Abstract
In barley (Hordeum vulgare), the Mla13 powdery mildew resistance gene confers Rar1-dependent, AvrMla13-specific resistance to Blumeria graminis f. sp. hordei (Bgh). We have identified cDNA and genomic copies of Mla13 and used this coiled-coil nucleotide-binding site leucine-rich repeat protein-encoding gene as a model for the regulation of host resistance to obligate biotrophic fungi in cereals. We demonstrate quantitatively that a rapid increase in the accumulation of Mla transcripts and transcripts of the Mla-signaling genes, Rar1 and Sgt1, is triggered between 16 and 20 h post inoculation, the same time frame that haustoria of avirulent Bgh make contact with the host cell plasma membrane. An abundance of Mla13 cDNAs revealed five classes of transcript leader regions containing two alternatively spliced introns and up to three upstream open reading frames (uORFs). Alternative splicing of introns in the transcript leader region results in a different number of uORFs and variability in the size of uORF2. These results indicate that regulation of Mla transcript accumulation is not constitutive and that induction is coordinately controlled by recognition-specific factors. The sudden increase in specific transcript levels could account for the rapid defense response phenotype conferred by Mla6 and Mla13.
Active defense against infection in plants is regulated by a myriad of responses that are induced upon detection of avirulence determinants presented by pathogens. In most cases, a resistance (R) gene in the plant confers the ability to recognize a pathogen expressing the corresponding avirulence (Avr) gene (Flor, 1971). Upon recognition, the plant activates a rapid signal cascade resulting in the induction of pathogenesis-related genes, the elicitation of systemic acquired resistance, and hypersensitive cell death (Staskawicz et al., 1995; Hammond-Kosack and Jones, 1996; Neuenschwander et al., 1996). However, the mechanism of regulation of R genes and R gene products before and after recognition is largely unknown. Even after pathogen challenge, mRNAs that encode many resistance proteins are virtually undetectable through total RNA-blot procedures and must be visualized using other methods (Parker et al., 1997; Ayliffe et al., 1999; Wang et al., 1999; Shen et al., 2002). However, despite low transcript levels, resistance proteins are likely maintained at a basal level in the plant cell because their primary function in resistance is to recognize signals signifying pathogen invasion (Shen et al., 2002). In some cases, R gene transcription can be induced (Yoshimura et al., 1998; Wang et al., 2001), but it has not previously been shown that transcript levels are increased only upon recognition of an avirulent pathogen.
Powdery mildew of barley (Hordeum vulgare), caused by Blumeria graminis f. sp. hordei (Bgh), is an ideal system for investigating the mechanisms of gene-for-gene interactions between large genome cereals and obligate fungal pathogens. An exceptionally large number of resistance specificities at the Mla locus govern gene-for-gene recognition to distinct isolates of Bgh (for review, see Jørgensen, 1994). Two Mla specificities, Mla6 and Mla1, encode resistance proteins with coiled-coil (CC), nucleotide (nt)-binding site (NBS), and Leu-rich repeat (LRR) motifs (Halterman et al., 2001; Zhou et al., 2001). Although the proteins encoded by these Mla variants are 92.2% similar (Halterman et al., 2001), they use different signaling pathways. To effect resistance specificity, the MLA6 protein requires the zinc-binding protein RAR1 (Shirasu et al., 1999a), and a subunit of the SCF (Skp1, Cullin, F-box) ubiquitin ligase complex, SGT1, whereas MLA1 does not (Zhou et al., 2001; Azevedo et al., 2002).
To date, searches for transcribed viral, bacterial, and fungal R genes have typically yielded 1 to 13 cDNAs per million plaques screened (Bent et al., 1994; Martin et al., 1994; Whitham et al., 1994; Parker et al., 1997; Wang et al., 1998, 1999; Tai et al., 1999). In contrast, screens for Mla6 cDNAs identified a relatively high incidence of clones (72 per million cDNAs), indicating that Mla6 transcripts may be more abundant (Halterman et al., 2001). In addition, the presence of long transcript leader regions (TLRs) and the identification of upstream open reading frames (uORFs) in mRNAs encoding Mla6 and Mla1 indicated that the TLRs may contain elements responsible for posttranscriptional regulation of protein levels (Halterman et al., 2001; Zhou et al., 2001). uORFs are present in up to 20% of plant mRNAs and function to down-regulate translation, alter mRNA stability, or control tissue-specific expression of the downstream cistron (Willis, 1999; Morris and Geballe, 2000; Kochetov et al., 2002).
In this report, we present the identification and functional characterization of the Rar1-dependent, Mla13 powdery mildew resistance gene. An abundance of Mla13 cDNAs revealed five classes of TLRs containing two alternatively spliced introns and up to three uORFs. Alternative splicing of the two TLR introns results in a different number of uORFs and variability in the size of uORF2. We show that recognition of avirulent isolates of Bgh results in the coordinate increase of Mla, Rar1, and Sgt1 transcripts. The timing of this induction and the splicing of TLR introns are correlated with previous investigations of attempted penetration by fungal appressoria in powdery mildew resistant plants.
RESULTS
The Barley Mla13 Gene Contains a Long TLR with Three uORFs and Alternatively Spliced Introns
We previously established that Mla13 resistance specificity cosegregates with three families of resistance gene homologs, RGH1a-e, RGH2a&b, and RGH3a&b, on the short arm of chromosome 1H (Wei et al., 1999, 2002). To identify expressed copies of Mla13, we screened 1.2 million plaque-forming units of an unamplified λ-Zap cDNA library with probes representing the LRRs of Mla-RGH1a/1e, 2a, and 3a from C.I. 16155, an accession that contains the Mla13 resistance specificity (Moseman, 1972). No plaques were identified with the RGH2a or RGH3a probes, however, 230 plaques hybridized to the RGH1a/1e probe mixture. One hundred and ten of these candidate Mla13 cDNAs were sequenced, and 23 possessed nearly full-length TLRs. All of the cDNAs contained identical open reading frames (ORFs) encoding a protein of 959 amino acids, with an estimated molecular mass of 107.5 kD. A COILS analysis (Lupas et al., 1991) revealed that the region between amino acids 27 and 47 of the predicted MLA13 protein forms a CC structure with greater than 95% probability (data not shown). The N-terminal one-half of the protein contains the motifs indicative of a NBS (Grant et al., 1995; van der Biezen and Jones, 1998). The C-terminal region of the protein encodes 11 imperfect LRRs with an average size of 26 amino acids.
