Abstract
Wild-type Arabidopsis roots develop a wavy pattern of growth on tilted agar surfaces. For many Arabidopsis ecotypes, roots also grow askew on such surfaces, typically slanting to the right of the gravity vector. We identified a mutant, wvd2-1, that displays suppressed root waving and leftward root slanting under these conditions. These phenotypes arise from transcriptional activation of the novel WAVE-DAMPENED2 (WVD2) gene by the cauliflower mosaic virus 35S promoter in mutant plants. Seedlings overexpressing WVD2 exhibit constitutive right-handed helical growth in both roots and etiolated hypocotyls, whereas the petioles of WVD2-overexpressing rosette leaves exhibit left-handed twisting. Moreover, the anisotropic expansion of cells is impaired, resulting in the formation of shorter and stockier organs. In roots, the phenotype is accompanied by a change in the arrangement of cortical microtubules within peripheral cap cells and cells at the basal end of the elongation zone. WVD2 transcripts are detectable by reverse transcriptase-polymerase chain reaction in multiple organs of wild-type plants. Its predicted gene product contains a conserved region named “KLEEK,” which is found only in plant proteins. The Arabidopsis genome possesses seven other genes predicted to encode KLEEK-containing products. Overexpression of one of these genes, WVD2-LIKE 1, which encodes a protein with regions of similarity to WVD2 extending beyond the KLEEK domain, results in phenotypes that are highly similar to wvd2-1. Silencing of WVD2 and its paralogs results in enhanced root skewing in the wild-type direction. Our observations suggest that at least two members of this gene family may modulate both rotational polarity and anisotropic cell expansion during organ growth.
The primary roots of Arabidopsis possess an intrinsic handedness to their growth, consistently forming counterclockwise coils as they elongate upon a horizontal surface of hard agar (Mirza, 1987). However, when the surface is positioned vertically, the downward growth behavior of roots dictated by gravitropism conflicts with this counterclockwise coiling tendency, resulting in a net direction of growth that is to the right of the gravity vector. It should be noted that, in this report, we follow the convention used in Simmons et al. (1995) and Rutherford and Masson (1996) to describe the direction of root coiling (clockwise or counterclockwise) and slanting (skewing: leftward or rightward of the vertical axis), as viewed through the agar medium.
While slanting on vertical surfaces, root tips of wild-type Arabidopsis seedlings also exhibit a largely left-handed rotation around the net axis of growth, resulting in a moderate, left-handed twisting of the discrete cell files that make up the root epidermal layer. This left-handed preference in root tip rotation is associated with and may be responsible for the counterclockwise bias in root coiling and, by extension, rightward root slanting on vertical surfaces (Simmons et al., 1995; Rutherford and Masson, 1996). Most mutants possessing an opposite (leftward root slanting phenotype accordingly display predominantly right-handed epidermal cell file rotation (CFR; Marinelli et al., 1997; Furutani et al., 2000), whereas mutants with exaggerated rightward root slanting often exhibit enhanced left-handed epidermal CFR (Rutherford and Masson, 1996; Sedbrook et al., 2002; Thitamadee et al., 2002).
Arabidopsis root tip rotation appears to be driven by circumnutation, an endogenous, elliptical movement pattern that all plant organs exhibit around their mean growth vector. Depending on the plant species in question, circumnutation can be left-handed, right-handed, or seemingly random (Hashimoto, 2002). Although the basis for circumnutational direction remains uncertain, several recent models have focused on the cortical microtubule arrangement within elongating cells (Liang et al., 1996; Furutani et al., 2000; Thitamadee et al., 2002). Cortical microtubules are thought to direct the anisotropic expansion of cells by guiding the deposition of cellulose microfibrils within the cell wall (Giddings and Staehelin, 1991). In rapidly elongating maize (Zea mays) and Arabidopsis root cells, the alignment of cortical microtubules is transverse with the axis of root elongation. As the cells enter the mature zone of roots, the microtubules consistently transition to a right-handed helical arrangement just before adopting the longitudinal configuration observed in cells that have ceased to expand (Liang et al., 1996; Sugimoto et al., 2000). The elongating cells of lefty1 and lefty2 mutant roots, which possess an exaggerated rightward-slanting phenotype, exhibit a primarily right-handed oblique arrangement of cortical microtubules (Thitamadee et al., 2002). The elongating root cells of the spiral1 mutant, which possesses an opposite (leftward) root-skewing phenotype, conversely contain predominantly left-handed, oblique cortical microtubule arrays (Furutani et al., 2000). On the basis of these observations, it has been proposed that the direction of cortical microtubules may specify the polarity of root tip rotation (Liang et al., 1996; Furutani et al., 2000).
Interestingly, although wild-type Arabidopsis roots embedded within a homogeneous agar-based environment grow straight downward, their tips still oscillate around the net growth axis. The degree of cell file twisting at the root tip does not differ significantly between roots growing in agar and roots growing on the surface of the agar medium (Sedbrook et al., 2002). Furthermore, the frequency of root tip oscillations under these two growth conditions does not differ (Mullen et al., 1998). These observations suggest that the coiling behavior of roots is surface-triggered and can be uncoupled from circumnutation in the absence of an asymmetric mechanical stimulus (Rutherford and Masson, 1996; Mullen et al., 1998; Sedbrook et al., 2002). Decreasing the agar content of the growth medium, or tilting the media-containing plates forward slightly so that gravitropism tends to pull the roots away from the surface, diminishes the slanting tendency of roots. These observations suggest that the coil-inducing stimulus may be tactile (Okada and Shimura, 1990; Rutherford and Masson, 1996).
In addition to the right-slanting phenotype, Arabidopsis roots exhibit an elaborate, wavy growth pattern on agar surfaces that have been tilted backward (Okada and Shimura, 1990). Root waving consists of a succession of left-handed and right-handed circumnutation-like movements, each accompanied by CFRs in the opposite direction, originating within the basal region of the root elongation zone (Okada and Shimura, 1990; Rutherford and Masson, 1996). This behavior can be described as a succession of leftward and rightward root slanting movements, which appears to result from a combination of gravity and touch stimulation (Okada and Shimura, 1990; Simmons et al., 1995; Rutherford and Masson, 1996; Rutherford et al., 1998). Other surface-derived stimuli may also be involved (Buer et al., 2000).
To further characterize the processes involved in determining root rotational polarity and helical growth, we have screened a collection of activation-tagged Arabidopsis lines for altered root-waving and/or root-skewing phenotypes. Here, we describe the isolation and characterization of a mutation (wvd2-1) that inhibits root waving and reverses root slanting on agar surfaces. Our analysis strongly suggests that WVD2 (GenBank accession no. AF548461) and at least one of its paralogs (WDL1) function as regulators of rotational polarity and anisotropic cell expansion during organ growth.
RESULTS
Isolation and Characterization of wvd2-1
The wvd2-1 mutant displayed altered root growth phenotypes on inclined agar surfaces. Whereas the roots of wild-type Arabidopsis seedlings (ecotype No-0) formed sinusoidal waves and skewed to the right of the gravity vector, wvd2-1 roots skewed leftward without waving when grown under these conditions (Fig. 1A). In addition, homozygous wvd2-1 roots elongated at a slower rate than wild type and were noticeably larger in diameter (Table I). wvd2-1 roots also possessed a higher density of root hairs than wild type, although no hairs were found at ectopic positions within atrichoblastic cell files (data not shown). Cells within the epidermal layer appeared to bulge out at random locations along the root (Fig. 2, C and H). The F1 progeny derived from out-crosses between homozygous wvd2-1 and wild-type plants also exhibited suppressed root waving, and had intermediate root-skewing, elongation, and radial expansion phenotypes (Fig. 1A; Table I). However, they lacked the hairy-root and epidermal cell-bulging phenotypes of homozygous wvd2-1 roots (Figs. 2, B and E). Genetic analysis of the subsequent F2 generation revealed that wvd2-1 segregated as a single, semidominant mutation (1:2:1 ratio; χ2 = 0.0169; n = 472).
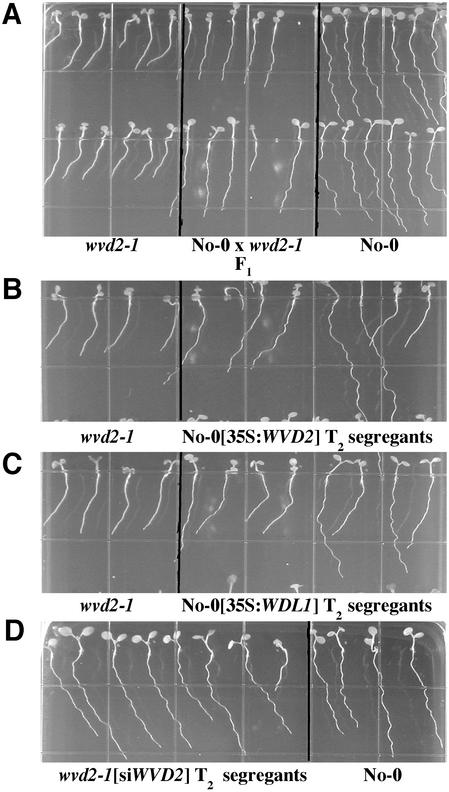
Figure 1.
