing 60 mm LiCl, 220 mm NaCl, or 500 mm mannitol along a nylon mesh, cultivated for 2 weeks, and photographed.
Physiological Assay of Transgenic Mature Leaves
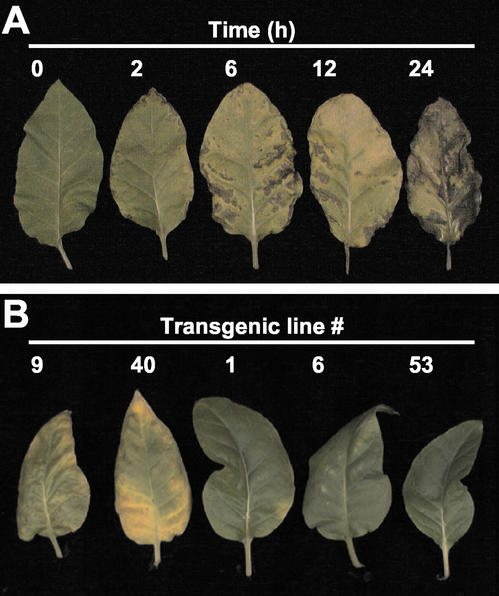
To determine whether or not mature plants also exhibited the tolerance, stress conditions were first determined. Healthy leaves of wild-type plants were cut out at the petiole, and treated with 500 mm mannitol solution by absorption for an appropriate time period. Leaves were then transferred to water and allowed to recover from wilting (Fig. 8A). Results showed that by as short as 2 h of treatment, leaves were already unable to recover from wilting, showing necrotic spots on the surface (Fig. 8A). Upon treatment for 12 h, leaf exhibited severe necrosis all over the surface, and ultimately died after 2 d. Based on these observations, transgenic leaves were assayed for recovery from wilting after 12 h of treatment (Fig. 8B). The same symptom as the control wild-type plants was observed with transgenic lines 9 and 40, which did not express the transgene (Fig. 7A). These leaves also died after 2 d. In contrast, transgenic lines 1, 6, and 53, actively expressing NtC7, rapidly recovered, showing apparently the same feature as untreated samples (Fig. 8B). These results were consistent with those of seedlings (Fig. 7B), and strongly suggested that NtC7 played an important role in tobacco response to osmotic stress.
Figure 8.
Effects of osmotic stress on tobacco mature leaves. A, Time course analysis of recovery from wilting in wild-type plants. A young, healthy leaf from approximately 2 month-old plants was cut out, put into a vessel containing 500 mm mannitol, and left at room temperature for indicated time period. B, Tolerance of transgenic lines against osmotic stress. A young, healthy leaf from each of approximately 2-month-old transgenic plants with indicated numbers was cut out, put into a vessel containing 500 mm mannitol, and left for 12 h at room temperature. Lines 9 and 40 contained the transgene without expression, and lines 1, 6, and 53 actively expressed the NtC7 (see Fig. 7A). After treatments, each sample was transferred to a vessel containing water to recover for 48 h and photographed.
DISCUSSION
This paper describes isolation and properties of a gene encoding a receptor-like membrane protein that functions in response to osmotic stress. The predicted NtC7 protein has a hydrophobic signal sequence at the N terminus (amino acid positions 1 to 23), a helix transmembrane region (amino acid positions 275 to 291), and a hydrophilic region at the C terminus (amino acid positions 292 to 308; Figs. 2A and 3A). A homology search with the predicted amino acid sequence indicated that NtC7 resembles the receptor domain of receptor-like kinases (RLKs). Plant RLKs are grouped into four types depending on amino acid sequences. Type 1 constitutes the so-called Leu-rich repeat proteins having 24 amino acid repeat units containing many Leu. Type 2 proteins have homology to the S locus glycoprotein. Type 3 proteins have lectin-like domains that are thought to bind oligosaccharides. The type 4 group demonstrates homology to epidermal growth factor repeated sequences (Hardie, 1999). The amino acid sequence of the NtC7 protein at the proximal N terminus showed highest similarity to the proximal N terminus of the receptor domain in rice homologs of wheat LRK10 (leaf rust disease resistant kinase), belonging to the type 2 group (Feuillet et al., 1997; Fig. 3, B and C). In contrast, the distal N terminus of the predicted NtC7 protein did not show any similarity to these proteins. Considering these structural properties, we conclude that NtC7 belongs to a new subfamily of RLKs. Because the NtC7-GFP fusion protein was shown to localize in the membrane fraction, it is highly probable that it is a membrane-associated receptor-like protein with the C terminus oriented to the cytoplasm.