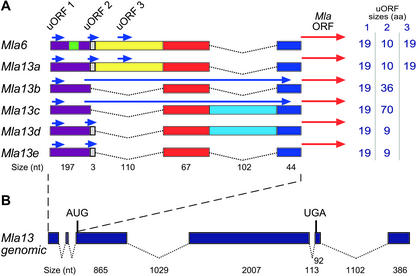
Candidate Mla13 mRNAs contain TLRs of up to 423 nt. As illustrated in Figure 1A, comparison of the 23 TLR sequences revealed two alternatively spliced introns that separate these cDNAs into five classes, designated Mla13a through Mla13e. The different classes of Mla13 cDNAs are likely not the products of separate genomic loci because, other than the alternatively spliced introns, the cDNA sequences are identical. Also, only one class of genomic sequence was identified in our screen. This was not the case for Mla6, where we also identified cosmids containing the related Mla6-2 gene (D.A. Halterman and R.P. Wise, unpublished data). Mla6-2 is a transcriptionally active paralog of Mla6 and encodes a truncated version (232 amino acids) of the 956-amino acid, MLA6 protein (Halterman et al., 2001).
Figure 1.
A, Representation of the Mla6 and Mla13 TLRs. Blue arrows represent the locations of the three uORFs. The corresponding sizes of each uORF are listed in the table on the right. The red arrows represent the Mla ORF. The gray box represents the GTG trimer that is spliced out of Mla13b and Mla13c cDNAs. Dashed lines represent the two alternatively spliced introns located within the TLRs. B, Representation of the genomic structure of Mla13. Introns are denoted by dashed lines.
Sequence analysis of the TLRs also revealed three potential uORFs encoding small peptides of varying sizes (Fig. 1A). The first, designated uORF1, is located 15 nt from the 5′ end of the longest cDNA isolated and encodes a 19-amino acid peptide with estimated molecular mass of 2.29 kD. Interestingly, although uORF1 is conserved between Mla6, Mla6-2, and Mla13 candidate cDNAs, it was not identified in Mla1 cDNAs (Zhou et al., 2001). However, uORF1 is conserved at the same position in the Mla1 genomic sequence, indicating that either the characterized Mla1 cDNAs are not full length or that Mla1 has a different transcription start site than Mla6 or Mla13. uORF2 begins 158 bases downstream of the start of uORF1 and is the most variable of the three uORFs. Depending on alternative intron splicing, uORF2 can encode four peptides ranging between 9 and 70 amino acids in length. The 10-amino acid (1.35-kD) uORF2 is found only in Mla6 and Mla13a. The complete splicing of an additional three bases at the start of intron 1, as in Mla13b/c, leads to a deletion of the uORF2 stop codon. The next in-frame stop codon for these cDNAs is located 29 nt upstream of the Mla13 ORF, and thus, the Mla13b and Mla13c uORF2s encode 36-amino acid (4.24 kD) and 70-amino acid (8.31 kD) peptides, respectively. The 34-amino acid difference in size between the predicted proteins encoded by uORF2 of Mla13b and Mla13c is due to the presence of the 102-nt intron 2 in Mla13c. Mla13d/e uORF2s encode a protein with only nine amino acids (1.16 kD) because the splicing of the first intron after the GTG leads to the formation of a stop codon. uORF3 is contained within the first TLR intron, which is not spliced in the previously isolated Mla6 cDNAs or Mla13a. uORF3 begins 218 bp from the start of uORF1 and encodes a 19-amino acid (2.24-kD) peptide.
Isolation of Mla13 Genomic Clones
To identify genomic clones containing Mla13 for use in a functional assay, a cosmid library was constructed from DNA of C.I. 16155. A PCR-based screen using primers designed from the ORF of the candidate Mla13 cDNA was used to identify 13 of 331 pools (10,000 clones/pool) with amplified products identical in size to those from C.I. 16155 genomic DNA. Sequence analysis of cosmid 10052-1 (35-kb) revealed a single CC-NBS-LRR type gene with an ORF identical to that of the candidate Mla13 cDNAs. Comparison of the cDNA and genomic copies revealed the presence of two introns (1,029 and 113 bp) within the ORF, one intron (1,102 bp) in the 3′-untranslated region (UTR) and two alternatively spliced introns (described above) within the TLR (Fig. 1B). The first intron within the ORF contains a small, simple sequence repeat made up of 28 AT repeats, similar to the functional Mla1 (14 AT) and Mla6 (10 AT) genes, as well as a paralog of Mla6, Mla6-2 (12 AT; Halterman et al., 2001; Zhou et al., 2001). Interestingly, of the 110 candidate Mla13 cDNAs, introns were alternatively spliced in the 5′-TLR, yet completely spliced within the ORF and the 3′-UTR.
Mla13 Confers Rar1-Dependent Resistance to Bgh Expressing AvrMla13
Cosmid 10052-1 and a 10-kb BamHI subclone (10052-1-10), both harboring the Mla13 candidate plus at least 3-kb of upstream sequence, were tested for function in an established single-cell transient assay using particle-mediated bombardment (Halterman et al., 2001; Zhou et al., 2001). In this assay, green fluorescent protein (GFP) fluorescing, leaf-epidermal cells are rendered susceptible to Bgh, due to the presence of wild-type Mlo, contained within the GFP-Mlo (pUGLUM) vector, whereas neighboring non-transformed cells retain broad-spectrum mlo resistance (Shirasu et al., 1999b; Zhou et al., 2001). The mlo resistance of non-transformed cells makes it possible to score the infection phenotypes on the few single-cell transformation events which otherwise would become masked by spreading fungal hyphae originating from neighboring susceptible cells.
