Abstract
Citrus tristeza virus (CTV) is the major virus pathogen causing significant economic damage to citrus worldwide, and a single dominant gene, Ctv, provides broad spectrum resistance to CTV in Poncirus trifoliata L. Raf. Ctv was physically mapped to a 282-kb region using a P. trifoliata bacterial artificial chromosome library. This region was completely sequenced to about 8× coverage using a shotgun sequencing strategy and primer walking for gap closure. Sequence analysis predicts 22 putative genes, two mutator-like transposons and eight retrotransposons. This sequence analysis also revealed some interesting features of this region of the P. trifoliata genome: a disease resistance gene cluster with seven members and eight retrotransposons clustered in a 125-kb gene-poor region. Comparative sequence analysis suggests that six genes in the Ctv region have significant sequence similarity with their orthologs in bacterial artificial chromosome clones F7H2 and F21T11 from Arabidopsis chromosome I. However, the analysis of gene colinearity between P. trifoliata and Arabidopsis indicates that Arabidopsis genome sequence information may be of limited use for positional gene cloning in P. trifoliata and citrus. Analysis of candidate genes for Ctv is also discussed.
Citrus is one of the most important fruit crops worldwide and citrus tristeza virus (CTV) is a major virus pathogen of citrus. Most citrus species and varieties are susceptible to CTV infection. However, Poncirus trifoliata, a close relative of citrus, is resistant to CTV. The resistance has been characterized and is controlled by a single dominant gene called Ctv (Gmitter et al., 1996). The genetic map around Ctv has been developed (Gmitter et al., 1996; Deng et al., 1997; Fang et al., 1998) and applied to map-based cloning of Ctv, an approach facilitated by the small size (382 Mb) of the citrus genome (Arumuganathan and Earle, 1991). A bacterial artificial chromosome (BAC) library with 9.6× genomic coverage was constructed from an individual P. trifoliata plant that was homozygous for Ctv. A contig of approximately 1.2 Mb was established after seven successful steps of chromosome walking from flanking markers. The Ctv gene was further delimited to a region of about 300 kb using DNA fragments from the 1.2-Mb contig as markers. This region is covered by four overlapping BAC clones 27A14, 20J24, 83D17, and 84F5 (Yang et al., 2001). Map-based cloning of the Ctv gene is also being undertaken elsewhere (Deng et al., 2001a, 2001b). In this case, a BAC library was constructed with 7× genomic coverage from an intergeneric citrus and P. trifoliata hybrid and two contigs were developed through chromosome walking. The contig encompassing the Ctv region was approximately 550 kb, and the other contig spanning the allelic susceptibility gene region was approximately 450 kb. The Ctv locus was further mapped to a region of 180 kb using DNA fragments from the 550-kb contig as markers (Deng et al., 2001a).
Plant disease resistance (R) genes have been identified in many plants (Hammond-Kosack and Jones, 1997), and they frequently occur in tightly linked clusters (Michelmore and Meyers, 1998). In citrus, Deng et al. (2000) identified 10 classes of citrus R gene candidate (RGC) sequences similar to the nucleic acid binding sequence (NBS)-Leu-rich repeat (LRR) class of R genes by PCR amplification from degenerate primers to the NBS domain. These PCR products were cloned, and pools of six and seven clones were hybridized to a BamHI based BAC library. Analysis of the BAC clones isolated gave an estimate of 80 to 140 unique NBS-containing sequences in the library (Deng et al., 2001b). In our previous work, we cloned three DNA fragments from BAC clones in the 1.2-Mb contig using a PCR approach. The hybridization of these DNA fragments with a HindIII-fingerprinting blot of the BAC clones indicated that there might be two disease R gene clusters in the 1.2-Mb contig. One cluster of disease R genes contains domains of a NBS and LRRs, and they are distributed in the 282-kb region where the Ctv gene is also located, whereas the second cluster is located about 175 kb away surrounding marker C19 (Yang et al., 2001). Because genes within a single cluster can determine resistance to different pathogens, the complete sequence of this region will lead to not only the cloning of the Ctv gene, but also presumably the cloning of other potential R genes.
Arabidopsis is the first flowering plant from which the genome has been completely sequenced (Arabidopsis Genome Initiative, 2001), and draft genome sequences of two rice (Oryza sativa) subspecies have been reported (Goff et al., 2002; Yu et al., 2002). The information generated from Arabidopsis and rice genes can be used for the study of other plant genomes through comparative genetics. Comparative mapping based on cross-hybridizing markers has demonstrated that gene content and order are highly conserved between different species within the grass family (Devos and Gale, 1997). The region between two markers on a genetic map usually comprises many genes. Microsynteny analysis investigates local gene repertoire, order, and orientation. Arabidopsis and closely related species Capsella rubella (Acarkan et al., 2000) and cauliflower (Brassica oleracea var alboglabra; O'Neill and Bancroft, 2000) and distantly related species such as tomato (Lycopersicon esculentum; Ku et al., 2000) are estimated to have diverged approximately 6.2 to 9.8, 12.2 to 19.2, and 112 to 156 million years ago, respectively. Microsynteny between Arabidopsis and these plants has been investigated (Acarkan et al., 2000; Ku et al., 2000; O'Neill and Bancroft, 2000; Mao et al., 2001; Rossberg et al., 2001). The genus Poncirus is a member of the Rutaceae, which is estimated to have diverged from Arabidopsis about 60 to 80 million years ago (Chase et al., 1993). Investigation of synteny between P. trifoliata and Arabidopsis will expand knowledge of microsynteny between Arabidopsis and other dicots.
In this paper, we present a complete sequence of about 282 kb that must contain Ctv. This is the first report of a large sequence contig in a tree species. The sequence analysis includes gene predictions, description of disease R genes and transposable elements, and an investigation of synteny between Arabidopsis and P. trifoliata. Analysis of candidate genes for Ctv is also discussed.