To identify the physiological role of NtC7, transgenic tobacco plants were constructed and analyzed for their stress responses. Mature leaves of transgenic plants showed a tolerance to osmotic stress, as clearly seen by rapid recovery from severe wilting caused by 500 mm mannitol, at which concentration control leaves suffered serious damages such as necrosis. Transgenic seedlings were also highly tolerant to the same stress. A notable finding, however, was that they were susceptible to salt stress, showing a similar sensitive response as the wild-type control. The simplest explanation for this is that transgenic plants produced some compatible solutes, which confer tolerance to osmotic stress, but not to sodium ion toxicity. Because major compatible solutes in tobacco plants are reported to be derivatives of sugars and amino acids like Pro (Yoshiba et al., 1997), it is conceivable that overexpressed NtC7 activates the production of such compounds. Judging from its structure, however, it is unlikely that NtC7 directly participates in their synthesis. Instead, it may be involved in the signaling pathway to activate osmotic stress responsive genes, functioning, for example, as part of the osmosensor system.
The best studied osmosensors are two-component His kinases, identified in E. coli, budding yeast, and Arabidopsis (Cai and Inouye, 2002; Hwang et al., 2002; Li et al., 2002). They are suggested to form homodimers, whose conformation is sensitive to changes in membrane architecture (Wurgler-Murphy and Saito, 1997; Yaku and Mizuno, 1997). Structurally, however, NtC7 distinctly differs from any known osmosensors, but resembles RLKs. Although it is unclear whether RLKs form dimers, Arabidopsis RLK5 has been proposed to form a homodimer through its Leu-rich regions and to interact with a kinase-associated protein phosphatase (Braun and Walker, 1996). By analogy, it may be possible that NtC7 forms dimers through the Leu-rich region near the C terminus (Fig. 2A). Another specific feature of NtC7 is the lack of a kinase domain, thus structurally resembling tomato Cf-9 (Fig. 3C), a transmembrane protein that confers resistance to tomato leaf mold and is considered to transmit the pathogen signal to the cytoplasmic protein through its cytoplasmic tail. One such cytoplasmic protein was proposed to be a protein kinase, typically represented by Pto (Hammond-Kosack and Jones, 1997; Fig. 3C), a cytoplasmic Ser/Thr kinase considered to play a critical role in several pathogen-signaling pathways (Braun and Walker, 1996; Hammond-Kosack and Jones, 1997). Proteins like Cf-9 and Pto have been repeatedly suggested to interact with each other in an analogous way with counterparts in the mammalian immune system (Braun and Walker, 1996; Hammond-Kosack and Jones, 1997). Taking account of structural similarities, it is conceivable that NtC7 interacts with partner protein(s) through its C-terminal tail region, thereby transmitting osmosignals to cytoplasmic components.
Transcripts of NtC7 spatially and transiently accumulated upon osmotic stress. Because a low level of transcripts was constitutively observed here even in the absence of stress, such a rapid induction may indicate that a relatively large amount of NtC7 is only needed upon stress. A similar pattern of transcript induction is observed for genes encoding; for example, the two-component signaling component Arabidopsis response regulator for cytokinine response (Kiba et al., 1999), RLKs for pathogen recognition (Du and Chen, 2000), MAP kinase (WIPK) in the wound response (Yap et al., 2002), and WRKY transcription factor (TIZZ) in the hypersensitive response (Yoda et al., 2002). All are involved in cellular signaling pathways, supporting the idea that NtC7 also functions in osmosignal transduction. Although more detailed analyses of the protein level are necessary, the temporal expression of these genes suggests that one of the mechanisms for activation and desensitization is strict control at the transcriptional level. Perhaps plants respond to environmental cues by rapid production and degradation of relevant proteins only as necessary, thereby best coping with severe biotic and abiotic stresses.