Root growth phenotypes on agar surfaces of wild type (ecotype Nossen-0 [No-0]), homozygous wvd2-1, or the F1 progeny of a cross between wvd2-1 and No-0 (A); of representative T2 plants segregating for either the 35S:WVD2 (B) or 35S:WDL1 (C) transgenes in a wild-type No-0 background; or for T2 plants segregating for the silencing transgene siWVD2 in a wvd2-1 background (D). Seedlings were grown for 7 d on 1.5% (w/v) agar-solidified medium, tilted backward 30° from vertical. For B, C, and D, the T2 seedlings were derived from selfing a transgenic plant hemizygous for the transgene. Pictures were taken from the bottom of the plates, through the medium.
Table I.
Biometrical analysis of root and shoot phenotypes
| Parameter | No-0 |
wvd2-1
|
|
|---|---|---|---|
| Heterozygous | Homozygous | ||
| Root | |||
| Elongation (mm) | 5.2 ± 0.7 | 3.8 ± 0.4 | 2.4 ± 0.2 |
| n = 68 | n = 41 | n = 69 | |
| Diameter (μm) | 99.2 μm ± 3.8 | 121.4 μm ± 4.5 | 148.3 μm ± 9.2 |
| n = 18 | n = 17 | n = 18 | |
| Skew angle (°) | 21.6 ± 6.9 | −15.0 ± 7.2 | −38.9 ± 9.4 |
| n = 66 | n = 57 | n = 69 | |
| Etiolated hypocotyl | |||
| Length (mm) | 17.5 ± 1.5 | n.d. | 8.3 ± 0.6 |
| n = 33 | n = 35 | ||
| Diameter (mm) | 0.42 ± 0.05 | n.d. | 0.59 ± 0.05 |
| n = 33 | n = 35 | ||
| Rosette leaf | |||
| Length (mm) | 29.2 ± 3.7 | 23.1 ± 3.1 | 18.0 ± 3.8 |
| n = 27 | n = 27 | n = 27 | |
| Width (mm) | 14.3 ± 1.7 | 13.5 ± 1.8 | 12.3 ± 2.4 |
| n = 27 | n = 27 | n = 27 | |
| Surface area (mm2) | 318.8 ± 65.0 | 248.8 ± 57.3 | 187.4 ± 67.2 |
| n = 26 | n = 26 | n = 26 | |
| Silique | |||
| Length (mm) | 13.85 ± 1.29 | 10.03 ± 0.96 | 7.85 ± 0.71 |
| n = 40 | n = 40 | n = 40 | |
| Diameter (mm) | 0.88 ± 0.18 | 1.36 ± 0.25 | 1.52 ± 0.19 |
| n = 40 | n = 40 | n = 40 | |
| Cell dimensions (roots) | |||
| Cortex length (μm) | 127 ± 18.5 | n.d. | 85.1 ± 15.8 |
| n = 45 | n = 45 | ||
| Width (μm) | 19.1 ± 3.6 | n.d. | 28.3 ± 3.7 |
| n = 45 | n = 45 | ||
| Epidermis length (μm) | 193.4 ± 34.8 | n.d. | 108.6 ± 14.3 |
| n = 27 | n = 27 | ||
| Width (μm) | 12.7 ± 1.9 | n.d. | 18.3 ± 3.1 |
| n = 27 | n = 27 | ||
Root phenotypes, Seedlings were grown on 1.5% (w/v) agar plates. Root elongation was measured over a 24-h interval, 4 to 5 d after germination (DAG). Root diameters were determined at the mature zone 7 DAG. In both cases, the plates were oriented vertically. The angles of root skewing were measured for seedlings grown on plates tilted backwards 30°, 7 DAG. Hypocotyls, Lengths and diameters are for etiolated seedlings grown on 0.8% (w/v) agar plates, 5 DAG. Rosette leaves, Values are for the first true leaf of each plant, measured within 24 h of bolt emergence. Siliques, Five fully elongated siliques were analyzed from eight wvd2-1 and eight wild-type No-0 plants. Measurement of epidermal and cortical cells within the mature zones of roots grown on vertical 1.5% (w/v) agar plates, 7 DAG. Five cortical and three epidermal cells were measured from nine wvd2-1 and nine wild-type No-0 roots. All results are mean values ± sd. n.d., Not determined.
Figure 2.
Epidermal CFR phenotypes. Close-up images of roots grown on a tilted agar surface (A-F) or embedded in agar (G and H). Images were taken either at the tip (A–C) or at the mature zone (D–H) of 7-d-old roots: A, D, and G, Wild-type No-0; B and E, heterozygous wvd2-1; C, F, and H, homozygous wvd2-1. Arrows indicate positions where bulging of epidermal cells are observed in homozygous wvd2-1 roots. Scale bars = 100 μm.
The mean angle of wvd2-1 root skewing varied according to the orientation of the agar surface. Mutant roots exhibited greater mean angles of leftward skewing on agar surfaces inclined backward than on vertically positioned surfaces, with the angle of leftward deviation increasing the further the agar plate was tilted backward (Table II). By comparison, the skewing angles of wild-type No-0 roots grown on tilted and untilted plates did not differ significantly, although plate tilting appeared to increase the amplitude of wild-type root waving (data not shown). The growth vectors of wvd2-1 and wild-type roots submerged within the agar-solidified growth medium did not deviate significantly from the vertical axis (Table II), indicating that the wvd2-1 skewed root growth phenotype neither is caused by nor results in an alteration of the root gravitropic set point angle. However, the bending kinetics of wvd2-1 roots lagged slightly behind that of wild-type roots upon gravistimulation (supplementary data can be viewed at www.plantphysiol.org), possibly because of the large difference in rate of root elongation between mutant and wild-type seedlings.
Table II.
Effect of the growth conditions on root skewing
| Conditions | Skew Angle
|
|
|---|---|---|
| No-0 | wvd2-1 (homozygous) | |
| ° | ||
| Plate conditions | ||
| 1.5% (w/v) Agar, 0° tilting | 23.8 ± 5.1 | −27.2 ± 9.2 |
| n = 53 | n = 50 | |
| 30° Tilting | 22.3 ± 6.5 | −36.2 ± 9.9 |
| n = 53 | n = 51 | |
| 45° Tilting | 24.9 ± 8.4 | −42.7 ± 10.5 |
| n = 43 | n = 51 | |
| 60° Tilting | 26.5 ± 11.4 | −47.7 ± 13.2 |
| n = 49 | n = 48 | |
| 0.8% (w/v) Agar 0° (surface) | 17.6 ± 5.6 | −16.9 ± 12.4 |
| n = 46 | n = 44 | |
| 0° (in Agar) | 0.1 ± 7.8 | 0.1 ± 5.7 |
| n = 85 | n = 85 | |
| Propyzamide | ||
| 0 μm | 21.5 ± 7.5 | −23.3 ± 13.3 |
| n = 70 | n = 68 | |
| 1 μm | 28.0 ± 7.0 | −44.9 ± 14.2 |
| n = 67 | n = 68 | |
| 3 μm | 40.2 ± 10.7 | −71.7 ± 21.9 |
| n = 62 | n = 65 | |
Plate conditions, Seedlings were grown on one-half-strength Murashige and Skoog media containing either 0.8% or 1.5% agar. The orientation of the plates (indicated next to and below the agar content) is relative to the vertical position (0°), with angles > 0° indicating backwards plate tilting. The angles of root skewing were measured for seedlings grown on the surface of the agar, except where denoted as being “in agar.” Propyzamide, Seedlings were grown for 7 d on 1.5% agar plates, tilted backwards 30°, with 0, 1, or 3 μm propyzamide added to the medium. Results are mean angles of root skewing ± sd. Positive and negative values indicate rightward and leftward skewing, respectively.
Under most conditions, directional growth on agar surfaces is associated with the polarity of epidermal cell file twisting at the root tip: Left-handed CFR occurs when the root coils in the counterclockwise direction, whereas right-handed CFR accompanies clockwise coiling (Okada and Shimura, 1990; Sedbrook et al., 2002). On tilted agar surfaces, wild-type roots of the No-0 ecotype alternated between left-handed and right-handed epidermal CFR, with a strong bias observed in favor of left-handed CFR (Fig. 2, A and D). Homozygous wvd2-1 roots, on the other hand, possessed a constitutive right-handed CFR phenotype (Fig. 2, C and F). The underlying cortical cell files of wvd2-1 also exhibited right-handed twisting (data not shown). A bias to-handed right-handed CFR was also observed in heterozygous wvd2-1 seedlings, although they periodically showed evidence of left-handed CFR as well (Fig. 2, B and E).