RESULTS
Sequence of BAC Clones in the Ctv Region
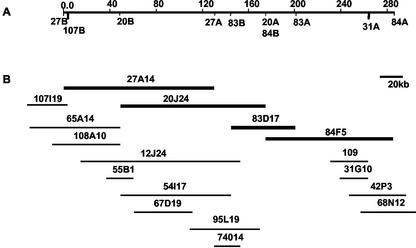
Our previous data indicated that Ctv mapped to a region between markers 31A and 107B (Yang et al., 2001). A set of overlapping BAC clones (27A14, 20J24, 83D17, and 84F5; Fig. 1) covering this region was chosen for shotgun sequencing. Ends of additional BAC clones in this region were sequenced and used as anchors for sequence assembly. A total of 3,455 reads were produced. These trimmed data gave 7.8× coverage of this region. Assembly of sequences from BAC clones 27A14 and 84F5 was completed by a combination of targeted cloning and PCR. For 27A14, after 850 sequence reads were assembled, inserts from subclones located in contig ends were isolated and hybridized to libraries to identify 81 additional clones for sequencing. An additional 46 clones were identified using five PCR products that spanned gaps as probes. One PCR product was sequenced by primer walking. For 84F5, after 750 sequence reads were assembled, inserts from subclone ends were used to identify 404 additional clones for sequencing. The five gaps in this sequence were filled by sequencing PCR products. For clones 20J24 and 83D17, assembly of shotgun sequences left seven gaps. Four gaps were filled by identification of long subclones, which were digested with restriction enzymes, subcloned, and sequenced. The other three gaps were filled by PCR. The complete sequence of the four BAC clones is 282,699 bp and has been deposited into GenBank under the accession no. AF506028. The sequences of BAC clones 27A14, 20J24, 83D17, and 84F5 correspond to nucleotide positions 1 to 130,352, 49,595 to 175,112, 145,563 to 201,202, and 175,107 to 282,699, respectively. The Ctv gene is located between markers 31A and 107B, which correspond to nucleotide positions 3,791 to 259,974 (Fig. 1).
Figure 1.
The contig around the Ctv locus based on the complete sequence. A, The physical map of the region between markers 107B and 31A that includes Ctv. B, The contig around the Ctv locus. The name of each BAC clone is shown above each line. The four BAC clones (27A14, 20J24, 83D17, and 84F5) chosen for shotgun sequencing are indicated in bold.
Gene Content of the Ctv Region
Genes in the Ctv locus were predicted by GenScan and further adjusted with the results of GeneMark. hmm, Glimmer A, BLAST searches, and sequence alignments. GenScan and GeneMark.hmm predicted four R genes (R2–R5) and one R gene (R5), respectively. The other three R genes were identified based on sequence alignments and BLAST searches. CTV.20 was predicted to contain three open reading frames (ORFs) by GenScan, but BLAST searches indicated that both the first and the third ORFs were highly homologous with petunia vein-clearing virus ORF1. Northern hybridization analyses using DNA fragments from the first and the third ORFs hybridized with the same band of about 9 kb (data not shown), the same size transcript as predicted by combining the ORFs predicted by GenScan. Thus, the three separate genes predicted by GenScan were combined to form CTV.20.
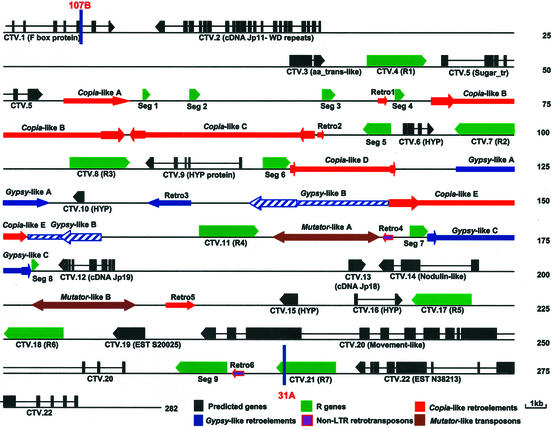
A total of 22 genes were predicted in this 282,699 bp region (Table I; Fig. 2). Three predicted genes were confirmed by isolation of corresponding cDNA clones and northern hybridization (CTV.2, CTV.12, and CTV.13), and three additional genes were confirmed by northern hybridization (CTV.3, CTV.14, and CTV.20). Of the 22 predicted genes, seven (R1-R7) are CC-NBS-LRR-type disease R genes similar to a putative Arabidopsis disease R gene At5g63020 and related genes. Six genes have significant similarity with other plant genes of known function. CTV.1 located at the beginning of this region (Fig. 2) contains a partial coding region. The predicted products of these six genes are similar to an Arabidopsis F-box protein that contains multiple LRRs (CTV.1), an Arabidopsis protein At1g15740 that contains a WD 40 repeat domain (CTV.2), a transmembrane amino acid transporter protein (CTV.3), a Glc transporter protein (CTV.5), a nodulin protein (CTV.14), and a plant virus movement-like protein (CTV.20; Table I). Five of the predicted genes are similar to unknown protein genes (CTV.9, CTV.12, and CTV.13) or ESTs (CTV.19, and CTV.22). The remaining four genes (CTV.6, CTV.10, CTV.15, and CTV.16) are hypothetical genes that have no significant sequence similarity with any other genes in the database or have sequence similarity to other hypothetical genes (Table I). CTV.9 and CTV.12 show considerable sequence similarity in coding regions, but their introns are quite different.
Table I.
Gene prediction in the Ctv region
| ID | Designation | Strand | Region | Best Protein Homolog | E Value | Predicted Gene Product |
|---|---|---|---|---|---|---|
| CTV.1 | F-box protein | + | 306–5,092 | At1g15740 | 1e-57 | F box protein with LRRs |
| CTV.2 | cDNA Jp11 | − | 7,554–15,436 | At1g15750 | 0 | Similar to Arabidopsis protein with WD40 repeats |
| CTV.3 | Aa_trans like | + | 39,061–40,533 | At1g80510 | e-165 | Transmembrane amino acid transporter protein |
| CTV.4 | R1 | + | 42,823–45,495 | At5g63020 | e-172 | CC-NBS-LRR disease resistance gene |
| CTV.5 | Sugar_tr | + | 46,474–51,670 | At1g11260 | e-46 | Monosaccharide transport protein |
| CTV.6 | HYP protein | + | 94,464–95,727 | – | No hits | Hypothetical protein |
| CTV.7 | R2 | − | 97,412–100,082 | At5g63020 | e-141 | CC-NBS-LRR disease resistance gene |
| CTV.8 | R3 | + | 103,146–105,845 | At5g63020 | e-159 | CC-NBS-LRR disease resistance gene |
| CTV.9 | HYP protein | − | 107,061–111,553 | AAF26974.1 | 3e-12 | Similar to Arabidopsis unknown protein |
| CTV.10 | HYP protein | − | 128,556–128,822 | – | No hits | Hypothetical protein |
| CTV.11 | R4 | + | 159,473–162,151 | At5g63020 | e-160 | CC-NBS-LRR disease resistance gene |
| CTV.12 | cDNA Jp19 | − | 177,791–180,306 | AAF26974.1 | 2e-48 | Similar to Arabidopsis unknown protein |
| CTV.13 | cDNA Jp18 | + | 191,847–192,452 | At1g15760 | 7e-47 | Similar to Arabidopsis unknown protein |
| CTV.14 | Nodulin-like | − | 193,527–198,154 | At1g80530 | e-173 | Nodulin-like protein |
| CTV.15 | HYP protein | − | 213,663–214,283 | At5g11090 | 1e-17 | Hypothetical protein |
| CTV.16 | HYP protein | + | 217,212–219,319 | – | No hits | Hypothetical protein |
| CTV.17 | R5 | − | 220,152–222,824 | At5g63020 | e-173 | CC-NBS-LRR disease resistance gene |
| CTV.18 | R6 | − | 225,244–227,929 | At5g63020 | e-154 | CC-NBS-LRR disease resistance gene |
| CTV.19 | Expressed sequence tag (EST) S20025 | − | 230,517–231,818 | BAA92411.1 | 1e-04 | Similar to rice EST S20025 |
| CTV.20 | Movement-like | − | 234,797–255,848 | AAK68664.1 | 4e-57 | Plant virus movement-like protein |
| CTV.21 | R7 | − | 263,612–266,302 | At5g63020 | e-105 | CC-NBS-LRR disease resistance gene |
| CTV.22 | EST N38213 | − | 267,393–278,222 | At1g15780 | e-120 | Similar to Arabidopsis EST N38213 |
Figure 2.