Powdery mildew resistant leaves containing the mlo-5 mutation were bombarded with the pUGLUM vector alone or in combination with candidate Mla13 genomic constructs to test whether they conferred AvrMla13-dependent resistance specificity. A 7-kb cosmid subclone harboring Mla6, 9589-5a-7, was used as a control in these experiments. Bombarded leaves were inoculated with Bgh isolate 5874, which contains AvrMla6 but not AvrMla13, isolate K1, which harbors AvrMla13 but not AvrMla6, or isolate 63.5, which contains both Avr genes. The combined results of three independent experiments are shown in Table I. Constructs that contained the Mla13 candidate gene significantly reduced the number of GFP-marked, mlo-5 cells that supported Bgh hyphal growth after inoculation with isolates K1 (0%) and 63.5 (7.3%) when compared with the Mla6 (57.6%) and pUGLUM (50.3%) controls, respectively. These results established that the candidate gene encoded within BamHI subclone, 10052-1-10, is able to confer AvrMla13-dependent resistance specificity.
Table I.
Results of the three-component transient assay in mlo-5 barley leaves inoculated with Bgh isolates 5874 or K1
| Test DNA | 5874 (AvrMla6, virMla13)
|
K1a (virMla6, AvrMla13) or 63.5b (AvrMla6, AvrMla13)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| GFP Cells with Conidia | GFP Cells with Hyphae | GFP Cells with Hyphae | P Value (test vs. pUGLUM)c | GFP Cells with Conidia | GFP Cells with Hyphae | GFP Cells with Hyphae | P Value (test vs. pUGLUM)c | P Value (5874 vs. K1/63.5)d | |
| n | % | n | % | ||||||
| pUGLUM | 267 | 132 | 49.4 | 464ab | 233ab | 50.2 | 0.9182 | ||
| pUGLUM/10052-1 (Mla13) | 59 | 34 | 57.6 | 0.2435 | 151b | 11b | 7.3 | <0.0001 | <0.0001 |
| pUGLUM/10052-1-10 (Mla13) | 433 | 218 | 50.3 | 0.8990 | 103a | 0a | 0.0 | <0.0001 | <0.0001 |
| pUGLUM/9589-5a-7 (Mla6) | 369 | 8 | 2.2 | <0.0001 | 250a | 135a | 54.0 | 0.5917 | <0.0001 |
Isolate K1 was used to inoculate leaves bombarded with pUGLUM alone, 10052-1-10/pUGLUM, and 9589-5a/pUGLUM.
Isolate 63.5 was used to inoculate leaves bombarded with pUGLUM alone and 10052-1/pUGLUM. For ease of presentation, results from bombardment of pUGLUM alone were combined with those from the K1 inoculation. Both isolates gave similar results on these leaves.
P values were obtained using a random effect model to test for a significant difference between the percentage of cells with hyphal colonies after cobombardment with test DNA in comparison to bombardment with pUGLUM alone. A P value < 0.05 indicates that the percentages are significantly different.
P values were obtained using a random effect model to test for a significant difference between the percentage of cells with hyphal colonies after inoculation with isolate 5874 in comparison with inoculation with isolate K1 or 63.5. A P value < 0.05 indicates that the percentages are significantly different.
MLA6 and MLA1 proteins are more than 90% identical at the amino acid level but differ in their dependence on other signaling pathway components. MLA6 requires RAR1 and SGT1, whereas MLA1 does not (Halterman et al., 2001; Zhou et al., 2001; Azevedo et al., 2002). Previous evidence indicated that Mla13 belongs to the subgroup of Ml genes that are dependent on the powdery mildew resistance-signaling gene Rar1 (Jørgensen, 1996; Schulze-Lefert and Vogel, 2000; Wise, 2000). We tested the Rar1-dependence of Mla13 using a rar1-2/mlo-31 double mutant that was used to show Rar1 dependence of Mla6 and Rar1 independence of Mla1 (Halterman et al., 2001; Zhou et al., 2001). The combined results of two independent experiments are shown in Table II. The number of GFP marked, rar1-2/mlo-31 cells that supported Bgh hyphal growth after bombardment with the 10-kb BamHI subclone, 10052-1-10 (44.8%), was not significantly different from the Mla6 (41.7%) or pUGLUM (46.1%) controls after inoculation with Bgh K1. These results indicate that the CC-NBS-LRR gene encoded within 10052-1-10 confers Mla13 resistance specificity only in a Rar1-dependent manner.
Table II.
Results of the three-component transient assay in mlo-5 and rar1-2/mlo-31 barley leaves inoculated with Bgh isolate K1
| Test DNA |
mlo-5 Leaves
|
rar1-2/mlo-31 Leaves
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| GFP Cells with Conidia | GFP Cells with Hyphae | GFP Cells with Hyphae | P Value (test vs. pUGLUM)a | GFP Cells with Conidia | GFP Cells with Hyphae | GFP Cells with Hyphae | P Value (test vs. pUGLUM)a | P Value (Rar1 vs. rar1)b | |
| n | % | n | % | ||||||
| pUGLUM | 158 | 75 | 47.5 | 228 | 105 | 46.1 | 0.8366 | ||
| pUGULM/10052-1-10 (Mla13) | 229 | 8 | 3.5 | <0.0001 | 194 | 87 | 44.8 | 0.8482 | <0.0001 |
| pUGLUM/9589-5a-7 (Mla6) | 193 | 99 | 51.3 | 0.5813 | 218 | 91 | 41.7 | 0.5169 | 0.1371 |
P values were obtained using a random effect model to test for a significant difference between the percentage of cells with hyphal colonies after cobombardment with test DNA in comparison with bombardment with pUGLUM alone. A P value < 0.05 indicates that the percentages are significantly different.
P values were obtained using a random effect model to test for a significant difference between the percentage of cells with hyphal colonies on mlo-5 leaves in comparison with mlo-31/rar1-2 leaves. A P value < 0.05 indicates that the percentages are significantly different.