When grown embedded within a homogenous environment of 1.5% (w/v) agar-solidified growth medium, both wild-type and wvd2-1 mutant roots grew straight down. However, wild-type roots still displayed left-handed CFR (Fig. 2G), whereas mutant roots displayed right-handed CFR (Fig. 2H). Furthermore, the cell-bulging phenotype of wvd2-1 roots was more evident than when grown on the surface of the medium.
Under our normal growth conditions, the roots of wvd2-1 seedlings appear to be both shorter and thicker than wild type. To determine whether defects in anisotropic cell expansion contribute to the altered morphology of wvd2-1 roots, measurements of the dimensions of cells within the mature zone of wild-type and mutant roots were obtained. As shown in Table I, both cortical and epidermal root cells were shorter in wvd2-1 (67% and 56% the length of wild type, respectively). The periclinal width of these cells conversely was significantly larger in the mutant (48% and 44% wider, respectively). Reduced cell anisotropy was also observed in cells of the endodermal layer of the root (data not shown). No increase in cell number was observed in any of the layers of mutant roots observed in cross section (data not shown). Thus, impaired cell anisotropy appears to account for a large portion of the root elongation and expansion phenotypes of wvd2-1.
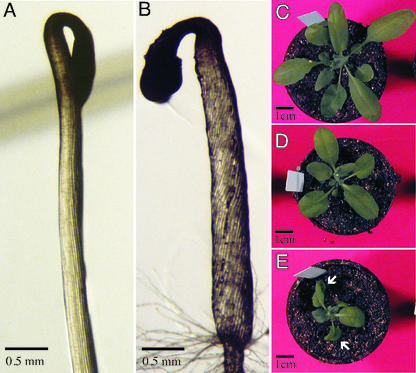
The wvd2-1 mutation also causes expansion-related defects in several shoot organs, including hypocotyls, siliques, and rosette leaves (Table I). However, seed size was not altered (data not shown). Moreover, defects in the polarity of axial rotation were also observed in shoot organs. The hypocotyls of etiolated wvd2-1 seedlings showed constitutive right-handed epidermal CFR (Fig. 3B), whereas the cell files of wild-type hypocotyls appeared to be linear with the axis of elongation (Fig. 3A). Interestingly, the petioles of homozygous wvd2-1 rosette leaves exhibited left-handed CFR, resulting in a consistent clockwise leaf curling (Fig. 3E). The rosette leaves of wild-type and heterozygous wvd2-1 plants generally did not display a tendency to curl (Fig. 3, C and D).
Figure 3.
Shoot phenotypes of wvd2-1. Close-up images of hypocotyls from wild-type No-0 (A) and wvd2-1 (B) seedlings grown in darkness on vertical 1.5% (w/v) agar plates, 4 d after germination (DAG). Rosette leaves of wild-type (C), heterozygous (D), and homozygous (E) wvd2-1 seedlings. Arrows indicate leaves showing prominent clockwise curling. Photographs were taken within 24 h of bolting.
Effect of wvd2-1 on Cortical Microtubules
Several mutants with defects in anisotropic cell expansion and root growth behavior display defects in cortical microtubule alignment within cells of the root elongation zone (Furutani et al., 2000; Thitamadee et al., 2002; Wasteneys, 2002). We examined the root growth behavior of wvd2-1 and wild-type seedlings in the presence of compounds that affect microtubule organization.
The addition of 1 to 3 μm propyzamide, a microtubule-destabilizing compound, to the growth medium caused an exaggerated rightward root-slanting phenotype in wild-type No-0 seedlings and enhanced leftward skewing of wvd2-1 roots (Table II). The increase in angle of root skewing was greater for wvd2-1 than for wild type at both concentrations (Table II). The addition of oryzalin, another microtubule-destabilizing compound (Baskin et al., 1994), to the growth medium also resulted in enhanced alteration of wvd2-1 mutant root elongation compared with wild type (supplementary data).
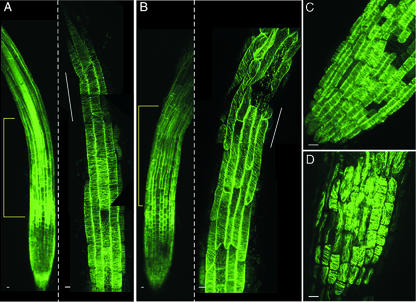
To determine how the arrangement of cortical microtubule arrays in elongating wvd2-1 root cells compared with those of wild-type, whole-mount immunolocalization experiments were performed on 4-d-old mutant and wild-type seedlings grown on vertical 0.8% (w/v) agar plates to diminish the effects of root waving. Consistent with the observations reported by Liang et al. (1996) and Sugimoto et al. (2000), epidermal cells in the elongation zone of wild-type No-0 roots possessed microtubule arrays that were predominantly transverse in alignment (Fig. 4A). On the other hand, a left-handed spiral arrangement of cortical microtubules was apparent within the elongating cells of wvd2-1 roots (Fig. 4B). This spiral arrangement was found in epidermal cells located on the distal side of CFR initiation, and became more pronounced as cells approached the region of the elongation zone where CFR became apparent. Although cells directly at the site of CFR initiation had a strongly left-handed cortical microtubule orientation (Fig. 4B), the cells located on the basal side of CFR initiation displayed a nearly longitudinal microtubular alignment (data not shown). Cortical microtubule organization was also altered in wvd2-1 peripheral root cap cells, with cells possessing transverse, longitudinal, or oblique arrays (Fig. 4D). Wild-type peripheral root cap cells, on the other hand, possessed uniformly transverse microtubule alignments (Fig. 4C).
Figure 4.
Arrangement of cortical microtubules. A and B, Localization of cortical microtubules within the primary roots of 4-d-old wild-type (A) and wvd2-1 (B) seedlings. The images shown to the right of the dashed lines in A and B correspond to the bracketed region of the roots shown to the left of the dashed lines, taken at a higher magnification. White lines along the roots indicate the site of CFR initiation. The images were constructed from overlapping confocal images taken along the length of the roots. C and D, Localization of cortical microtubules within the peripheral root cap cells of wild-type (C) and wvd2-1 (D) seedlings. Scale bar = 10 μm.
WVD2 Encodes a Small Hydrophilic Protein
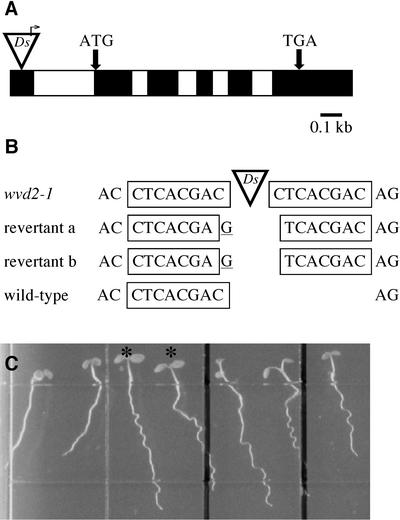
Ds(HYG 35S), the transposon used to generate the activation-tagged population from which wvd2-1 was isolated, harbors both a hygromycin resistance gene (HPT) and an outward-directed cauliflower mosaic virus (CaMV) 35S promoter (Wilson et al., 1996). Cosegregation analysis of the wvd2-1 and hygromycin resistance phenotypes in 114 F3 families (derived from crosses between homozygous wvd2-1 and wild-type plants) indicated complete linkage between the mutant phenotype and a Ds(HYG 35S) insertion. Using inverse PCR, the genomic sequence immediately flanking one end of the Ds element was obtained. This sequence was found to match a segment on the upper arm of chromosome 5, near the nga76 marker. A cDNA clone (p6D1) matching the Ds-disrupted locus was isolated from an Arabidopsis cDNA library (CD4-7) and sequenced. Subsequent primer extension analysis revealed that full-length transcripts possess an additional 64 bases upstream of the 5′ end of p6D1. A comparison of the full-length cDNA and genomic sequences revealed that the identified gene consists of six exons and five introns, and encodes a transcript 1.0 kb in length. The Ds(HYG 35S) element cosegregating with the wvd2-1 phenotype is inserted within the first exon of this gene, 362 bp upstream of the first potential start codon, with its CaMV 35S promoter directed toward the gene's open reading frame (Fig. 5A). The WVD2 gene was predicted by genome annotations (At5g28646 [gi:22327159]), although the annotated sequence omits the first and fourth exons depicted in Figure 5A.
Figure 5.
The wvd2-1 mutation is caused by the insertion of a Ds(HYG 35S) element within the 5′-untranslated region of WVD2. A, Structure of the wvd2-1 allele. The dark boxes indicate exons, whereas the light boxes represent introns. The site of Ds(HYG 35S) insertion within the wvd2-1 allele is denoted by ▿, with the direction of CaMV 35S promoter transcriptional activation represented by an arrow. The positions of the predicted start and stop codons of the WVD2 open reading frame are indicated by ATG and TGA, respectively. B, Genomic sequence immediately flanking the Ds insertion site. Insertion of the Ds element is accompanied by an 8-bp target site duplication (boxed sequence). In two independently isolated revertant lines, excision of the Ds element also caused a C→G transversion and a single-base pair deletion, resulting in a 7-bp footprint. C, Root-waving phenotype of a wvd2-1 revertant line. On the left, progeny derived from the self-fertilization of a plant heterozygous for wvd2-1 and revertant allele a. Note the wavy appearance and rightward skewing of the roots of seedlings denoted with an asterisk. Wild-type No-0 seedlings are in the middle (between the two black bars), and a Landsberg erecta (Ler) seedling is located at the right. This picture was taken from below the plate.