The present findings further suggest that one wound signal could be associated with osmotic change. To date, many signal molecules that induce transcription of so-called wound-responsive genes have been identified, including JA, ABA, ethylene, small peptides, oligosaccharides, and reactive oxygen (Kessler and Baldwin, 2002). In addition, physical signals such as hydraulic pressure, electric currents (Leon et al., 2001), and pH change (Hara et al., 2000) have also been suggested to play a role. The induction profile of NtC7 transcripts, featuring rapid accumulation in both local and systemic leaves and independent of JA, ABA, and ethylene supports the idea that the hydraulic status is one of the factors underlying wound signaling.
Salt and drought tolerance is one of the most important traits for crops because world arable lands are continuously being injured from salt accumulation and desiccation. Transgenic technology has been expected to be helpful to solve such problems. Because salt induces both ion toxicity and osmotic stress, introduction of multiple genes that cope with these stresses would be practical. In this context, our NtC7 may be useful if utilized in the combination with genes involved in salt ion homeostasis, such as HKT that encodes a Na+/K+ symporter (Maser et al., 2002).
MATERIALS AND METHODS
Plant Materials and Wound and Chemical Treatments
Tobacco (Nicotiana tabacum cv Xanthi nc) plants were grown in soil in a growth cabinet at 23°C under a 14-h-light/10-h-dark photocycle. Wound stress was applied by cutting mature leaves with a pair of scissors. Wounded (local) and adjacent upper unwounded (systemic) leaves were harvested at appropriate time points. After being put in water for 4 h to diminish cutting stress, samples were transferred into a solution containing one of the following chemicals: 200 mm NaCl, 60 mm LiCl, 500 mm mannitol, or 100 μm ABA. For treatment with volatile chemicals, samples were exposed to 50 μm methyl ester JA or 100 μm ethephon (ethylene) in a sealed box.
FDD
The FDD and comigration tests were essentially performed as described earlier (Hara et al., 2000). In brief, total RNAs were isolated from treated or untreated sample leaves, digested with DNase I, and cDNAs were synthesized and subjected to PCR using rhodamine-labeled 3′-anchored primers (Takara, Kyoto) and 12-mer arbitrary primers. The reactions were carried out with 25 cycles of 94°C for 30 s, 40°C for 1 min, 72°C for 1 min, and 72°C for 5 min for a final extension. After PCR amplification, samples were fractionated by 5% (w/v) denaturing PAGE, and migration patterns were analyzed with an image analyzer (FM-BIO, Hitachi, Tokyo). cDNAs differentially amplified were eluted, reamplified by PCR with the same pair of primers as used for the first amplification, and subcloned into the pT7blue vector (Novagen, Madison, WI).
cDNA Library Construction and Screening
Total RNA was isolated from wound-treated leaves of tobacco by the acid guanidium thiocyanate-phenol-chloroform method (Chomczynski and Sacchi, 1987) with a slight modification, and used for cDNA library construction with the λZapII vector (Stratagene, La Jolla, CA). In brief, cDNAs were ligated to the vector at EcoRI and XhoI sites. After transformation, the library was screened with a 32P-labeled NtC7 fragment obtained by FDD, and positive clones were rescued in the pBluescript SK-phagemid vector by in vivo excision. After amplification in Escherichia coli JM109, nucleotide sequences were determined by the dideoxynucleotide chain termination method (PRISM BigDye Terminator, ABI, Sunnyvale, CA). Sequence editing, prediction of amino acid sequences, hydropathy plots and multiple alignment, and similarity and localization searches were carried out with appropriate computer software (GeneWorks, National Center for Biotechnology Information, and PSORT server).