Accumulation of Rar1-Dependent Resistance-Signaling mRNAs Is Up-Regulated upon Specific Recognition of Bgh
The cDNA libraries used to identify transcribed copies of Mla6 and Mla13 were constructed with mRNA isolated from leaves 20 and 24 h after challenge with avirulent isolates of Bgh. Our ability to isolate a large number of cDNAs from both the C.I. 16151 (Mla6) and C.I. 16155 (Mla13) libraries as compared with other cloned R genes (Bent et al., 1994; Martin et al., 1994; Parker et al., 1997; Wang et al., 1998; Tai et al., 1999; Wang et al., 1999) suggested that either Mla genes are transcribed constitutively at a high level or that Mla transcript accumulation is induced after recognition of Bgh infection. To test these hypotheses, we performed reciprocal inoculations with avirulent or virulent isolates of Bgh onto plants harboring Mla6 or Mla13 and assayed specific transcript levels 0, 8, 16, 20, and 24 h after inoculation (hai). mRNA levels were quantitated by real-time reverse transcription-PCR (RT-rtPCR) using a real-time PCR detection system. The primers used to amplify a portion of the Mla6 and Mla13 gene were designed within a region downstream of any similarity to Mla6-2. As an internal standard, the mRNA level of each gene was calculated relative to that of actin mRNA at each time point. As an additional control, actin mRNA was also compared with the level of 18S ribosomal RNA. No significant increase of 18S or actin RNA levels was observed at any time point (data not shown), indicating that actin mRNA levels were suitable as a steady-state control for these experiments. The -fold change of Mla transcript accumulation was calculated by comparison with the basal level of mRNA measured at 0 hai.
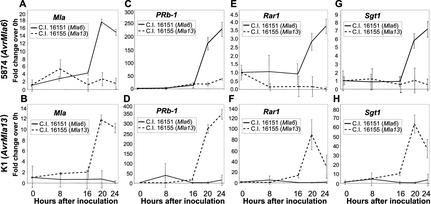
As illustrated in Figure 2, Mla transcripts accumulated to a higher level in incompatible compared with compatible interactions with Bgh. The level of Mla6 transcripts increased dramatically between 16 and 20 hai with the incompatible Bgh isolate 5874 (Fig. 2A). Sixteen to 20 hai is the time frame in which membrane to membrane contact is made between the host plasma membrane and developing Bgh haustoria (Ellingboe, 1972; Boyd et al., 1995). At 20 hai, the amount of Mla6 transcripts was 4.3-fold higher than at 16 hai and was 10.9-fold higher than Mla13 transcripts detected in C.I. 16155 plants compatible with Bgh isolate 5874. C.I. 16155 (Mla13) plants inoculated with 5874 did not show a significant accumulation of Mla transcripts at any time point, indicating that this increase is dependent on recognition of the invading Bgh. In the reciprocal experiment, a 12.5-fold increase in Mla13 mRNA levels was observed between 16 and 20 hai with Bgh isolate K1 (AvrMla13; Fig. 2B). This level of expression was 17.2-fold higher than that of Mla6-containing plants inoculated with the same isolate. There was no significant increase in Mla6 expression after inoculation with K1.
Figure 2.
Transcript profiles of Mla, PRb-1, Rar1, and Sgt1. Quantitative, RT-rtPCR was carried out on RNA from C.I. 15151 (Mla6) or C.I. 16155 (Mla13) plants after heavy inoculation with compatible or incompatible Bgh spores. Tissue samples were harvested immediately after inoculation (0 hai), and 8, 16, 20, and 24 hai. The quantity of RNA for each gene was calculated relative to actin mRNA. The -fold change was calculated by comparing the relative expression levels at each time point to the expression level at 0 hai. A, C, E, and G, RNA was isolated from plants inoculated with Bgh isolate 5874 (AvrMla6; virMla13). B, D, F, and H, RNA was isolated from plants inoculated with Bgh isolate K1 (AvrMla13; virMla6). Note differences in scale among the graphs.
To compare the regulation of Mla transcript levels with other genes critical to Mla-mediated defense, we used the same RNA samples to assay the kinetics of PRb-1, Rar1, and Sgt1 mRNA accumulation. PRb-1 was monitored in our experiments as a control because previous data has shown that the extent of PRb-1 transcripts is regulated differently in resistant and susceptible barley/Bgh interactions (Muradov et al., 1993). Rar1 and Sgt1 are required for resistance of some Mla genes, including Mla6 (Halterman et al., 2001; Azevedo et al., 2002). In our experiments, a 13-fold increase in PRb-1 mRNAs was observed between 16 and 20 hai in the resistant interaction of Mla6 with Bgh 5874 (AvrMla6; Fig. 2C). Between 20 and 24 hai, the quantity of PRb-1 transcripts increased even further. PRb-1 transcript accumulation was also induced in a similar manner during the Mla13/AvrMla13 interaction (Fig. 2D). A slight increase of PRb-1 mRNAs was observed during the compatible interactions, although this took place later—between 20 and 24 hai.
Because Mla6 and Mla13 are Rar1 dependent, we were interested in determining whether mRNAs of related resistance-signaling genes are also up-regulated upon recognition of Bgh. In C.I 16151 (Mla6) plants, Rar1 transcript levels increased 3.1-fold between 16 and 20 hai with 5874 (AvrMla6; Fig. 2E). During this same time period, Sgt1 mRNAs increased 6.4-fold (Fig. 2G). Neither gene showed increased transcript levels in C.I 16155 plants inoculated with 5874. In C.I 16155 (Mla13) plants inoculated with K1 (AvrMla13), an increase in Rar1 transcripts was observed between 16 and 20 hai with a 6.8-fold increase over the 16-h time point (Fig. 2F). Sgt1 transcript accumulation mirrored this pattern with a 6.3-fold increase between 16 and 20 hai (Fig. 2H). mRNA levels of both of these genes were reduced somewhat between 20 and 24 hai, although this reduction was not observed in the Mla6/AvrMla6 interaction. No increase in transcript levels of either gene was observed in the compatible interactions over this 24-h time course. The overall -fold change of Rar1 and Sgt1 transcripts is noticeably different between Mla6- and Mla13-containing plants inoculated with avirulent Bgh (Figs. 2, E–H). However, for these experiments, the fact that the -fold change of Rar1 and Sgt1 transcripts between 16 and 20 hai is equivalent within the two genotypes appears more significant. Because this calculation was made using the level of accumulated transcripts at 0 hai, the difference could be due to a low level of Rar1 and Sgt1 mRNAs in C.I 16155 (Mla13) plants at 0 hai. Taken together, these results indicate that up-regulation of transcript levels during gene-for-gene interactions is not exclusive to defense-related genes but also includes the CC-NBS-LRR type Mla genes and their resistance-signaling pathway components.