Two genetically stable revertant lines were identified among the progeny of wvd2-1 plants harboring the Nae35S-Ac element (Fig. 5C). PCR amplification of the genomic sequence flanking the original Ds insertion site, followed by sequencing of the amplification products, revealed that the reversions were accompanied by Ds excision events, which left behind an identical 7-bp footprint in both cases (Fig. 5B). Our revertant analysis indicates that a Ds(HYG 35S) element insertion within this gene, which we have designated as WVD2, is responsible for the phenotype of wvd2-1 plants.
On the basis of the sequence of its cDNA, the deduced WVD2 protein (Fig. 6A) is 23 kD in size, is highly hydrophilic, and is predicted to localize in the cytoplasm. Its peptide sequence lacks significant stretches of similarity to proteins of known function. The amino acid content of WVD2 is biased toward Glu, Lys, and Ser, which constitute 14% (Glu) and 11% each (Lys and Ser) of all amino acids within the protein. WVD2 contains a short region (residues 89–172) that shares homology with seven predicted proteins from Arabidopsis (Fig. 6, B and C). With the exception of the predicted gene product of At1g70950, which possesses 50% identity to only a subregion of the conserved domain of WVD2 (residues 89–149), the degree of identity between WVD2 and the remaining six proteins within this conserved domain ranges from 50% to 88% (Fig. 6C). We have termed this region of conservation the KLEEK domain, after the five contiguous residues invariantly found in all eight Arabidopsis proteins (Fig. 6C). According to the PAIRCOIL program (Berger et al., 1995), a portion of the KLEEK domain is predicted to form a coiled-coil structure (WVD2 residues 92–136 [P = 0.892]). BLAST searches performed with the KLEEK domain of WVD2 have identified additional KLEEK-containing proteins from a wide range of plant species, including barley (Hordeum vulgare), corn, cotton (Gossypium hirsutum), pine (Pinus spp.), potato (Solanum tuberosum), rice (Oryza sativa), rye (Secale cereale), sorghum (Sorghum bicolor), soybean (Glycine max), and tomato (Lycopersicon esculentum). However, we were unable to identify any non-plant proteins possessing this motif (data not shown).
Figure 6.
The gene product of WVD2 contains the novel KLEEK domain. A, The deduced amino acid sequence of WVD2. Residues corresponding to the conserved domain are underlined. B, Pair-wise similarity between predicted Arabidopsis proteins containing the KLEEK domain. Each predicted protein is assigned an identification number (1–8), which is defined on the right of the table, and is represented in the first and last lanes and columns of the table. Percent identity between two proteins is represented in the corresponding table box. C, Alignment of a region surrounding the KLEEK domain for all eight predicted KLEEK-carrying Arabidopsis proteins. Residues matching the consensus are shaded. In B and C, alignment was performed using MegAlign (ClustalV method: gap penalty = 10, gap length penalty = 10, PAM250 Matrix; DNASTAR, Inc., Madison, WI).
The product encoded by At3g04630, which we hereafter refer to as WVD2-Like1 (WDL1), shares the most similarity to WVD2 over its entire length (59.9% amino acid identity; Fig. 6B). The sequence of full-length WDL1 cDNA (Ceres cDNA clone 39706) indicates that the predicted WDL1 protein has a molecular mass of 32 kD and is also highly hydrophilic. The two next most similar proteins to WVD2 from Arabidopsis (encoded by At1g54460 and At3g23090) share 41.1% and 40.1% identity, respectively, with WVD2 over its length. The other four predicted proteins share only limited sequence similarity with WVD2 (25.7%–34.2% identity), essentially limited to the KLEEK domain.
Altering Expression of WVD2 and WDL1 Modulates Organs' Growth Behavior
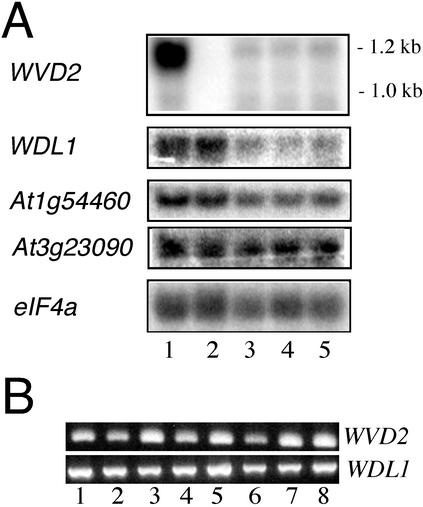
Because Ds(HYG 35S) carries an outward-transcribing CaMV 35S promoter on one of its flanks, northern-blot analysis was performed to assess whether WVD2 was up-regulated in wvd2-1 tissues. As shown in Figure 7A, a highly abundant 1.2-kb transcript was detected by WVD2-specific probes in total RNA extracted from mutant (lane 1) but not wild-type (lane 2) tissues. On the other hand, no differences in level of expression were detected for three other genes showing similarity to WVD2, relative to an eIF4A loading control, indicating that the wvd2-1 phenotype was most likely the result of increased expression of WVD2 rather than silencing of a related gene.
Figure 7.
A, Northern-blot analysis of WVD2 expression. Total RNA was prepared from wvd2-1 (lane 1), wild-type No-0 (lane 2), and silencing lines wvd2[siWVD2]-1 (lane 3), wvd2[siWVD2]-3 (lane 4), and wvd2[siWVD2]-4 (lane 5). Seedlings were grown for 5 d on 1.5% (w/v) agar plates, tilted backward 30°. Blots were hybridized with gene-specific probes, as indicated. Note that the expected size of wild-type WVD2 transcripts is 1.0 kb, but transcripts from the wvd2-1 mutant allele are 1.2 kb in length because of sequences corresponding to the border of the Ds(HYG 35S) element. The coding potential of mutant transcripts is unchanged (data not shown). B, Tissue-specific RT-PCR analysis of WVD2 and WDL1 expression. Using gene-specific primers, RT-PCR analysis was performed with 0.5 μg of total RNA extracted from light-grown whole seedlings (lane 1), hypocotyls and cotyledons (lane 2), roots (lane 3), dark-grown whole seedlings (lane 4), flowers (lane 5), rosette leaves (lane 6), siliques (lane 7), and stems (lane 8). All tissues were derived from wild-type Columbia (Col) plants. Tissue samples were collected from plants grown on tilted 1.5% (w/v) agar plates, 4 to 5 DAG (lanes 1–3); from plants grown on vertical 1.5% (w/v) agar plates, 4 d after germination (lane 4); or from soil-grown plants, 4 to 6 weeks after germination (lanes 5–8). Both primer pairs flank a single intron of their respective genes. Sequencing of WVD2 and WDL1 RT-PCR amplification products confirmed that they did not include intron-related sequences (data not shown).
To verify that the increased expression of WVD2 was responsible for the wvd2-1 mutant phenotype, we transformed wild-type No-0 plants with a construct designed to express the open reading frame of WVD2 under the control of the CaMV 35S promoter (p35S:WVD2). Of 20 transgenic seedlings recovered, 17 independent T2 lines segregated for root-waving, root-skewing, and root elongation phenotypes. Although some phenotypic variability existed between transgenic lines, presumably because of position-mediated effects on transgene expression, the majority possessed phenotypes strongly reminiscent of wvd2-1, including occasional bulging of epidermal cells, increased root hair production, and clockwise curling rosette leaves (data not shown). Similarly to heterozygous wvd2-1 seedlings, the three transgenic lines with the weakest wvd2-like phenotypes still exhibited predominantly right-handed CFR in roots, but periodically switched to left-handed CFR when grown on tilted agar surfaces (data not shown). The root growth pattern of a representative line on tilted agar surfaces is shown in Figure 1B. Northern-blot analysis confirmed the expression of the transgene in these lines (data not shown).
To determine whether CaMV 35S-driven expression of WDL1 would also result in phenotypes similar to those observed in wvd2-1, wild-type No-0 seedlings were transformed with the p35S:WDL1 construct (see “Materials and Methods”). In the T2 generation, 14 of the 17 transgenic lines isolated segregated for altered root growth phenotypes, the majority of which possessed short roots that did not wave and slanted to the left on tilted agar surfaces (Fig. 1C). The shoot morphology of 35S:WDL1 transgenic plants was also highly similar to wvd2-1 (data not shown).