DNA and RNA Gel-Blot Analysis
Genomic DNA was isolated from green leaves by the cetyl-trimethyl-ammonium bromide method (Murray and Thompson, 1980) with a slight modification, and 20-μg aliquots were digested with one of the restriction enzymes (BamHI, EcoRI, HnidIII, or XhoI), separated by electrophoresis on 0.6% (w/v) agarose gels, and transferred onto nylon membranes (Hybond N+, Amersham, Buckinghamshire, UK). After cross linking using a UV cross linker (RPN 2501, Amersham), the membranes were subjected to hybridization with appropriate 32P-labeled probes at 65°C for 16 h. After successive washing with 0.1× SSC, 0.1% (w/v) SDS at 65°C, they were used to expose either BAS (Fuji Photo Film, Tokyo) or x-ray film (Eastman-Kodak, Rochester, NY). Total RNAs were isolated by the aurintricarboxylic acid method (Gonzalez et al., 1980) with a slight modification, and used for RNA gel-blot analysis. A 20-μg aliquot per lane was fractionated by formaldehyde/agarose gel electrophoresis and transferred to a nylon membrane (Hybond N+, Amersham). Hybridization was performed in the same way as for the DNA hybridization described above, except the hybridizing temperature was 42°C.
Plasmid Construction and Histochemical Analysis
The coding region of NtC7, with the stop codon deleted, was amplified by PCR with ExTaq DNA polymerase (Takara) using forward (5′-GTCGACATGTTGACAAGAGGGCTGC-3′) and reverse (5′-CCATGGAACAGTTCTGTTCATCGGAGG-3′) primers containing an SalI site upstream and a NcoI site downstream of the deleted stop codon, respectively. This PCR fragment was first introduced into the pGEM-Teasy (Promega, Madison, WI) vector for amplification in the E. coli strain JM109. The plasmid was digested with SalI and NcoI, and the resulting fragment was introduced into the SalI/NcoI site of the psGFP(S65T) vector by fusing the coding region in-frame to that of GFP. Particle bombardment was performed according to the manufacturer's instructions (PDS-1000, Bio-Rad Laboratories, Hercules, CA), with 1-μm diameter gold particles coated with plasmids. A 3-cm square onion (Allium cepa) scaly leaf fragment was placed under the stopping screen at a distance of 6 or 9 cm and bombarded twice per sample in a vacuum of 28 inches of mercury using a helium pressure of 1,100 psi to accelerate the macrocarrier. Bombarded leaves were kept in the dark for 12 h at 25°C before analysis. DAPI staining and GFP epifluorescence assay were performed essentially as described (Nishiyama et al., 2002).
Plant Transformation
For plant transformation, PCR-amplified NtC7 using forward (5′-GCTCTAGAGAACATGTTGACAAGAGGGC-3′) and reverse (5′-GAGCTCTTAACAGTTCTGTTCATCGG-3′) primers containing XbaI and SacI sites at the 5′ and 3′ ends, respectively, was introduced into the pGEM-Teasy vector as described above. Digested fragments were ligated to XbaI/SacI sites of pBI121 vector (CLONTECH Laboratories, Palo Alto, CA), and introduced into the Agrobacterium tumefaciens strain LBA4404 cells. Tobacco transformation was performed as described previously (Yap et al., 2002).
Bioassay
Transgenic NtC7 plants were grown to maturity to yield progeny seeds. For estimation of stress tolerance, approximately 10 T3 seeds were sown on a one-half-strength Murashige and Skoog agar plate containing appropriate concentrations of salt or mannitol and cultured under continuous light at 23°C. After appropriate time periods, germinated seedlings were counted and measured for growth. Healthy, unwounded leaves from wild-type and T0 transgenic plants were cut out with a sharp razor blade at petioles, and put in a vessel containing 500 mm mannitol. After standing for appropriate time intervals at room temperature under continuous light, samples were transferred to water, allowed to recover from wilting for additional 48 h, and photographed.