Mla13 TLR Intron Splicing Is Variable during Challenge by Bgh
The presence of alternatively spliced introns within the TLR of Mla13 led us to investigate whether these splicing events are altered upon pathogen recognition. Using quantitative RT-rtPCR, we monitored intron splicing within the TLR of Mla13 after inoculation with Bgh isolate K1 (AvrMla13). Using primer pairs with one member entirely within an intron or one primer spanning the intron-splicing site (Table III), we were able to detect either the presence or absence of introns over time. Two different reverse primers were used to detect the absence of Mla13 intron 1. The first was designed to specifically amplify TLRs retaining the GTG trimer at the 5′ end of the intron-splicing site (Fig. 1). The second was designed to work only when this GTG was spliced out.
Table III.
Primers used in RT-rtPCR reactions
| Gene Amplified | Primer Sequences (5′→3′) | Fragment Size |
|---|---|---|
| bp | ||
| Mla | TGCGATGCTGGATGCCAATC GCAAAGTTGATGCCTTCCCAC | 120 |
| Mla13 + TLR intron1 | GAAATCTTTCACGTCCACACGTTC TTCACGGCGGAGCAGAACACAG | 138 |
| Mla13 − TLR intron1 − GTG | GAGTACTTGCATAGTAGCTCGCCC GGATCGCTCCAATCGATCGC | 114 |
| Mla13 − TLR intron1 + GTG | GAGTACTTGCATAGTAGCTCGCCC GAGGATCGCTCCAATCGATCAC | 119 |
| Mla13 + TLR intron2 | CCGCCGTGAAGAATCAAGGTG GGAGCTGTAACGCAATCCAAATTG | 100 |
| Mla13 − TLR intron2 | CCGTGAAGAATCAAGGCTTCC TGAGTAGCTCCCCCAACTTGG | 115 |
| Rar1 | AACTCCTCAAAGGCAACCCCAC GAACAAAAGAAACCCTGACGGC | 104 |
| Sgt1 | GGCTGTTGCTCCTGCTACATCTTC TGGGCTTGCTTGGCACTTCTAC | 116 |
| PRb-1 | TCATCGTCTCTCGTCCCTAATCTC CCTTTTATTTACTCGCTCGGTCC | 77 |
| actin | TCGCAACTTAGAAGCACTTCCG AAGTACAGTGTCTGGATTGGAGGG | 130 |
| 18S | GACAGACTGAGAGCTCTTTCTTGA GCATAGCTAGTTAGCAGGCTGAG | 123 |
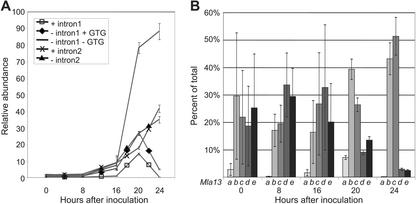
As shown in Figure 3A, the first intron within the TLR was present at low levels at all the time points tested with only a small increase at 20 hai. Mla13 mRNAs that lacked intron 1 were more abundant, and of these, the majority also lacked the GTG trimer. At 20 and 24 hai, mRNAs containing the second intron, representing Mla13c and Mla13d, were present in similar amounts when compared with mRNAs with this intron spliced out. Both of these variants increased in concentration at these time points, although the -fold increase of mRNAs containing intron 2 was somewhat higher at 24 hai (10.6 versus 6.6). Using the data gathered from these experiments we were able to calculate the percentage that each cDNA class represents in the pool of all Mla13 mRNAs (Fig. 3B). With the exception of Mla13a, the cDNA classes were essentially equally represented up to and including 16 hai. Because only low levels of all Mla13 mRNAs were present at these early time points, the calculated sd for the percentages of each class was large. At 20 and 24 hai, however, Mla13b and Mla13c are significantly over-represented in the pool, indicating that these are the most prevalent forms of the TLR at these times.
Figure 3.
Preferential splicing of Mla13 TLR introns after pathogen recognition. A, This graph represents the intron-splicing events taking place within the TLR of Mla13 after inoculation with Bgh K1 (AvrMla13). Quantitative RT-rtPCR was used to monitor the presence or absence of introns at each time point. The relative abundance is a ratio of test mRNA levels over actin mRNA levels. Error bars represent the sd from three replicates of two independent inoculations. B, Each bar represents the percentage of the Mla13 cDNA class present at each time point after inoculation. The letters under each bar represent the corresponding Mla13 mRNA class (Fig. 1). The abundance of each intron, derived from quantitative RT-rtPCR, was used to calculate the percentage of each cDNA class present at each time point (see “Materials and Methods”). Error bars represent the sd.