In plants, the simultaneous expression of sense and antisense RNA can induce gene-specific silencing at high frequency (Waterhouse et al., 1998). Because wild-type WVD2 transcripts were not detectable via northern-blot analysis, we used the wvd2-1 mutant background to examine the effects of transgene-mediated silencing of WVD2 on root skewing, thereby allowing us to confirm transcriptional suppression through northern-blot analysis. Transgenic wvd2-1 seedlings harboring the p-siWVD2 silencing construct, which expresses inverted copies of the WVD2 cDNA under the CaMV 35S promoter, exhibited suppression of the mutant phenotype (Fig. 1D). The onset of silencing varied among different lines, with some wvd2[siWVD2] lines appearing noticeably different from wvd2-1 seedlings 4 d after germination, whereas other lines took as long as 10 d after germination to exhibit suppression of the wvd2-1 phenotype. In the latter case, roots would initially skew leftward on a tilted agar surface and then switch to rightward slanting (data not shown). Interestingly, quantification of the mean root-skewing angles for four representative lines displaying early onset of wvd2-1 suppression indicated that the rightward root skewing of these lines was increased over wild-type No-0 seedlings. Root skewing varied between 27° ± 8° and 32° ± 7° from the vertical in these silenced lines, compared with 20° ± 7° from the vertical for untransformed No-0 roots. Differences in average root-skewing angle between silenced wvd2-1 and wild-type lines were all statistically meaningful (t test P values varying between 3.5 × 10−8 and 2.7 × 10−20). Transcripts derived from the wvd2-1 allele were diminished in the silenced lines, reaching levels barely detectable by northern-blot analysis (Fig. 7A). The levels of WDL1 transcripts also were substantially diminished in all silenced lines analyzed, whereas those of At1g54460 changed only slightly. On the other hand, the levels of At3g23090 transcripts remained unchanged in these lines. Hence, silencing appeared to affect expression of the genes with the highest similarity to WVD2 at the cDNA sequence level (Fig. 7A; data not shown).
WVD2 and WDL1 Are Expressed in Multiple Plant Tissues
We performed reverse transcriptase (RT)-PCR on RNA extracted from a variety of organs to determine the wild-type expression pattern of WVD2 and WDL1. For both sets of experiments, the gene-specific primer pairs amplified a region spanning an intron in genomic DNA, thereby allowing us to distinguish between PCR products derived from reverse transcription of mRNA and amplification products arising from genomic DNA contamination. As shown in Figure 7B, using WVD2-specific primers, RT-PCR products of the expected size (550 bp) were detected in 0.5 μg of total RNA from seedlings grown under the standard root-wave assay conditions (lane 1) or etiolated seedlings grown on vertically positioned plates (lane 4). Furthermore, WVD2 transcript was detected in both cotyledon/hypocotyl-specific (lane 2) and root-specific (lane 3) RNA preparations. Transcript was also detected in tissue-specific RNA extracts from the flowers, rosette leaves, siliques, and inflorescence stems of wild-type plants (lanes 5–8). Similar results were obtained when WDL1-specific primers were used (Fig. 7B). Therefore, both WVD2 and WDL1 appear to be expressed in most organs of wild-type Arabidopsis.
DISCUSSION
We have identified wvd2-1, a mutant that exhibits defects in both the expansion and rotational polarity of plant organs. The mutant allele contains an activation-tagging Ds element inserted within the 5′-untranslated region of WVD2, driving increased expression of transcripts with full-coding potential. Because the wvd2-1 mutation can be phenocopied by the 35S:WVD2 transgene, the mutant phenotype is most likely the result of ectopic expression and/or overexpression of WVD2. Furthermore, the effects of WVD2 expression appear to be dosage dependent, because heterozygous wvd2-1 seedlings possess an intermediate phenotype. Reduction of WVD2, WDL1, and At1g54460 transcript levels in a wvd2-1 background reverses the phenotypes and results in enhanced root slanting in the wild-type direction.
The wvd2-1 mutation also causes roots to be shorter and thicker than wild type under our growth conditions. This appears to result primarily from impaired anisotropic cell expansion resulting in increased radial expansion and decreased elongation in all layers of the root (Table I) and in most organs of the plant (data not shown). However, the smaller average length of cells in the mature zone of mutant roots compared with wild type (56% for epidermal cells, and 66% for cortical cells) is not sufficient to account for the full reduction in root growth (46%) also associated with the mutation (Table I; data not shown). Root growth is conditioned by a combination of cell division and cell expansion, and we have not excluded the possibility that the mutation might affect both processes.
Even though several phytohormones have been found to modulate anisotropic cell expansion in plants (for review, see Shibaoka, 1994), it appears that the cell expansion defect of wvd2-1 is not associated with dramatic changes in sensitivity to phytohormones. Slight increases in root growth resistance to indole-3-acetic acid (IAA), naphthalene acetic acid (NAA), 1-aminocyclopropane-1-carboxylic acid (ACC), and 6-benzyladenine (BA) were observed in mutant seedlings (supplementary data). However, these changes in root growth sensitivity were much smaller than those typically found in phytohormone-resistant mutants (Chang et al., 1993; Leyser et al., 1993; Chen et al., 1998) and were similar in all phytohormone response assays. These results suggest that the altered phytohormone sensitivity of wvd2-1 may not be specific and could result indirectly from the profound effect of wvd2-1 on anisotropic cell expansion.
On the other hand, increasing Suc concentrations in the medium exacerbated radial expansion phenotype of wvd2-1 roots, whereas wild-type root expansion was not modified under these conditions (supplementary data). This Suc effect was highly specific, because mannitol had little or no effect on this aspect of the phenotype (supplementary data). Therefore, the radial expansion defect of wvd2-1 roots may involve pathways or processes similar to those responsible for the phenotypes of the Arabidopsis conditional root expansion mutants, which also exhibited increased radial expansion in the presence of high concentrations of Suc (Hauser et al., 1995). We hypothesize that the Suc effect on radial cell expansion in roots may be related to its activity as a signaling molecule (Smeekens, 2000).
Whereas wild-type roots exhibited a wavy, rightward-slanting growth pattern on tilted agar surfaces, the roots of wvd2-1 seedlings did not wave significantly and slanted to the left under these conditions. This altered growth behavior of mutant roots on agar surfaces was accompanied by an apparent shift in the polarity of root directional growth bias, as evidenced by their epidermal CFR phenotype. Homozygous wvd2-1 roots possessed constitutive righthanded CFR when grown on tilted agar plates, which is typically associated with clockwise coiling on horizontal agar surfaces (Okada and Shimura, 1990; Simmons et al., 1995; Rutherford and Masson, 1996; Sedbrook et al., 2002). The leftward-skewing phenotype of wvd2-1 roots may therefore derive from the combined influence of gravitropism and a surface-dependent clockwise coiling behavior in the mutant (Rutherford and Masson, 1996).
The net growth vector of wvd2-1 roots was influenced by the angle at which the agar surface was positioned, with the smallest leftward deviation from the gravity vector occurring when the agar plates were positioned vertically. Backward plate tilting is believed to enhance the interaction between roots and the surface, because of gravitropism directing root growth into the medium (Okada and Shimura, 1990). Thus, one possible interpretation of the increased leftward skewing of wvd2-1 roots on inclined surfaces is that an increase in surface-derived tactile stimulation causes an enhanced root-coiling phenotype. Because wvd2-1 roots appear to coil constitutively in the clockwise direction, this enhancement of root coiling results in a more dramatic left-slanting phenotype. Wild-type No-0 root skewing, on the other hand, appeared to be less dependent upon the orientation of the surface. This may be attributable to the ability of wild-type roots to alternate between clockwise and counterclockwise coiling. If the increase in tactile stimulation caused by backward plate tilting affects coiling in both directions equally, the net change in angle of root skewing may be negligible. However, increased coiling in both directions would be anticipated to result in an increased frequency and/or amplitude of root waving. Backward plate tilting has previously been shown to enhance the wavy growth pattern of wild-type roots (Okada and Shimura, 1990; Simmons et al., 1995; Mullen et al., 1998).
It is also possible that wvd2-1 displays a constitutive or enhanced thigmoresponse phenotype. Under the alternating circumnutation model of root waving, formation of the wavy growth pattern involves touch-induced reversions of root tip rotational polarity, resulting in back-and-forth changes in the direction of root coiling (Okada and Shimura, 1990; Rutherford and Masson, 1996). On the basis of the WVD2 and WDL1 overexpression phenotypes, the wild-type function of these genes may be to promote a switch from the default (left-handed) direction of wild-type root-tip rotation to right-handed rotation in response to a perceived stimulus. The thicker root phenotype of wvd2-1 may alternatively result in increased contact of the root with the agar surface, thereby allowing for enhanced stimulation of the root (and thus, exaggerated coiling). Finally, we cannot eliminate the possibility that the increased diameter of mutant roots also contributes to the suppression of wvd2-1 root waving. In general, plant species that develop relatively thick roots do not exhibit strong waving patterns on agar surfaces (Simmons et al., 1995). We have similarly observed that several mutants characterized by root expansion defects similar to wvd2-1, including the eto (Guzman and Ecker, 1990) and erh (Schneider et al., 1997) mutants, also lack root waving on tilted agar plates (C. Yuen and P.H. Masson, unpublished data). However, it is interesting to note that, unlike wvd2-1, these other mutants do not skew significantly to either the left or right under these conditions (C. Yuen and P.H. Masson, unpublished data).