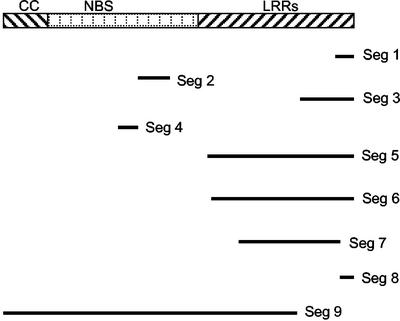
Gene and repetitive element map of the 282,699-bp segment of the P. trifoliata genome surrounding the Ctv gene. The designation of each of the putative genes and transposable/repetitive elements is described in Tables I and II. Seg 1 through 9, Partial R genes. Retro 1–6 represents partial retrotransposable elements. The region that must contain the Ctv gene is between markers 107B and 31A as indicated by vertical blue lines.
Two relatively large regions, from about 15 to 39 kb and from 180 to 192 kb, contain no predicted genes or other sequences with high similarity to those in GenBank (Fig. 2). These regions have low GC content (28.5% and 26.9%) in comparison with the entire sequenced region (34.8%).
To obtain cDNA clones in the Ctv locus, a cDNA library was constructed from the midrib of leaves and bark tissues collected from the plant used for the BAC library construction (Yang et al., 2001). BAC clones 108A10 and 83D17 (Fig. 1) were used to screen the cDNA library. Three cDNA clones (Jp11, Jp18, and Jp19) were isolated, and sequence comparisons indicated that Jp11 (2.3 kb) is encoded by CTV.2, Jp19 by CTV.12, and Jp18 by CTV.13.
Jp18 is a full-length cDNA encoded by the single exon of CTV.13. Both GenScan and GeneMark.hmm correctly predicted this gene. On the basis of the comparison between the Jp19 cDNA sequence and the CTV.12 genomic sequence, CTV.12 contains seven exons, all correctly predicted by GeneMark.hmm. However, one 5′ splice site was not predicted correctly by GenScan. On the basis of the comparison between a partial cDNA sequence of Jp11 and the CTV.2 genomic sequence, this region of CTV.2 contains 13 exons. GeneMark.hmm predicted 13 exons with one 3′ splice site and one 5′ splice site not predicted correctly. GenScan predicted 12 exons and missed one exon located between nucleotide positions 10,225 and 10,239. Of the 12 exons, one 3′ splice site and one 5′ splice site were also not predicted correctly. For these three genes, GenScan correctly predicted 36 exons and GeneMark 39 exons of 41 total exons.
Disease R Gene Cluster
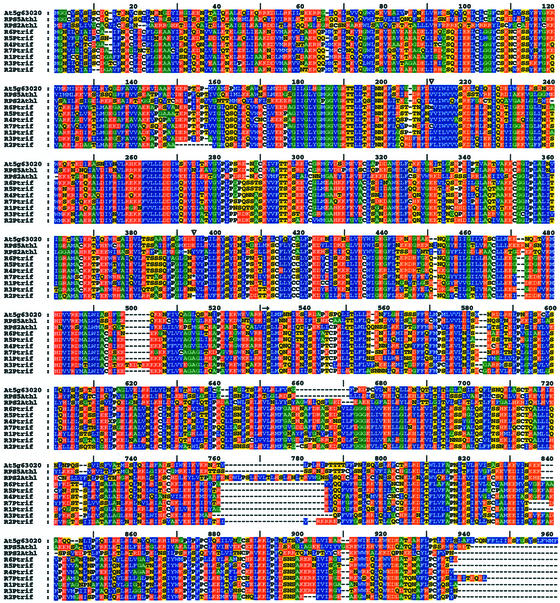
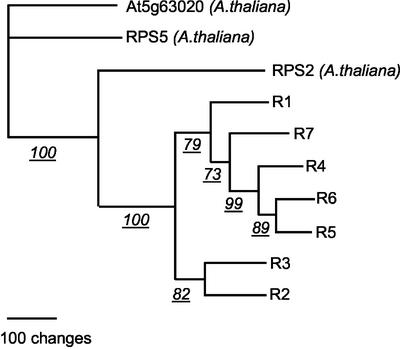
A total of seven CC-NBS-LRR type disease R genes (which lack the toll/interleukin receptor [TIR] domain) were identified from gene prediction and sequence alignments (Table I). All of the predicted R genes are highly homologous with At5g63020, a putative Arabidopsis disease R gene with a single exon (Table I). The R6 gene contains a frameshift in the 5′ region as indicated with an “X” at position 211, and the R7 gene has a stop codon at position 395 (Fig. 3), therefore, these two genes are probably pseudogenes. The other five R genes (R1–R5) contain complete ORFs of about 2.7 kb. Sequence comparisons among predicted proteins coded by the Arabidopsis disease R gene RPS2 (Mindrinos et al., 1994) and these R genes indicated that they contain 14 LRRs in the 3′ region (Fig. 3). The putative amino acid sequences encoded by these R genes have 68.9% to 84.1% similarity and 62.3% to 81.5% identity (data not shown). Parsimony analysis of entire predicted amino acid sequences shows that these genes are more closely related to each other than to Arabidopsis R genes in this class (Fig. 4). R4 to R6 clustered together in the single most parsimonious tree. R1 and R7 clustered with this group, but with R7 closer in most trees. R2 and R3 clustered together and were somewhat divergent from the other genes. Similar results were obtained from analysis of nucleic acid sequences from the coding regions. The PCR products (pY65 and pY28) used as probes to hybridize with the HindIII-fingerprinting blot of the BAC clones in the region in our previous work (Yang et al., 2001) are located in R1 and R7, respectively. Marker 31A is located in the 3′ end of R7 and the other R genes are located between markers 107B and 31A where the Ctv gene is delimited.