DISCUSSION
Mla6 and Mla13 Transcript Accumulation Is Induced upon Recognition of Bgh
The molecular mechanisms governing the regulation of plant disease-resistance genes are largely unknown. Recent reports established that these genes are transcribed at a basal level in healthy plants (Ayliffe et al., 1999; Wang et al., 1999; Shen et al., 2002), however, most of these studies concluded that transcription was constitutive. The flax L6 gene, which encodes the subclass of NBS-LRR proteins with an amino-terminal toll interleukin receptor domain, has been shown to be constitutively expressed even after inoculation with an avirulent rust pathogen (Ayliffe et al., 1999). Two other resistance genes, Xa1 and Pib, are inducible after pathogen inoculation, however induction does not appear to be gene-for-gene specific (Yoshimura et al., 1998; Wang et al., 1999). The barley Mlo gene is transiently induced by both biotic and abiotic stresses approximately 6 h after treatment (Piffanelli et al., 2002). Furthermore, the increase in Mlo transcript levels is observed in resistant, susceptible, and non-host pathogen interactions. Here, we present evidence of isolate-specific induction of Mla transcript accumulation. Our results show that transcripts of Mla6 and Mla13 are induced between 16 and 20 hai and only upon recognition of Bgh isolates expressing the corresponding avirulence gene over the 24-h time course. The timing of this increase in Mla transcripts correlates with previous studies of Mla6- and Mla13-mediated responses to Bgh infection (Ellingboe, 1972; Wise and Ellingboe, 1983) and defense-related gene transcripts in plants challenged with Bgh (Boyd et al., 1995). Messenger RNAs of other Mla-signaling pathway components, Rar1 and Sgt1, also increased in an R-gene-dependent manner during this time period along with the defense related gene PRb-1, used as a control in our experiments. Varet et al. (2002) recently showed that transcripts of the Arabidopsis NDR1/HIN1-like genes, NHL25 and NHL3, accumulate specifically during infection with Pseudomonas syringae pv. tomato DC3000 that possessed any of the four avirulence genes avrRpm1, avrRpt2, avrB, or avrRps4, but not during infection with virulent P. syringae. However, Bgh-induced Rar1 and Sgt1 expression profiles contrast with those of other R gene-signaling components, such as NDR1, EDS1, and PAD4, which show increased transcript levels during both incompatible and compatible interactions (Century et al., 1997; Feys et al., 2001; Peart et al., 2002a).
Functional homologs of Rar1 and Sgt1 are present in Arabidopsis and tobacco (Nicotiana tabacum) and play a part in a multitude of R gene-signaling pathways (Austin et al., 2002; Liu et al., 2002; Muskett et al., 2002; Peart et al., 2002b; Tör et al., 2002; Tornero et al., 2002). It has also been shown that, depending on the R gene involved, Rar1 can function independently of NDR1 in Arabidopsis (Tornero et al., 2002). The expression kinetics for barley Rar1 and Sgt1 further indicate that their function is specific for R gene signaling and is not critical during compatible interactions, at least within the first 24 h. Bgh begins its invasion of the host plasma membrane approximately 16 h after contact between the conidia and the leaf surface (Ellingboe, 1972). This suggests that signal transduction leading to defense gene induction occurs rapidly because a coordinated increase in mRNA levels was observed during this time frame. In most cases, the transcript profiles were similar not only within the same plant background but also after recognition of different Bgh isolates. However, in contrast to the other host-pathogen gene pairs, Rar1 and Sgt1 transcript levels appear to decrease between 20 and 24 hai in C.I. 16155 (Mla13) plants inoculated with K1 (AvrMla13). Also, the -fold change in Rar1 and Sgt1 mRNAs was quite different in C.I. 16155 plants, which is likely due to a relatively low level of transcripts at the 0-hai time point. Because inoculations with K1 (AvrMla13) were maintained in a growth chamber separate from 5874 (AvrMla6), it is possible that different environmental factors (however slight) had an effect on the induction of Mla13 by Bgh harboring AvrMla13 as opposed to induction of Mla6 by Bgh harboring AvrMla6. Also, it is important to note that a direct comparison of transcript levels among different genes is not possible because the primer annealing temperature, secondary structure of primer targets, and PCR product size differs for each gene target.
The coordinate increase in defense-related gene transcripts takes place well after contact between conidia and the leaf surface, but before the formation of elongating secondary hyphae. Therefore, we predict that recognition occurs during the invasion of the host cell and the formation of Bgh haustoria. This supports previous data that indicate resistance is elicited only after intimate membrane-to-membrane contact between the host and pathogen (Ellingboe, 1972). In our model, the haustorium is responsible for sending a signal across the host plasma membrane where it is recognized directly or indirectly by a host factor, possibly MLA. This recognition leads to a coordinated defense response that includes an induction of Mla, Rar1, and Sgt1 transcription. There are two possible scenarios that could explain the increase in expression of these genes. The escalation of Mla mRNA levels could be due to increased transcript accumulation in cells surrounding the site of infection and not in the attacked cell itself. Because the attacked cell most likely dies quickly via the hypersensitive cell death and because only a portion of the leaf cells came into contact with Bgh conidia, it is possible that a signal sent out from the attacked cell induced the surrounding cells to heighten their sensitivity to the avirulence signal through the activation of Mla, Rar1, Sgt1, and PRb-1 transcription. This is supported by the fact that haustoria formation is not required for induction of pathogenesis-related genes in mlo resistant plants (Peterhänsel et al., 1997). Mla transcripts could alternatively be induced to a high level in cells that come into direct contact with invading Bgh haustoria. The recognition of the invading pathogen could lead to the activation of one or more transcription factors responsible for the increase in Mla, Rar1, and Sgt1 transcription.
Mla13 TLR Introns Are Alternatively Spliced
The presence of introns within the TLRs of plant genes appears to be rare. In detailed investigations of full-length mRNAs, only 15% of 5′-TLRs and 4% of 3′-UTRs contain introns (Haas et al., 2002; Mignone et al., 2002). Mla6 and Mla13 contain multiple introns in the 5′-TLR as well as a large intron in the 3′-UTR. Although the introns within the TLR of Mla13 appear to be alternatively spliced, the introns located within the ORF and 3′-UTR are completely spliced. At 24 hai with the incompatible Bgh K1 isolate, mRNAs with the second intron spliced from the TLR are present in about equal amounts to those retaining it. At the same time, almost all of Mla13 mRNAs lack the first TLR intron and, therefore uORF3. Relatively few mRNA retain the GTG trimer at the beginning of intron 1, which leads to a majority of mRNAs encoding relatively long uORF2s. In vivo data using human cells has shown that in some cases the length of a uORF is proportional to inhibition of translation of the downstream gene (Luukkonen et al., 1995). Therefore, in the case of Mla, mRNA belonging to Mla13b or Mla13c have the potential to produce less MLA protein than the other classes. Because these two classes of transcripts are most prevalent at 20 and 24 hai, when Mla13 mRNAs are abundant, this could counteract an escalation of MLA protein levels brought about by an increase in translation templates. It is also possible that the longer peptides encoded by uORF2 actively function to alter reinitiation through direct interaction with the ribosome, as has been shown to occur in some cases (for review, see Morris and Geballe, 2000). The combination of alternatively spliced introns and uORFs within the 5′-TLR of Mla suggests that translational control of MLA protein synthesis, if present, could be coordinated by intricate biological feedback loops. In maize (Zea mays), the translation of an anthocyanin regulatory gene, Sn, is regulated by an upstream ORF (Procissi et al., 2002). Alternative splicing within the 5′-TLR, which contains three uORFs, removes uORF2 and uORF3 and results in a decrease in anthocyanin synthesis in 75% of transformed protoplasts (Procissi et al., 2002). This appears to be due to the efficiency of ribosome re-initiation after translation of uORF1.