A growing body of evidence suggests that the direction of cortical microtubules is a major determinant in the rotational polarity of roots (Liang et al., 1996; Furutani et al., 2000; Thitamadee et al., 2002). Similar to the leftward-skewing spr1 mutant, the elongating root cells of wvd2-1 contain microtubule arrays with a left-handed oblique organization. This left-handed arrangement is observed in root cells apical to the position where cell file twisting is first observed in wvd2-1, but becomes more prominent in cells proximal to the site of CFR initiation (Fig. 4). Cells basal to this position appear to adopt a longitudinal alignment (Fig. 4), similar to that observed in nonelongating cells at the mature zone of wild-type roots (Liang et al., 1996; Sugimoto et al., 2000). Cortical microtubules have long been hypothesized to regulate anisotropic cell expansion by defining the orientation of cellulose microfibril deposition within the cell wall (Giddings and Staehelin, 1991) and may also be involved in hemicellulose deposition (Burk et al., 2001). Therefore, the observed misalignment of cortical microtubules in wvd2-1 cells may potentially account for both the reduced cell anisotropy and clockwise root-coiling phenotypes of wvd2-1.
On the basis of the preceding discussion, we hypothesize that WVD2 interacts directly or indirectly with microtubules, altering their dynamics and consequently affecting anisotropic cell expansion and root growth behavior on surfaces. This model is consistent with the fact that wvd2-1 enhances root growth sensitivity to intermediate concentrations of oryzalin, a drug that destabilizes cortical microtubules (see above), and with the observation that propyzamide, another microtubule-destabilizing compound, enhances the root slanting of wvd2-1 to a greater degree than wild type (Table II). Both WVD2 and WDL1 are predicted to possess coiled-coil domains, a structural configuration implicated in a number of protein-protein interactions. It is notable that plants highly expressing a transgene that encodes a green fluorescent protein-MAP4 chimeric protein also possess a strong, leftward root-skewing phenotype (Hashimoto, 2002). MAP4 is a micro-tubule-associated protein that is believed to assist in the polymerization and stabilization of microtubules (Olsen et al., 1995). Moreover, mutations within two genes encoding α-tubulins (lefty1 and lefty2) were shown to enhance root slanting in the wild-type direction (Thitamadee et al., 2002). It should, however, be cautioned that it remains formally possible that the altered cytoskeleton arrangement of elongating wvd2-1 cells is caused by changes in the architecture of the cell wall (Fisher and Cyr, 1998; Sugimoto et al., 2001; Takeda et al., 2002).
Further characterization is necessary to determine whether the phenotypes associated with wvd2-1 are the consequence of overexpression in functionally relevant cells or ectopic expression in cells that do not normally transcribe WVD2 and/or WDL1. Interestingly, although transgene-mediated cosuppression of genes encoding WVD2 and WVD2-like proteins restores rightward root slanting and wild-type organ expansion and morphology in the wvd2-1 mutant background, the angle of root skewing is significantly greater than that of wild-type controls. Although expression of the wvd2-1 mutant allele is reduced to almost undetectable levels in silenced lines, these seedlings still possess a greater abundance of WVD2 transcripts than wild type. However, the siWVD2 transgene also reduced the expression level of WDL1 and At1g54460 in silenced lines (Fig. 7A). These observations suggest that a reduction in the net abundance of transcripts from both WVD2 and WVD2-like genes results in a phenotype (increased rightward root slanting) that is the opposite of the wvd2-1 mutation. This is consistent with our hypothesis that WVD2 and its paralogs may promote clockwise root coiling in wild-type plants. Expression of both genes is detected in a broad range of wild-type plant organs, indicating that they may modulate helical growth and/or anisotropic cell expansion processes throughout wild-type Arabidopsis plants. Interestingly, whereas plants overexpressing either WVD2 or WDL1 were characterized by right-handed helical growth phenotypes in roots and etiolated hypocotyls, they also exhibited left-handed petiole twisting and clockwise leaf curling (Fig. 3E; data not shown). This may indicate that the specific activity of WVD2 and WDL1 is dependent upon the organ in which it is expressed. By contrast, the spr2, lefty1, and lefty2 mutations cause organ polarity defects of the same direction in both roots and petioles (Hashimoto, 2002).
In summary, the root growth behavior exhibited by wvd2-1 (this study) and spr1 (Furutani et al., 2000) mutant seedlings indicates that mutations that reverse the direction of the axial rotational bias promote opposite directional preference of root coiling on horizontal surfaces and opposite slanting on tilted or vertical media. Our characterization of WVD2 and WDL1 strongly suggests that both genes function as positive regulators of right-handed helical growth in roots and etiolated hypocotyls, possibly by affecting the dynamics of cortical microtubules within elongating cells. The latter effect appears distinct from that of spr1 (Furutani et al., 2000), considering the differential slanting response displayed by these mutants to intermediate concentrations of propyzamide (reversal of root skewing for spr1 [Furutani et al., 2000] and enhancement for wvd2-1 [Table II]). In any case, the role of WVD2 and WDL1 in Arabidopsis roots may be to modulate the polarity of circumnutation. Such adjustments in the direction and of root coiling in response to asymmetric tactile stimulation may facilitate the ability of roots to maneuver in natural soil environments by allowing them to avoid impediments to growth. The stems of twining plants also exhibit coiled growth patterns allowing them to grow around objects for structural support. Although Arabidopsis stems do not coil, it is tempting to speculate that modulation of root coiling in Arabidopsis and stem coiling in twining plants may involve similar processes.
MATERIALS AND METHODS
Plant Stocks and Manipulation
Seeds of the Arabidopsis ecotypes Col, Ler, and No-0 were obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus). Transgenic Ler seeds carrying the Ds(HYG 35S) transposon (Wilson et al., 1996) were supplied by George Coupland (John Innes Centre, Norwich, UK), and Nina Fedoroff (Pennsylvania State University, University Park) provided seeds from an No-0 line containing the disabled (non-transposing) Ac construct Nae35S-Ac (Fedoroff and Smith, 1993). All techniques and conditions used to surface sterilize and germinate Arabidopsis seeds and to grow seedlings and plants were as described by Rutherford and Masson (1996), except where noted otherwise.
Assays pertaining to root elongation in the presence of phytohormones, oryzalin, mannitol, or increased levels of Suc were conducted as described by Sedbrook et al. (1999). IAA, NAA, and ACC were purchased from Sigma-Aldrich (St. Louis) and prepared as 0.1 mm IAA or 1 mm NAA and ACC stock solutions in ethanol. Oryzalin (Chem Service, West Chester, PA) and BA (Sigma-Aldrich) were prepared as 0.5 mm and 10 mm stock solutions, respectively, in dimethyl sulfoxide (DMSO). These compounds were added to the pH-buffered medium at the concentrations defined in the text. All plates in one experiment contained the same amount of solvent (0.05% [v/v] ethanol for experiments involving IAA, NAA, and ACC; 0.1% [v/v] DMSO for experiments involving BA; and 0.05% [v/v] DMSO for experiments involving oryzalin). The kinetics of root growth in the presence or absence of drugs and the kinetics of root tip reorientation in response to gravistimulation were quantified as described by Rutherford et al. (1998). Data were subjected to statistical analysis (t and F tests) using the Microsoft Excel program (MS Office 98, Microsoft, Redmond, WA).
For assays of root waving/skewing, we used square petri dishes containing one-half-strength Murashige minimal organics medium (Invitrogen, Carlsbad, CA) solidified with 1.5% (w/v) agar (type E, Sigma-Aldrich). The Suc content within the medium was 1.5% (w/v). After sowing seeds, the plates were wrapped with paper surgical tape (Micropore, 3M, St. Paul). The plates were then kept in darkness, at 4°C, for 2 to 4 d. Afterward, the plates were transferred to a growth chamber (22°C, 100% relative humidity, and 16-h/8-h light/dark cycle; TC16, Conviron, Winnipeg, Manitoba, Canada). Plates were positioned vertically for the first 3 d of seedling growth, then inclined backward 30° (unless otherwise specified), and returned to the growth chamber. Seedlings were photographed with a digital camera (Coolpix800, Nikon, Tokyo) 6 to 7 DAG, and the digitized images were analyzed as described by Rutherford and Masson (1996). The effect of propyzamide on root slanting was tested by germinating and growing wild-type No-0 and wvd2-1 mutant seeds on media containing propyzamide (Chem Service) at the concentrations defined in the text.
Ds Transposition and Activation Screening for Arabidopsis Root-Waving Mutants
Activation-tagging mutagenesis was performed essentially as described by Wilson et al. (1996), except that crosses to plants harboring the Nae35S-Ac element were performed to induce Ds(HYG 35S) element transposition. Approximately 1,800 independent transposition lines were obtained in the F2 generation. Seedlings from the subsequent F3 generation were screened for abnormal root-waving and/or -skewing phenotypes on tilted agar surfaces. The wvd2-1 mutant was among those identified in the screen. Lines derived from at least four cycles of wvd2-1 introgression into the No-0 ecotype were used to obtain all quantitative data for wvd2-1.