Figure 3.
Multiple alignment of deduced amino acid sequences from seven putative disease R genes (R1–R7) in the Ctv region and three related Arabidopsis genes. The alignment was generated using ClustalX. The locations of R1 to R7 are indicated in Figure 2. Triangles indicate locations of frameshift (x) and stop codon (*) at positions 211 of R6 and 395 of R7, respectively. The arrow represents the beginning of the LRR region.
Figure 4.
Parsimony phenogram from parsimony analysis of amino acid sequences of seven P. trifoliata R genes and three Arabidopsis R genes. The tree shown is the single most parsimonious tree. Accession numbers for the Arabidopsis genes RPS2 and RPS5 are At4g26090 and At1g12220, respectively.
Besides the R genes described above, a total of nine DNA segments that are very similar to disease R genes were identified in the intergenic sequence of the Ctv region (Seg 1–9; Fig. 2). These DNA fragments are in the same orientations as their closest R genes such as Seg 1 to 4 with R1; Seg 5 with R2; Seg 6 with R3; Seg 7 and 8 with R4; and Seg 9 with R7 (Fig. 2). Because they are similar to different NBS-LRR type R genes of about 2.7 kb, we can align these DNA segments with R genes and infer their origin. Most of these DNA segments (Seg 1, Seg 3, Seg 5, Seg 6, and Seg 8) derive from the 3′ end of R genes (Fig. 5). Seg 7 is most likely from Seg 8 because of the insertion of Gypsy-like C (Figs. 2 and 5). Seg 2 and Seg 4 are from the NBS region, and Seg 9 contains the most complete R gene sequence.
Figure 5.
Identified partial R genes in the Ctv locus and their relative regions in a typical CC-NBS-LRR class disease R gene. Seg 1 through 9, Partial R genes (see Fig. 2 for locations of Seg 1–9).
Repetitive Sequences
Apart from the 22 genes and R gene segments identified, repetitive sequences including simple sequence repeats (SSRs), class I (retrotransposons), and class II (transposons) transposable elements were also found (Table II). A total of 61 SSRs with each sequence repeated at least five times were identified in the Ctv region. Most of the SSRs are dimer repeats, and eight are trimer repeats. (AT) n and (TA) n types are the most common class of SSRs. Overall, these SSRs give a density of one SSR per 4.3 kb.
Table II.
Transposons and retrotransposons identified in the Ctv region.
| Designation | Positiona | 5′-LTR | 3′-LTR | Main Features | Size |
|---|---|---|---|---|---|
| bp | |||||
| Copia-like A | <52,962>–56,156 | ND | ND | LTRs not defined; homologous to Arabidopsis putative retroelement gb AAD19773.1 (BlastX E = 0) | 3,195 |
| Copia-like B | 70,974–80,959 | 70,974–72,123 | 79,788–80,959 | Five-nucleotide direct repeats (CATAC), 98% identical LTRs; homologous to soybean (Glycine max) gag-pol polyprotein gb AAC64917.1 (BlastX E = 0) | 9,986 |
| Copia-like C | 81,054–90,224 | 89,493–90,224 | 81,054–81,730 | Direct repeats not defined, 82.3% identical LTRs; homologous to Arabidopsis putative polyprotein gb AAK43485.1 (BlastX E = 5e-59) | 9,171 |
| Copia-like D | 114,081–119,204 | 114,081–114,329 | 118,956–119,204 | Five-nucleotide direct repeats (TAATG), 95.6% identical LTRs; homologous to potato retroelement Tst1 gb X52387 (BlastX E = e-130) | 5,124 |
| Copia-like E | 143,900–151,112 | 143,900–145,393 | 149,597–151,112 | Five-nucleotide direct repeats (TGTTG), 98.6% identical LTRs; homologous to rice putative gag-pol polyprotein gb AAK50132.1 (BlastX E = 9e-17) | 7,213 |
| Gypsy-like A | <122,185>–127,238 | ND | ND | LTRs not defined; homologous to Arabidopsis gb AAF79618.1 (BlastX E = 0) | 5,054 |
| Gypsy-like B | 136,944–153,359 | 151,350–153,359 | 136,944–139,269 | Five-Nucleotide direct repeats (GTAAA), 97.1% identical LTRs; homologous to sorghum (Sorghum bicolor) gypsy-like element gb AAD22153.1 (BlastX E = 0); Copia-like E has inserted between positions 143,900 to 151,112. | 9,203 |
| Gypsy-like C | 170,779–176,386 | 170,779–171,224 | 175,941–176,386 | Four-nucleotide direct repeats (TCCC), 99.8% identical LTRs; homologous to tomato polyprotein gb AAD13304.1 (BlastX E = 0) | 5,608 |
| Mutator-like A | 162,659–168,354 | 162,659–162,786 | 168,228–168,354 | Four-nucleotide direct repeat (TTTT), 88.9% identity between TIR A(127) and TIR B(126); homologous to Arabidopsis mutator-like transposase gb AAF04891.1 (BlastX = e-44) | 5,696 |
| Mutator-like B | 201,351–205,931 | 201,351–201,429 | 205,853–205,931 | Three-nucleotide direct repeat (AAA), 91.1% identical TIRs (79 bp); homologous to rice putative transposon gb AAK63883.1 (BlastX E = 2e-71) | 4,581 |
Angle brackets indicate that the sequence range is an approximation.
Numerous retroelements were identified including five copia-like and three gypsy-like retroelements (Fig. 2; Table II). These retroelements are not dispersed in this region, but are clustered in the region of 52,962 to 176,386 where relatively very few other genes were identified (Fig. 2). Copia-like A and Gypsy-like A were identified by their high similarity to Arabidopsis copia-like and gypsy-like retroelements, although the long terminal repeats (LTRs) could not be determined. The LTRs of Copia-like C are 82.3% identical, however, the putative target duplication sequences cannot be defined. All the other copia-like and gypsy-like retroelements contain LTRs and four to five nucleotide direct repeats around each element, which serve as integration sites in the genome (Table II). The size of LTRs ranges from 249 bp for Copia-like D to 2,326 bp for Gypsy-like B. Sequence comparison between the LTRs of Gypsy-like B indicates that there is a deletion of 316 bp in the left LTR although they are 97.1% identical. Inside the Gypsy-like B, another transposable element (Copia-like E) was identified (Fig. 2). No complete ORFs have been identified inside any of these retroelements, indicating that all of them may be inactive.