ACKNOWLEDGMENTS
We thank Drs. Yasuo Niwa (University of Shizuoka, Japan) and Miho Tekemura (Nara Institute of Science and Technology, Japan) for the generous gifts of psGFP(S65T) and pIMK3-GFP plasmids, respectively. We are also grateful to Drs. Hideki Nakayama and Kazuya Yoshida and Ms. Yuko Tatsumi (Nara Institute of Science and Technology) for valuable advice and suggestion and technical assistance, respectivley, and to Dr. Malcolm Moore (Intermal, Nagoya, Japan) for critical reading of the manuscript.
Footnotes
This work was supported by the Japan Society for the Promotion of Science (Research for the Future Program grant no. JSPS–RFTF 00L01604 and Research Fellowship for Young Scientist no. 06586 to T.T.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.011007.
LITERATURE CITED
- Albertyn J, Hohmann S, Thevelein JM, Prior BA. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bohnert HJ, Nelson DE, Jensen RG. Adaptations to environmental stresses. Plant Cell. 1995;7:1099–1111. doi: 10.1105/tpc.7.7.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DM, Walker JC. Plant transmembrane receptors: new pieces in the signaling puzzle. Trends Biochem Sci. 1996;21:70–73. [PubMed] [Google Scholar]
- Cai SJ, Inouye M. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J Biol Chem. 2002;277:24155–24161. doi: 10.1074/jbc.M110715200. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Du L, Chen Z. Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salycylic acid-induced WRKY DNA-binding proteins in Arabidopsis. Plant J. 2000;24:837–847. doi: 10.1046/j.1365-313x.2000.00923.x. [DOI] [PubMed] [Google Scholar]
- Feuillet C, Keller B. High gene density is conserved at syntenic loci of small and large grass genomes. Proc Natl Acad Sci USA. 1999;96:8265–8270. doi: 10.1073/pnas.96.14.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet C, Schachermayr G, Keller B. Molecular cloning of a new receptor-like kinase gene encoded at the Lr10disease resistance locus of wheat. Plant J. 1997;11:45–52. doi: 10.1046/j.1365-313x.1997.11010045.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez RG, Haxo RS, Schleich T. Mechanism of action of polymeric aurintricarboxylic acid, a potent inhibitor of protein-nucleic acid interactions. Biochemistry. 1980;19:4299–4303. doi: 10.1021/bi00559a023. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JDG. Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- Hara K, Yagi M, Koizumi N, Kusano T, Sano H. Screening of wound-responsive genes identifies an immediate-early expressed gene encoding a highly charged protein in mechanically wounded tobacco plants. Plant Cell Physiol. 2000;41:684–691. doi: 10.1093/pcp/41.6.684. [DOI] [PubMed] [Google Scholar]
- Hardie DG. Plant protein serine/threonine kinases: classification and functions. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:97–131. doi: 10.1146/annurev.arplant.50.1.97. [DOI] [PubMed] [Google Scholar]
- Hiraga S, Ito H, Sasaki K, Yamakawa H, Mitsuhara I, Toshima H, Matsui H, Honma M, Ohashi Y. Wound-induced expression of a tobacco peroxidase is not enhanced by ethephon and suppressed by methyl jasmonate and coronatine. Plant Cell Physiol. 2000;41:165–170. doi: 10.1093/pcp/41.2.165. [DOI] [PubMed] [Google Scholar]
- Hwang I, Chen HC, Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002;129:500–515. doi: 10.1104/pp.005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JDG. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fuluvumby transposon tagging. Science. 1994;266:789–793. doi: 10.1126/science.7973631. [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT. Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol. 2002;53:299–328. doi: 10.1146/annurev.arplant.53.100301.135207. [DOI] [PubMed] [Google Scholar]