Intron splicing appears to be a critical element in resistance mediated by at least some plant R genes (Jordan et al., 2002). It is yet unknown whether alternative splicing within the TLR of Mla6 and Mla13 is required for full resistance to Bgh. The tobacco N gene, which confers resistance to tobacco mosaic virus, also undergoes alternative splicing (Whitham et al., 1994). The alternative splicing of the N gene is initiated by tobacco mosaic virus signals, which lead to an increase in the transcript encoding the truncated N protein 4 to 8 hai (Dinesh-Kumar and Baker, 2000). Alternative intron splicing results in the production of various transcripts of the flax L6 and M genes as well (Lawrence et al., 1995; Anderson et al., 1997). In both of these cases, the introns reside within the coding region of the gene and the alternatively spliced transcripts encode truncated proteins. It is not known whether pathogen recognition affects the abundance of alternative L6 transcripts, however, transgenic flax plants incapable of producing truncated L6 proteins confer resistance indistinguishable from that of wild-type plants (Ayliffe et al., 1999). Our results indicate that the splicing of introns within the Mla13 TLR may be regulated after recognition of Bgh because the majority of transcripts belong to the Mla13b and Mla13c classes 24 hai. It is also plausible that the alternative splicing of introns leads to changes in mRNA stability. Mla13b and Mla13c transcripts may be more stable, leading to an abundance of these transcripts relative to the others at 24 hai.
MATERIALS AND METHODS
Fungal Isolates
Bgh isolates 5874 (AvrMla6, virMla13), K1 (virMla6, AvrMla13), and 63.5 (AvrMla6, AvrMla13) were propagated on barley (Hordeum vulgare cv Manchuria) in separate growth chambers at 18°C (16 h of light/8 h of darkness).
cDNA Library Screening and Sequencing
A C.I. 16155 cDNA library was constructed from mRNA isolated from seedlings inoculated with Bgh isolate A27 (AvrMla13) by D.-W. Choi in the T.J. Close laboratory (University of California, Riverside) using the Uni-ZAP XR library kit (Stratagene, La Jolla, CA). Tissue was harvested 20 and 24 hai and snap-frozen in liquid nitrogen. DNA sequencing and oligonucleotide synthesis was performed by the Iowa State University DNA Sequencing and Synthesis Facility.
Cosmid Library Construction, Screening, and Sequencing
Cosmid library construction was done in cooperation with Cell & Molecular Technologies, Inc. (Phillipsburg, NJ). High-Mr genomic DNA from C.I. 16155 was partially digested with Sau3A, size selected for fragments ranging between 50 and 75 kb, and ligated into the BamHI site of digested cosmid SuperCos-1 (Stratagene).
Single-Cell Transient Assay
Biolistic bombardment of leaves was carried out according to Shirasu et al. (1999b) using a biolistic PDS-1000/He system (Bio-Rad, Hercules, CA). Detached leaves of 7-d-old barley seedlings were placed onto 1% (w/v) agarose (BioWhittaker Molecular Applications, Rockland, ME) plates supplemented with 10% (w/v) Suc and allowed to recover for 1 h at room temperature. Gold particles (Bio-Rad) were coated with plasmid and/or cosmid DNA at a plasmid:cosmid molar ratio of 2:3 and delivered to the leaves using 450-psi rupture discs. The leaves were then incubated at room temperature for 4 h and transferred to 1% (w/v) agarose before fungal inoculation. The inoculated leaves were incubated at 18°C (16 h light/8 h darkness) for 5 d. Barley cells expressing GFP were visualized 5 d after fungal inoculation using a microscope with an excitation filter of 450 to 490 nm (Chroma 41001).
Quantitative RT-rtPCR
Flats of 10 cm of C.I. 15151 and C.I. 16155 barley seedlings were inoculated simultaneously with either Bgh isolate 5874 or K1 and maintained under controlled conditions in growth chambers at 18°C (16 h of light/8 h of darkness). Ten to 15 seedlings were harvested and snap-frozen in liquid nitrogen at each time point (0, 8, 16, 20, and 24 hai) for RNA preparation. RNA from each time point was isolated from two independent replications of each time course. RT-rtPCR was performed using the SuperScript RT-rtPCR with Platinum Taq System (Invitrogen, Carlsbad, CA) with the iCycler real-time PCR detection system (Bio-Rad). RNA was DNase treated using DNA-free (Ambion, Austin, TX) and tested for DNA contamination using PCR with Taq alone. Reactions were prepared according to the manufacturer's instructions and included 0.1× SYBR green (Molecular Probes, Eugene, OR) and 10 nm fluorescein. Primers used for each gene are shown in Table III. Reverse transcription was carried out at 50°C for 30 min followed by 2 min at 95°C. Amplification of this product was carried out by 45 cycles of PCR (95°C for 15 s and 58°C for 1 min). A melting curve (80 cycles of 30 s at 55°C + 0.5°C/cycle) was carried out on each sample to ensure all amplification products had the same melting temperature. Amplification products were cloned and sequenced for verification. The starting quantity (SQ) of each sample was calculated using a standard curve derived from a 5-fold dilution series of RNA taken at 24 hai from resistant barley (C.I. 16151 [Mla6] plants inoculated with Bgh 5874 or C.I. 16155 [Mla13] plants inoculated with Bgh K1). The correlation coefficient for each standard curve was greater than 0.99. RT-rtPCR reactions were performed in triplicate, and the SQ mean was calculated for each time point for each primer pair. The expression level of each gene compared with actin was calculated by dividing the SQ mean of one primer set to the actin SQ mean from the same RNA sample.