This region also contains class II (transposon) transposable elements. Mutator-like A and B are overall most similar to Arabidopsis mutator-like transposase (AAF04891.1) and rice mutator-like transposase (AAK63883.1), respectively. The TIRs of the two mutator like transposons are 126 and 79 bp, respectively.
Six DNA segments similar to parts of other known transposable elements also were identified (Fig. 2). Retro1, Retro2, and Retro5 are similar to copia-like elements, Retro3 is similar with gypsy-like elements, and Retro4 and Retro6 are similar with non-LTR like elements (Fig. 2).
Using the FINDMITE program (Tu, 2001) we searched for MITE-like sequences of 30 to 700 bp with at least 11 bp TIRs and 2- to 8-bp target site duplications (TSD). This search identified 299 putative MITEs with 2-bp TSD, 89 with 3-bp TSD, 38 with 4-bp TSD, 10 with 5-bp TSD, 6 with 6-bp TSD, and 2 with 8-bp TSD. Thirty-five TA and two TAA TSD were found among the sequences with 2- and 3-bp TSD, respectively. The MITE-like sequences showed various secondary structures including hairpins. However, we did not find Stowaway or Tourist-like structures, which may indicate that new types of MITEs are found in this region. Overall, these MITE-like sequences have a density of one per 1.57 kb.
Gene Colinearity between P. trifoliata and Arabidopsis
Because all R genes in the Ctv locus are very similar to the putative Arabidopsis R gene At5g63020 (Table II) and they are similar to each other, these genes were not used to study synteny with Arabidopsis genes. The other genes in the Ctv region were used to search the Arabidopsis sequences in GenBank using TBLASTN. Seven genes had no significant sequence similarity with Arabidopsis genes with an expectation value of E < e-20. The remaining nine genes have significant sequence similarity with Arabidopsis genes as shown in Table III. CTV.5 and CTV.15 have more than five Arabidopsis matches with an E value less than e-21, suggesting that they are members of various gene families. The orthologs of P. trifoliata genes in the Ctv locus are distributed over all five Arabidopsis chromosomes (Table III).
Table III.
TBLASTN sequence comparisons of genes in the Ctv locus with the Arabidopsis genome
| Gene ID | No. of Matches with E < e-20 | Syntenic Homologsa | Chromosome Locationb | TBLASTN E Value |
|---|---|---|---|---|
| centiMorgans | ||||
| CTV.1 | 3 | At1g15740 | I (15.7) | 7e-71 |
| CTV.2 | 8 | At1g15750 | I (15.7) | 0.0 |
| At1g80490 | I (125.4) | e-104 | ||
| At3g15880 | III (21.0) | 0.0 | ||
| CTV.3 | 6 | At1g80510 | I (125.4) | e-167 |
| At2g40420 | II (75.0) | 7e-29 | ||
| At3g30390 | III (51.0) | 1e-32 | ||
| At5g38820 | V (80.8) | 6e-36 | ||
| CTV.5 | >10 | At1g11260 | I (12.0) | 5e-44 |
| At4g21480 | IV (65.6) | 2e-41 | ||
| At3g19940 | III (26.0) | 6e-30 | ||
| At5g23270 | V (42.5) | 5e-28 | ||
| CTV.13 | 4 | At1g15760 | I (15.7) | 5e-49 |
| At1g80520 | I (125.4) | 3e-45 | ||
| CTV.14 | >10 | At1g80530 | I (125.4) | 0.0 |
| At2g28120 | II (53.6) | 4e-59 | ||
| At3g01930 | III (4.9) | 3e-41 | ||
| At4g34950 | IV (83.5) | 4e-48 | ||
| At5g14120 | V (30.4) | 2e-62 | ||
| CTV.15 | 1 | At5g11090 | V (25.3) | 2e-31 |
| CTV.20 | >10 | At2g01034 | II (0.0) | 5e-33 |
| At1g36590 | I (60.0) | 7e-33 | ||
| At3g11970 | III (2.3) | 7e-33 | ||
| At4g10580 | IV (32.5) | 8e-33 | ||
| CTV.22 | 5 | At1g15780 | I (15.7) | 1e-60 |
| At2g10440 | II (19.2) | 3e-51 |
Only the syntenic homolog with the lowest E value is listed for each chromosome. All syntenic homologs on Arabidopsis BAC clones F7H2 and T21F11 are listed.
The locations of clones containing syntenic homologs are indicated in parentheses based on the Arabidopsis RI genetic map.
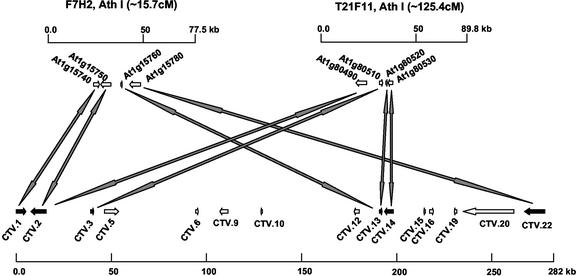
Microsynteny was observed between two Arabidopsis DNA segments (F7H2 and T21F11) and the Ctv region (Fig. 6). Arabidopsis BACs F7H2 and T21F11 are located in the duplicated regions of chromosome I at positions of about 15.7 and 125.4 centiMorgans, respectively. A total of six genes from the Ctv region correspond to eight Arabidopsis genes in the two BAC clones. Four genes, CTV.1, CTV.2, CTV.13, and CTV.22, from the Ctv region correspond to four genes (At1g15740, At1g15750, At1g15760, and At1g15780) from BAC clone F7H2, and genes CTV.2, CTV.3, CTV.13, and CTV.14 correspond to four genes (At1g80490, At1g80510, At1g80520, and At1g80530) from BAC clone T21F11. The six genes in the Ctv region and their orthologs in Arabidopsis are in the same order and transcription orientation. However, the physical distances encompassing the genes in P. trifoliata and their orthologs in Arabidopsis are very different. CTV.1, CTV.2, CTV.13, and CTV.22 are located in a region that spans 280 kb, and CTV.2, CTV.3, CTV.13, and CTV.14 are located in a region that spans 191 kb. However, their orthologs are located in 25- and 20-kb regions of Arabidopsis BAC clones F7H2 and T21F11, respectively.
Figure 6.