To isolate revertant alleles of WVD2, the progeny of plants identified as being homozygous for the wvd2-1 Ds insertion allele and also containing the Nae35S-Ac element were germinated on tilted agar plates. DNA was extracted from the cotyledons of revertant plants showing a wild-type phenotype under the root-waving assay (Klimyuk et al., 1993), and WVD2-specific primers flanking the Ds insertion site (5′-GCTCACCGACTCAACAACTTCT-3′ and 5′-AATATCCAACAACTTAACAAACCA-3′) were used to screen for putative excision alleles via PCR. The amplification products were then sequenced to check for the presence of excision footprints. Putative revertant plants were allowed to self-pollinate, and the resulting progeny were genotyped and phenotyped to determine the heritability of detected excision events.
Cloning WVD2 and Isolation of cDNA
The sequence immediately flanking the Ds insertion site of wvd2-1 was obtained by inverse PCR (Long et al., 1993) using ligated fragments of XhoI-digested genomic DNA as a template. The first round of PCR used primers DL6 (Long et al., 1993) and Ds-a1 (Bancroft et al., 1993), whereas the second round used primers DL6 and DL3 (Long et al., 1993). Employing the plaque screening method described by Ausubel et al. (1994), a partial WVD2 cDNA clone (p6D1) was isolated from the CD4-7 Arabidopsis λ-PRL2 cDNA library (ABRC). Probes used in this screening were PCR-amplified from Wassilewskija genomic DNA with the primer pair 5′-TGGTGAAATATGACAGTAAGTTGGG-3′ and 5′-CTCAGTGCATTCTTTCTCCTCAT-3′ and 32P-labeled using the Multiprime labeling system (Amersham Biosciences AB, Uppsala).
Generation of T-DNA Constructs and Plant Transformation
The p35S:WVD2 construct was generated by cloning a PCR-amplified copy of the WVD2 open reading frame between the unique NcoI and BstEII restriction sites of pCAMBIA1302, immediately downstream of a CaMV 35S promoter cassette. An NcoI restriction site overlapping the start codon of WVD2 was added to the forward primer (5′-GAAATATGACAGTAAGTTGCCATGGGAAGAGA-3′), and a BstEII site immediately following the stop codon was added to the reverse primer (5′-GGAAGCTTTTTGGGTCACCTCATTCTACCACAC-3′). The p35S:WDL1 construct was generated in a similar manner, using WDL1 primers (5′-ATGTCCAGTGCCATGGGAAGAGAAGTTGTTG-3′ and 5′-ATCATCTTTGGTCACCTCAAGCTTCTTCTGA-3′).
The WVD2 cosuppression construct (p-siWVD2) was created by inserting an antisense-oriented copy of the WVD2 coding region between the NcoI and SpeI restriction sites of pCAMBIA1381 (upstream of the GUS gene), and a sense-oriented copy at the BstEII site (downstream of GUS). Both antisense and sense WVD2 fragments were PCR-amplified, using the p6D1 cDNA clone as template (antisense primers, 5′-CTTAAACTAGTATGAGAAGAGAAGTTGTTGAG-3′ and 5′-GAGACCATGGATT-CTACCACACTCTGGCGATCC-3′; sense primers, 5′-TTAGGTGACCATGAGAAGAGAAGTTGTTGAG-3′ and 5′-GAGGGTCACCATTCTACCACACTCTGGCGATTC-3′). A 2× CaMV 35S promoter fragment (obtained by XmaI/SpeI digestion of pCAMBIA1302) was subsequently inserted between the XmaI and SpeI sites of the vector, directly upstream of the antisense WVD2 cassette, to initiate transgene transcription in planta. Finally, to allow for kanamycin resistance selection in transformed plants, the HPTII cassette of pCAMBIA1381 was replaced with the NPTII cassette of pCAMBIA2300 through XmaI/SacII digestion, followed by vector/insert ligation.
The aforementioned pCAMBIA binary vectors (CAMBIA, Canberra, Australia) were kindly provided by Anthony Bleecker (University of Wisconsin, Madison). Constructs were introduced into plants via Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998).
Northern-Blot and RT-PCR Analysis
Total RNA was isolated from plant tissues using the RNeasy Plant Mini kit (Qiagen USA, Valencia, CA) and subjected to northern-blot analysis, as described in the QIAGEN Guide to Analytical Gels, Parts IV-VI (Qiagen USA, 2000a, 2000b, 2000c). 32P-labeled probes were generated by random priming (Multiprime labeling system, Amersham Biosciences AB). The template for WVD2 probe generation was an EcoRI/XbaI fragment liberated from the vector backbone of our isolated WVD2 cDNA, p6D1. Template for WDL1 probes was obtained by EcoRI/XbaI digestion of a partial WDL1 cDNA clone, 170L2 (ABRC). Probes specific to At1g54460 and At3g23090, which are both predicted to encode products with sequence similarity to WVD2 and WDL1, were amplified from a cDNA library (CD4-22, ABRC) by PCR (At1g54460 primers, 5′-GACAAACACATGGATAAGAAAGCGAATAGT-3′ and 5′-ATAAGAAAAGATCATACGAAAATAGAGCA-3′; At3g23090 primers, 5′-AAAGAGTGTACTAGTGAGATCCCCGTTGGT-3′ and 5′-TGGTTTATCTGGTCAGCATTCCGTGTGG-3′). Hybridization was performed in ULTRAhyb buffer (Ambion, Austin, TX), using the hybridization conditions and washes recommended by the manufacturer.
The Qiagen OneStep RT-PCR kit was used for all RT-PCR analyses. The following primers were used for detection of WVD2 expression: 5′-GCTTATCAAGAAATCATTGTTCAATCAGAC-3′ and 5′-CTTAGGACGCGTCAGA-GGAAACTTCT-3′. The primers used for RT-PCR detection of WDL1 transcripts were: 5′-CATCATCTTTTGAAATTTCAAGCTTCTTCTGAG-3′ and 5′-GAATGGGAAGA-GAAGTTGTTGAGGTGCT-3′. We defined the 5′ end of full-length wild-type WVD2 transcripts by primer-extension analysis (Yamada et al., 1998), using a WVD2-specific primer (5′-TTCTTTCTCCTCATAATCTTCTTCCTCTGC-3′) and poly(A) RNA extracted with the PolyATract System 1000 kit (Promega, Madison, WI) from 4-d-old Ler seedlings grown on tilted agar plates.
Immunofluorescence
For visualization of microtubules within cells of the root cap, we used the protocol described by Goodbody and Lloyd (1994), with modifications. Cell wall digestion was performed with 0.1% (w/v) Pectolyase Y-23 and 0.5% (w/v) Macerozyme (Karlan Research Products Corporation, Santa Rosa, CA), dissolved in microtubule-stabilizing buffer (MTSB; Goodbody and Lloyd, 1994). After cell wall digestion, a 15-min membrane permeabilization step was performed (10% [v/v] DMSO and 3% [v/v] NP-40, in MTSB), followed by three 15-min washes in MTSB. Before incubation with primary or secondary antibodies, the samples were blocked in a solution of 1% (v/v) normal goat serum and 3% (w/v) bovine serum albumin (in MTSB) for 30 to 60 min. The antibodies were also diluted in this blocking solution. Detection of microtubules within cells of the root elongation zone, on the other hand, was performed as described by Sugimoto et al. (2000), except that the samples were blocked in a solution of 1% (v/v) normal goat serum and 3% (w/v) bovine serum albumin (in PEM buffer [25 mm PIPES, 0.5 mm MgSO4, and 2.5 mm EGTA {pH 7.2}]) and the cold methanol extraction step was omitted. In both cases, rat anti-tubulin antibodies (clone YOL1/34, Harlan Sera-Lab Ltd., Indianapolis), diluted 1:100, were used for primary immunolabeling. Fluorescein isothiocyanate-conjugated goat anti-rat IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) was used as a secondary antibody, also diluted 1:100.
Image Analysis and Microscopy
Pictures of seedlings on plates, of plants in soil, and of rosette leaves and siliques were obtained with either a Nikon Coolpix800 digital camera or a Nikon 8008S camera. The Nikon 8008S camera was attached to a dissecting microscope (Wild M3Z, Leica, Wetzlar, Germany) to acquire images of root CFR. Images used for the quantitative analysis of the diameters of roots and etiolated hypocotyls and for lengths and periclinal widths of root cells were obtained by a SPOT RT Slider digital camera (National Diagnostics, Atlanta) attached to a Nikon Optiphot-2 microscope equipped with Nomarski optics. For cellular measurements, whole seedlings were chemically cleared (Malamy and Benfey, 1997) before being mounted on microscopic slides. For confocal microscopy, we used a laser scanning confocal microscope (MRC-1024, Bio-Rad, Hercules, CA) at the W.M. Keck Laboratory for Biological Imaging (University of Wisconsin, Madison). Quantitative analysis of images was performed using the public domain NIH Image program (v1.62; developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image), with obtained data exported to Microsoft Excel spreadsheets for statistical calculations.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Nina Fedoroff, George Coupland, and the Arabidopsis Biological Resource Center for providing several of the Arabidopsis lines used in this study, and Dr. Anthony Bleecker for providing the pCAMBIA binary vectors. We also thank Dr. Sebastian Bednarek for assistance in immunodetection of cytoskeleton elements in root cells, the W.M. Keck Laboratory for Biological Imaging for allowing access to and technical assistance in use of the confocal microscope, and members of the Masson laboratory for fruitful discussion.