- Kiba T, Taniguchi M, Imamura A, Ueguchi C, Mizuno T, Sugiyama T. Differential expression of genes for response regulators in response to cytokinins and nitrate in Arabidopsis thaliana. Plant Cell Physiol. 1999;40:767–771. doi: 10.1093/oxfordjournals.pcp.a029604. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Leon J, Rojo E, Sanchez-Serrano JJ. Wound signalling in plants. J Exp Bot. 2001;52:1–9. doi: 10.1093/jexbot/52.354.1. [DOI] [PubMed] [Google Scholar]
- Li S, Dean S, Li Z, Horecka J, Deschenes RJ, Fassler JS. The eukaryotic two-component histidine kinase Sln1p regulates OCH1via the transcription factor, Skn7p. Mol Biol Cell. 2002;13:412–424. doi: 10.1091/mbc.01-09-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann J, Harter K. Plant two-component signaling systems and the role of response regulators. Plant Physiol. 2002;128:363–369. doi: 10.1104/pp.010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Mäser P, Hosoo H, Goshima S, Horie T, Eckelman B, Yamada K, Yoshida K, Bakker EP, Shinmyo A, Oiki S et al. Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc Natl Acad Sci USA. 2002;99:6428–6433. doi: 10.1073/pnas.082123799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T. His-Asp phosphotransfer signal transduction. J Biochem. 1998;123:555–563. doi: 10.1093/oxfordjournals.jbchem.a021972. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nishiyama R, Ito M, Yamaguchi Y, Koizumi N, Sano H. A chloroplast-resident DNA methyltransferase is responsible for hypermethylation of chloroplast genes in Chlamydomonasmaternal gametes. Pro Natl Acad Sci USA. 2002;99:5925–5930. doi: 10.1073/pnas.082120199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Reiser V, Salah SM, Ammerer G. Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat Cell Biol. 2000;2:620–627. doi: 10.1038/35023568. [DOI] [PubMed] [Google Scholar]
- Takemura M, Tani E, Takeda Y, Hasegawa M, Nemoto K, Yokota A, Kohchi T. Functional analysis of the receptor-like kinase gene, IMK3 expressed in Arabidopsisshoot and root meristems. Plant Cell Physiol. 2000;41:s69. [Google Scholar]
- Tao W, Malone CL, Ault AD, Deschenes RJ, Fassler JS. A cytoplasmic coiled-coil domain is required for histidine kinase activity of the yeast osmosensor, SLN1. Mol Microbiol. 2002;43:459–473. doi: 10.1046/j.1365-2958.2002.02757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 1998;118:1–7. doi: 10.1104/pp.118.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11:1743–1754. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, Saito H. Two-component signal transducers and MAPK cascades. Trends Biochem Sci. 1997;22:172–176. doi: 10.1016/s0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- Yaku H, Mizuno T. The membrane-located osmosensory kinase, EnvZ, that contains a leucine zipper-like motif functions as a dimer in Escherichia coli. FEBS Lett. 1997;417:409–413. doi: 10.1016/s0014-5793(97)01329-x. [DOI] [PubMed] [Google Scholar]
- Yap YK, Kakamu K, Yamaguchi Y, Koizumi N, Sano H. Promoter analysis of WIPK, a gene encoding a tobacco MAP kinase, with reference to wounding and tobacco mosaic virus infection. J Plant Physiol. 2002;159:77–83. [Google Scholar]
- Yoda H, Ogawa M, Yamaguchi Y, Koizumi N, Kusano T, Sano H. Identification of early-responsive genes associated with the hypersensitive response to tobacco mosaic virus and characterization of a WRKY-type transcription factor in tobacco plants. Mol Genet Genomics. 2002;267:154–161. doi: 10.1007/s00438-002-0651-z. [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol. 1997;38:1095–1102. doi: 10.1093/oxfordjournals.pcp.a029093. [DOI] [PubMed] [Google Scholar]
- Zhu JK, Hasegawa PM, Bressan RA. Molecular aspects of osmotic stress in plants. Crit Rev Plant Sci. 1997;16:253–277. [Google Scholar]