A schematic display of the Arabidopsis segments syntenic to the Ctv region. The two BAC clones F7H2 and T21F11 are located at 15.7 and 125.4 centiMorgans of Arabidopsis chromosome I (Ath I). The arrows below these BAC clones represent the ORFs in the corresponding BAC clones that have significant sequence similarity with genes in the Ctv region.
DISCUSSION
Our previous work established a 1.2-Mb contig around the Ctv locus and further mapped this gene to a region between markers 31A and 107B, which is covered by four BAC clones (Yang et al., 2001). In this work, we have completely sequenced these BAC clones, and the entire sequence of the four BAC clones spans 282,699 bp. The physical distance between markers 31A and 107B where Ctv is located is 259,974 bp, somewhat smaller than our previous estimate of 300 kb. The contig in Figure 1 is based on the new sequence data, and therefore, the relationship of all BAC clones is to scale.
Genomic Organization
The region sequenced has a gene density of one gene per 12.8 kb. If this average gene density is extrapolated to the entire 382-Mb citrus genome, the total number of genes is predicted to be 29,844, a value fairly consistent with the values reported for Arabidopsis (25,498; Arabidopsis Genome Initiative, 2001), and rice (32,000–50,000; Goff et al., 2002). Therefore, the Ctv region apparently has average gene density.
The sequence analyses indicate that there is a disease R gene cluster in the Ctv region including possibly five functional R genes, two pseudogenes, and nine partial R gene segments. The clustering of disease R genes is a common occurrence in plant genomes (Michelmore and Meyers, 1998), and genes within a single cluster can determine resistance to very different pathogens. This disease R gene cluster may supply a resource for P. trifoliata and citrus resistance to different pathogens including CTV.
Unequal crossing-over plays an important role in disease R gene cluster evolution, and it has been observed in the L alleles of flax (Linum usitatissimum; Ellis et al., 1997), Rp1 alleles of maize (Zea mays; Hulbert, 1997), and the major cluster of R genes in lettuce (Lactuca sativa; Chin et al., 2001). In our work, a total of nine partial R gene segments (Seg 1–9) have been identified around the R1, R2, R4, and R7 genes. These DNA segments are in the same orientations as their closely linked R genes. This suggests that partial R gene segments may be a result of intragenic unequal crossing-over or of intergenic unequal crossing-over followed by deletion events. In the Cf4/9 haplotypes that originated from different tomato species, all of the paralogs in each haplotype are oriented in the same direction (Parniske et al., 1997). In our work, R1, R3, and R4 are in the same orientation, and the other R genes (R2, R5, R6, and R7) are in another orientation. This indicates that there may be other mechanisms to duplicate genes besides the unequal crossing-over if these R genes are considered to originate from a common ancestor or that they originated from different ancestors.
Another interesting feature in the Ctv region is the clustered transposable elements. In the 282-kb Ctv region, the eight retrotransposons are clustered in a 119-kb region (nucleotide positions 52,962–171,224). Arabidopsis has a relatively small genome size (130 Mb) and a relatively low proportion of repetitive sequences; the retrotransposons primarily occupy the centromere. The centromeres usually contain repetitive arrays, including the 180-bp repeats (Arabidopsis Genome Initiative, 2000). It is not known whether the Ctv locus is near the centromere of a P. trifoliata chromosome. For many plants with large genomes, retrotransposons contribute most of the nucleotide content (San Miguel et al., 1996). Retrotransposons are nested in the intergenic regions of the maize genome (San Miguel et al., 1996) and dispersed around the rice Adh1-Adh2 region (Tarchini et al., 2000). In dicots, very few genomic sequences larger than 100 kb have been reported except in Arabidopsis. In the 119-kb (Mao et al., 2001) and the 105-kb (Ku et al., 2000) tomato genomic sequences, only two copia-like retrotransposons were found (Mao et al., 2001).
Synteny
Arabidopsis and its closely related species show extensive conservation of gene repertoire, order, and orientation (Acarkan et al., 2000; O'Neill and Bancroft, 2000). The synteny between Arabidopsis and tomato showed limited conservation (Ku et al., 2000; Mao et al., 2001), although a remarkable degree of conserved microsynteny between these two plants can also be found (Rossberg et al., 2001). The Ctv region is about 282 kb, however, only two Arabidopsis genomic DNA fragments (BAC clones F7H2 and T21F11) have been identified that contain more than one ortholog of P. trifoliata genes in the Ctv region (Fig. 6). In this region, synteny between these two plants is less conserved than that between the sequenced regions of Arabidopsis and tomato, despite evidence that P. trifoliata and Arabidopsis diverged much later than tomato and Arabidopsis did (Chase et al., 1993). The Ctv region contains a disease R gene cluster and clustered retrotransposable elements. The disease R gene cluster region might evolve rapidly (Michelmore and Meyers, 1998), and retrotransposable elements tend to insert in this region. These processes would increase the rate of evolution in this region, but they do not fully explain the limited synteny observed. In this genome region, considerable structural reorganization has occurred since P. trifoliata and Arabidopsis diverged. Analysis of this region in additional taxa will be necessary to clarify the timing and mechanism of these genomic changes. This comparison of P. trifoliata and Arabidopsis suggests that the rate and type of evolution and resulting synteny varies over the genome.
Putative Ctv Gene
The target of our project is to clone the Ctv gene, which is a virus disease R gene. Several virus R genes have currently been identified. The tobacco (Nicotiana tabacum) mosaic virus R gene N (Whitham et al., 1994), tomato tospovirus R gene Sw-5 (Brommonschenkel et al., 2000), and potato (Solanum tuberosum) virus X R gene Rx (Bendahmane et al., 1999) are NBS-LRR type disease R genes. In this work, five R genes (R1–R5) with complete ORFs have been identified and can be considered as candidates for Ctv. We used reverse transcriptase-PCR to study expression of several of these R genes. Primers specific to four R genes within the contig were designed and used to amplify from RNA isolated from CTV-challenged bark and leaf tissue of resistant and susceptible genotypes. Primers for R1, R2, and R3 amplified PCR products of the expected size in several resistant genotypes (data not shown). Primers for R4 did not amplify any detectable products from RNA samples but did amplify products of the expected size from DNA templates, suggesting that R4 is not expressed. Sequence alignments show a 10-bp deletion in the putative promoter region of the R4 gene on the chromosome carrying the Ctv-resistant allele.