Footnotes
This work was supported by the Fundamental Space Biology Program of the National Aeronautic and Space Administration (grant nos. NAG2–1189 and NAG2–1492), by Wisconsin Hatch funds (no. WIS04310), and by the National Institutes of Health (genetics training grant no. 5T32GMO7133).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.015966.
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994. [Google Scholar]
- Bancroft I, Jones JDG, Dean C. Heterologous transposon tagging of the DRL1 locus in Arabidopsis. Plant Cell. 1993;5:631–638. doi: 10.1105/tpc.5.6.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI, Wilson JE, Cork A, Williamson RE. Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol. 1994;35:935–942. [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Masle J, Wasteneys GO. Growth conditions modulate root-wave phenotypes in Arabidopsis. Plant Cell Physiol. 2000;41:1164–1170. doi: 10.1093/pcp/pcd042. [DOI] [PubMed] [Google Scholar]
- Burk DH, Liu B, Zhong R, Morrison WH, Ye Z-H. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell. 2001;13:807–827. [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson PH. The Arabidopsis thaliana AGRAVITROPIC1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc Natl Acad Sci USA. 1998;95:15112–15117. doi: 10.1073/pnas.95.25.15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV, Smith DL. A versatile system for detecting transposition in Arabidopsis. Plant J. 1993;3:273–289. doi: 10.1111/j.1365-313x.1993.tb00178.x. [DOI] [PubMed] [Google Scholar]
- Fisher DD, Cyr RJ. Extending the microtubule/microfibril paradigm: Cellulose synthesis is required for normal cortical microtubule alignment in elongating cells. Plant Physiol. 1998;116:1043–1051. doi: 10.1104/pp.116.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani I, Watanabe Y, Prieto R, Masukawa M, Suzuki K, Naoi K, Thitamadee S, Shikanai T, Hashimoto T. The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development. 2000;127:4443–4453. doi: 10.1242/dev.127.20.4443. [DOI] [PubMed] [Google Scholar]
- Giddings TH, Staehelin LA. Microtubule-mediated control of microfibril deposition: a re-examination of the hypothesis. In: Lloyd CW, editor. The Cytoskeletal Basis of Plant Growth and Development. New York: Academic Press; 1991. pp. 85–89. [Google Scholar]
- Goodbody KC, Lloyd CW. Immunofluorescence techniques for analysis of the cytoskeleton. In: Harris N, Oparka KJ, editors. Plant Cell Biology: A Practical Approach. Oxford: IRL Press; 1994. pp. 221–243. [Google Scholar]
- Guzman P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T. Molecular genetic analysis of left-right handedness in plants. Phil Trans R Soc Lond B. 2002;357:799–808. doi: 10.1098/rstb.2002.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M-T, Morikami A, Benfey PN. Conditional root expansion mutants of Arabidopsis. Development. 1995;121:1237–1252. doi: 10.1242/dev.121.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk VI, Carroll BJ, Thomas CM, Jones JDG. Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J. 1993;3:493–494. doi: 10.1111/j.1365-313x.1993.tb00169.x. [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Lincoln CA, Timpte CS, Lammer D, Turner JC, Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Liang BM, Dennings AM, Sharp RE, Baskin TI. Consistent handedness of microtubule helical arrays in maize and Arabidopsis primary roots. Protoplasma. 1996;190:8–15. [Google Scholar]
- Long D, Martin M, Sundberg E, Swinburne J, Puansomlee P, Coupland G. The maize transposable element system Ac/Ds as a mutagen in Arabidopsis: identification of an albino mutation induced by Ds insertion. Proc Natl Acad Sci USA. 1993;90:10370–10374. doi: 10.1073/pnas.90.21.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinelli B, Gomarasca S, Soave C. A pleiotropic Arabidopsis thaliana mutant with inverted root chirality. Planta. 1997;202:196–205. doi: 10.1007/s004250050119. [DOI] [PubMed] [Google Scholar]
- Mirza JI. The effects of light and gravity on the horizontal curvature of roots of gravitropic and agravitropic Arabidopsis thaliana L. Plant Physiol. 1987;83:118–120. doi: 10.1104/pp.83.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JL, Turk E, Johnson K, Wolverton C, Ishikawa H, Simmons C, Söll D, Evans ML. Root-growth behavior of the Arabidopsis mutant rgr1. Plant Physiol. 1998;118:1139–1145. doi: 10.1104/pp.118.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Shimura Y. Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science. 1990;250:274–276. doi: 10.1126/science.250.4978.274. [DOI] [PubMed] [Google Scholar]
- Olsen KR, McIntosh JR, Olmstead JB. Analysis of MAP4 function in living cells using green fluorescent protein (GFP) chimeras. J Cell Biol. 1995;130:639–650. doi: 10.1083/jcb.130.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiagen USA. The QIAGEN guide to analytical gels. Part IV: Preparing formaldehyde agarose gels for RNA analysis. QIAGEN News. 2000a;2:20–24. [Google Scholar]
- Qiagen USA. The QIAGEN guide to analytical gels. Part V: Running and analyzing formaldehyde agarose gels for RNA analysis. QIAGEN News. 2000b;3:18–21. [Google Scholar]
- Qiagen USA. The QIAGEN guide to analytical gels. Part VI: RNA analysis by northern blotting. QIAGEN News. 2000c;4:22–25. [Google Scholar]
- Rutherford R, Masson PH. Arabidopsis thaliana sku mutant seedlings show exaggerated surface-dependent alteration in root growth vector. Plant Physiol. 1996;111:987–998. doi: 10.1104/pp.111.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford R, Gallois P, Masson PH. Mutations in Arabidopsis thaliana genes involved in the tryptophan biosynthesis pathway affect root waving on tilted agar surfaces. Plant J. 1998;16:145–154. doi: 10.1046/j.1365-313x.1998.00279.x. [DOI] [PubMed] [Google Scholar]
- Schneider K, Wells B, Dolan L, Roberts K. Structural and genetic analysis of epidermal cell differentiation in Arabidopsis primary roots. Development. 1997;124:1789–1798. doi: 10.1242/dev.124.9.1789. [DOI] [PubMed] [Google Scholar]
- Sedbrook JC, Chen R, Masson PH. ARG1 (altered response to gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proc Natl Acad Sci USA. 1999;96:1140–1145. doi: 10.1073/pnas.96.3.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR. The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell. 2002;14:1635–1648. doi: 10.1105/tpc.002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibaoka H. Plant hormone-induced changes in the orientation of cortical microtubules: alterations in the cross-linking between microtubules and the plasma membrane. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:527–544. [Google Scholar]
- Simmons C, Söll D, Migliaccio F. Circumnutation and gravitropism cause root waving in Arabidopsis thaliana. J Exp Bot. 1995;46:143–150. [Google Scholar]
- Smeekens S. Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:49–81. doi: 10.1146/annurev.arplant.51.1.49. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO. New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 2000;124:1493–1506. doi: 10.1104/pp.124.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO. Wall architecture in the cellulose-deficient rsw1 mutant of Arabidopsis thaliana: Microfibrils but not microtubules lose their transverse alignment before microtubules become unrecognizable in the mitotic and elongation zones of roots. Protoplasma. 2001;215:172–183. doi: 10.1007/BF01280312. [DOI] [PubMed] [Google Scholar]
- Takeda T, Furuta Y, Awano T, Mizuno K, Mitsuishi Y, Hayashi T. Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc Natl Acad Sci USA. 2002;99:9055–9060. doi: 10.1073/pnas.132080299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee S, Tuchihara K, Hashimoto T. Microtubule basis for left-handed helical growth in Arabidopsis. Nature. 2002;417:193–196. doi: 10.1038/417193a. [DOI] [PubMed] [Google Scholar]
- Wasteneys GO. Microtubule organization in the green kingdom: chaos or self-order? J Cell Sci. 2002;115:1345–1354. doi: 10.1242/jcs.115.7.1345. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang M-B. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G. A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETELA2. Plant Cell. 1996;8:659–671. doi: 10.1105/tpc.8.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Izu H, Nitta T, Kurihara K, Sakurai T. High-temperature, nonradioactive primer extension assay for determination of a transcription-initiation site. Biotechniques. 1998;25:72–78. doi: 10.2144/98251st02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.