There are also some virus disease R genes without NBS and LRRs (Chisholm et al., 2000; Kachroo et al., 2000; Whitham et al., 2000), and it is possible that Ctv belongs to this class of R genes. In the Ctv region, CTV.20 contains a domain similar (score = 45.3; E = 3e-05) to a plant virus movement protein. CTV.20 also contains domains with high amino acids identities (score = 97.3; E = 1e-20) to retroelement and caulimovirus reverse transcriptases. Another domain contains a region similar to the integrase proteins of retroviruses and retrotransposons. Northern hybridization indicated that CTV.20 and its ortholog are highly expressed in P. trifoliata and sweet orange leaves and in P. trifoliata bark tissues but are relatively lowly expressed in the phloem of sweet orange (data not shown). The ortholog of CTV.20 in sweet orange is about 8.5 kb, which is slightly smaller than CTV.20 (9 kb) in P. trifoliata. CTV tends to accumulate in phloem tissue of infected plants, which suggests that CTV.20 could also be considered as a candidate gene for Ctv. For the five other genes (CTV.1, CTV.2, CTV.12, CTV.13, and CTV.14) we have examined to date, we have not seen differences in expression patterns that correlate with Ctv resistance (data not shown).
MATERIALS AND METHODS
DNA Sequencing
In our previous work, the Ctv gene was mapped to a contig between markers 107B and 31A (Yang et al., 2001). BAC clones 27A14, 20J24, 83D17, and 84F5 from the contig (Fig. 1) were chosen for shotgun sequencing (Bodenteich et al., 1993). BAC DNA was isolated using a large-construct kit (Qiagen USA, Valencia, CA). Subcloning libraries were constructed using a TOPO shotgun cloning kit from Invitrogen (Carlsbad, CA) with BAC DNA sheared by nebulization to approximately 2 kb. After transformation, recombinant clones were randomly picked and grown in 5 mL of Luria-Bertani medium containing 50 mL L−1 kanamycin at 37°C overnight. DNA was isolated by either Concert High Purity Plasmid Miniprep System from Invitrogen or Wizard Plus Minipreps DNA Purification System from Promega (Madison, WI). Shotgun clones from BAC 27A14 and 84F5 were sequenced on ABI Prism 377 or 3700 sequencers (Applied Biosystems, Foster City, CA) at Iowa State University, whereas shotgun clones from BAC 20J24 and 83D17 were sequenced on a LI-COR 4200 sequencer (LI-COR, Lincoln, NE) at the University of California, Riverside.
After sequence assembly, gaps were filled by isolating DNA fragments located in the gaps using an LA PCR kit (Takara Shuzo, Kyoto). Primers were designed based upon the assembled contig sequences. PCR products were used as probes to screen subcloning libraries to obtain subclones located in the gaps for sequencing or were cloned using a TOPO TA cloning kit from Invitrogen and sequenced using a primer walking method. In some cases, subclone inserts located in the end of contigs were also used to screen subcloning libraries to obtain clones located in the gap. In some regions with low coverage, the internal regions of subclones were also sequenced using a primer walking method.
Analysis of Sequence Data
Sequences were assembled with Seq Man II from DNASTAR, Inc. (Madison, WI). Genes were identified by a combination of several methods. The genes in this region were predicted by GenScan+ (Burge and Karlin, 1997; http://genes.mit.edu/GENSCAN.html). The modeling of exon structure was adjusted with the prediction result of GeneMark (Lukashin and Borodovsky, 1998; http://genemark.biology.gatech.edu/GeneMark/) and GlimmerA (a variant of GlimmerM; http://www.tigr.org/softlab/). The Arabidopsis settings were chosen for all programs. For the identification of putative disease R genes, the programs Pileup and Gap (Genetics Computer Group, Madison, WI; Devereux et al., 1984) and ClustalX (Thompson et al., 1997) were used to align the uncertain sequence regions with identified R genes. The DOTTER program (Sonnhammer and Durbin, 1995) was used to identify and classify repeat families. Intergenic sequence was also divided into 3-kb segments with 1-kb overlap and used for BLASTN and BLASTX homology searches (Altschul et al., 1997) against the GenBank database as described (Tarchini et al., 2000). The SSRs were identified with the program “SSRIT” (http://ars-genome.cornell.edu/rice/tools.html). MITE-like sequences were identified using FINDMITE as described (Tu, 2001). Phylogenetic relationships between the R genes were analyzed using parsimony with the PAUP* program (Phylogenetic Analysis Using Parsimony, version 4.0 b8a, Sinaur Associates, Sunderland, MA).
cDNA Library Construction and Screening
Total RNA was extracted from leaves or bark of Poncirus trifoliata cv Pomeroy and sweet orange as described (Jones et al., 1985). The mRNA was then purified from the total RNA using an Oligotex mRNA Kit from Qiagen USA as recommended by the manufacturer. The mRNA purified from CTV-challenged P. trifoliata was also used to construct a cDNA library. cDNA was synthesized using a SMART PCR cDNA library Construction Kit according to the user manuals (BD Biosciences Clontech, Palo Alto, CA). After PCR amplification, SfiI digestion, and size fractionation, cDNA was ligated to λTriplEx2 and packaged with GigapackIII Gold Packaging Extract from Stratagene (La Jolla, CA) according to the instruction manual. A total of 350,000 phages was screened essentially as described (Sambrook et al., 1989).
ACKNOWLEDGMENT
We thank Julieta G. Plancarte for helping with cDNA library screening and cDNA clone sequencing.
Footnotes
This work was supported by the California Citrus Research Board (grant no. CTV–009 to M.L.R.), by the U.S. Department of Agriculture-Agricultural Research Service (grant no. 59–0790–8-51 to T.E.M. and M.L.R.), and by the U.S. Department of Agriculture-Cooperative State Research, Education, and Extension Service (grant nos. 99–34399–8460, 00–34399–9343, and 01–34399–10748 to T.E.M. and M.L.R.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.011262.
LITERATURE CITED
- Acarkan A, Rossberg M, Koch M, Schmidt R. Comparative genome analysis reveals extensive conservation of genome organization for Arabidopsis thaliana and Capsella rubella. Plant J. 2000;23:55–62. doi: 10.1046/j.1365-313x.2000.00790.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2001;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- Bendahmane A, Kanyuka K, Baulcombe DC. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell. 1999;11:781–791. doi: 10.1105/tpc.11.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenteich A, Chissoe S, Wang YF, Roe BA. Shotgun cloning as the strategy of choice to generate templates for high-throughput dideoxynucleotide sequencing. In: Venter JC, editor. Automated DNA Sequencing and Analysis Techniques. London: Academic Press; 1993. pp. 42–50. [Google Scholar]
- Brommonschenkel SH, Frary A, Frary A, Tanksley SD. The broad-spectrum tospovirus resistance gene Sw-5 of tomato is a homolog of the root-knot nematode resistance gene Mi. Mol Plant-Microbe Interact. 2000;13:1130–1138. doi: 10.1094/MPMI.2000.13.10.1130. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structure in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Chase MW, Soltis DE, Olmstead RG, Morgan D, Les DH, Mishler BD, Duvall MR, Price RA, Hills HG, Qui YL et al. Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Ann MO Bot Gard. 1993;80:528–580. [Google Scholar]
- Chin DB, Arroyo-Garcia R, Ochoa OE, Kesseli RV, Lavelle DO, Michelmore RW. Recombination and spontaneous mutation at the major cluster of resistance genes in lettuce (Lactuca sativa) Genetics. 2001;157:831–849. doi: 10.1093/genetics/157.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Mahajan SK, Whitham SA, Yamamoto ML, Carrington JC. Cloning of the ArabidopsisRTM1 gene, which controls restriction of long-distance movement of tobacco etch virus. Proc Natl Acad Sci USA. 2000;97:489–494. doi: 10.1073/pnas.97.1.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Huang S, Ling P, Chen C, Yu C, Weber CA, Moore GA, Gmitter FG., Jr Cloning and characterization of NBS-LRR class resistance gene candidate sequences in citrus. Theor Appl Genet. 2000;101:814–822. [Google Scholar]
- Deng Z, Huang S, Ling P, Yu C, Tao Q, Chen C, Wendell MK, Zhang HB, Gmitter FG., Jr Fine genetic mapping and BAC contig development for the citrus tristeza virus resistance gene locus in Poncirus trifoliata(Raf.) Mol Genet Genomics. 2001a;265:739–747. doi: 10.1007/s004380100471. [DOI] [PubMed] [Google Scholar]
- Deng Z, Huang S, Xiao S, Gmitter FG., Jr Development and characterization of SCAR markers linked to the citrus tristeza virus resistance gene from Poncirus trifoliata. Genome. 1997;40:697–704. doi: 10.1139/g97-792. [DOI] [PubMed] [Google Scholar]
- Deng Z, Tao Q, Chang YL, Huang S, Ling P, Yu C, Chen C, Gmitter FG, Jr, Zhang HB. Construction of a bacterial artificial chromosome (BAC) library for citrus and identification of BAC contigs containing resistance gene candidates. Theor Appl Genet. 2001b;102:1177–1184. [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Gale MD. Comparative genetics in the grasses. Plant Mol Biol. 1997;35:3–15. [PubMed] [Google Scholar]
- Ellis J, Lawrence G, Ayliffe M, Anderson P, Collins N, Finnegan J, Frost D, Luck J, Pryor T et al. Advances in the molecular genetic analysis of the flax-flax rust interaction. Annu Rev Phytopathol. 1997;35:271–291. doi: 10.1146/annurev.phyto.35.1.271. [DOI] [PubMed] [Google Scholar]
- Fang DQ, Federici CT, Roose ML. A high-resolution linkage map of the citrus tristeza virus resistance gene region in Poncirus trifoliata(L.) Raf. Genetics. 1998;150:883–890. doi: 10.1093/genetics/150.2.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmitter FG, Xiao SY, Huang S, Hu XL, Garnsey SM, Deng Z. A localized linkage map of the citrus tristeza virusresistance gene region. Theor Appl Genet. 1996;92:688–695. doi: 10.1007/BF00226090. [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan T, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JDG. Plant disease resistance genes. Annu Rev Plant Physiol Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- Hulbert SH. Structure and evolution of the rp1 complex conferring rust resistance in maize. Annu Rev Phytopathol. 1997;35:293–310. doi: 10.1146/annurev.phyto.35.1.293. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dunsmuir P, Bedbrook J. High level expression of introduced chimeric genes in regenerated transformed plants. EMBO J. 1985;4:2411–2418. doi: 10.1002/j.1460-2075.1985.tb03949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P, Yoshioka K, Shah J, Dooner HK, Klessig DF. Resistance to turnip crinkle virus in Arabidopsisis regulated by two host genes and is salicylic acid dependent but NPR1, ethylene, and jasmonate independent. Plant Cell. 2000;12:677–690. doi: 10.1105/tpc.12.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku HM, Vision T, Liu J, Tanksley SD. Comparing sequenced segments of the tomato and Arabidopsis genomes: Large-scale duplication followed by selective gene loss creates a network of synteny. Proc Natl Acad Sci USA. 2000;97:9121–9126. doi: 10.1073/pnas.160271297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene-finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Begum D, Goff SA, Wing RA. Sequence and analysis of the tomato JOINTLESSlocus. Plant Physiol. 2001;126:1331–1340. doi: 10.1104/pp.126.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM. The A. thaliana disease resistance gene RPS2encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- O'Neill CM, Bancroft I. Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J. 2000;23:233–243. doi: 10.1046/j.1365-313x.2000.00781.x. [DOI] [PubMed] [Google Scholar]
- Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/g locus of tomato. Cell. 1997;91:821–832. doi: 10.1016/s0092-8674(00)80470-5. [DOI] [PubMed] [Google Scholar]
- Rossberg M, Theres K, Acarken A, Herrero R, Schmitt T, Schumacher K, Schmitz G, Schmidt R. Comparative sequence analysis reveals extensive microcolinearity in the lateral suppressor regions of the tomato, Arabidopsis, and Capsellagenomes. Plant Cell. 2001;13:979–988. doi: 10.1105/tpc.13.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- San Miguel P, Tikhomov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–767. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:1–10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Tarchini R, Biddle P, Wineland R, Tingey S, Rafalski A. The complete sequence of 340 kb of DNA around the rice Adh1-Adh2 region reveals interrupted colinearity with maize chromosome 4. Plant Cell. 2000;12:381–391. doi: 10.1105/tpc.12.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z. Eight novel families of miniature inverted repeat transposable elements in the African malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci USA. 2001;98:1699–1704. doi: 10.1073/pnas.041593198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B. The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell. 1994;78:1011–1015. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- Whitham SA, Anderberg RJ, Chisholm ST, Carrington JC. ArabidopsisRTM2 gene is necessary for specific restriction of tobacco etch virus and encodes an unusual small heat shock-like protein. Plant Cell. 2000;12:569–582. doi: 10.1105/tpc.12.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZN, Ye XR, Choi SD, Molina J, Moonan F, Wing RA, Roose ML, Mirkov TE. Construction of a 1.2-Mb contig including the citrus tristeza virus resistance gene locus using a bacterial artificial chromosome library of Poncirus trifoliata(L.) Raf. Genome. 2001;44:382–393. [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]