The percentage of each class of Mla13 TLRs was determined by totaling the relative abundances of each amplification product at each time point for each intron. We then determined the ratio of the calculated abundance of each individual amplification product versus the total for the intron. This calculates the percentage of each intron variant separately. To calculate the percentage of each TLR as a whole, we multiplied the percentages of each intron present within the TLR. For example, the relative abundance of intron1 absent but retaining the GTG at 20 hai is 26.75 of a total of 119.00, which equals 22.48%. The percentage of TLRs with intron 2 present at 20 hai is 40.14%. Therefore, the percentage of Mla13d at 20 hai is 9.02% (0.2248 × 0.4014).
GenBank Accession Numbers
The accession number for the Mla13 genomic sequence is AF523678. The accession numbers for the five classes of Mla13 cDNAs are AF523679 (Mla13a), AF523680 (Mla13b), AF523681 (Mla13c), AF523682 (Mla13d), and AF523683 (Mla13e).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Drs. A. Miller, S. Whitham, and A. Bogdanove for critical review of the manuscript, D.-W. Choi of the Close lab for assistance in preparation of the C.I. 16155 cDNA library, and J. Orme of the Schulze-Lefert lab for the rar1-2/mlo-31 double mutant.
Footnotes
This work was supported by U.S. Department of Agriculture-National Research Initiative/Competitive Grants Program (grant nos. 98–35300–6169 and 00–35300–9213 to R.P.W.). D.A.H. was supported in part by a U.S. Department of Agriculture-Agricultural Research Service Postdoctoral Research Associateship. This paper is a joint contribution of the Corn Insects and Crop Genetics Research Unit, U.S. Department of Agriculture-Agricultural Research Service, and the Iowa Agriculture and Home Economics Experiment Station.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.014407.
LITERATURE CITED
- Anderson PA, Lawrence GJ, Morrish BC, Ayliffe MA, Finnegan EJ, Ellis JG. Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell. 1997;9:641–651. doi: 10.1105/tpc.9.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JDG, Parker JE. Regulatory role of SGT1 in early R gene-mediated plant defense. Science. 2002;295:2077–2080. doi: 10.1126/science.1067747. [DOI] [PubMed] [Google Scholar]
- Ayliffe MA, Frost DV, Finnegan EJ, Lawrence GJ, Anderson PA, Ellis JG. Analysis of alternative transcripts of the flax L6 rust resistance gene. Plant J. 1999;17:287–292. doi: 10.1046/j.1365-313x.1999.00377.x. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Friealdenhoven A, Shirasu K, Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Boyd LA, Smith PH, Foster EM, Brown JKM. The effects of allelic variation at the Mla resistance locus in barley on the early development of Erysiphe graminis f. sp. hordei and host responses. Plant J. 1995;7:959–968. [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ. NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science. 1997;278:1963–1965. doi: 10.1126/science.278.5345.1963. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Baker BJ. Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci USA. 2000;97:1908–1913. doi: 10.1073/pnas.020367497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingboe AH. Genetics and physiology of primary infection by Erysiphe graminis. Phytopathology. 1972;62:401–406. [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20:5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Volfovsky N, Town CD, Troukhan M, Alexandrov N, Feldmann KA, Flavell RB, White O, Salzberg SL. Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol. 2002;3:RESEARCH0029. doi: 10.1186/gb-2002-3-6-research0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman D, Zhou F, Wei F, Wise RP, Schulze-Lefert P. The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 2001;25:335–348. doi: 10.1046/j.1365-313x.2001.00982.x. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JD. Resistance gene-dependent plant defense responses. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan T, Schornack S, Lahaye T. Alternative splicing of transcripts encoding Toll-like plant resistance proteins: What's the functional relevance to innate immunity? Trends Plant Sci. 2002;B:392–398. doi: 10.1016/s1360-1385(02)02311-7. [DOI] [PubMed] [Google Scholar]
- Jørgensen JH. Genetics of powdery mildew resistance in barley. Crit Rev Plant Sci. 1994;13:97–119. [Google Scholar]
- Jørgensen JH. Effect of three suppressors on the expression of powdery mildew resistance genes in barley. Genome. 1996;39:492–498. doi: 10.1139/g96-063. [DOI] [PubMed] [Google Scholar]
- Kochetov AV, Sirnik OA, Rogosin IB, Glazko GV, Komarova ML, Shumny VK. Contextual features of higher plant mRNA 5′-untranslated regions. Mol Biol. 2002;36:510–516. [PubMed] [Google Scholar]
- Lawrence GJ, Finnegan EJ, Ayliffe MA, Ellis JG. The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell. 1995;7:1195–1206. doi: 10.1105/tpc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Serino G, Deng X-W, Dinesh-Kumar SP. Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to tobacco mosaic virus. Plant Cell. 2002;14:1483–1496. doi: 10.1105/tpc.002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein structures. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Luukkonen BGM, Tan W, Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GBM, Frary A, Wu T, Brommonschenkel S, Chunwongse J, Earle ED, Tanksley SD. A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell. 1994;6:1543–1552. doi: 10.1105/tpc.6.11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome. 2002;Biol:3. doi: 10.1186/gb-2002-3-3-reviews0004. : REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseman JG. Isogenic barley lines for reaction to Erysiphe graminis f. sp. hordei. Crop Sci. 1972;12:681–682. [Google Scholar]
- Muradov A, Petrasovits L, Davidson A, Scott KJ. A cDNA clone for a pathogenesis-related protein 1 from barley. Plant Mol Biol. 1993;23:439–442. doi: 10.1007/BF00029021. [DOI] [PubMed] [Google Scholar]
- Muskett PR, Kahn K, Austin MJ, Moisan LJ, Sadanandom A, Shirasu K, Jones JD, Parker JE. Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell. 2002;14:979–992. doi: 10.1105/tpc.001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenschwander U, Lawton K, Ryals J. Systemic acquired resistance. In: Stacey G, Keen NT, editors. Plant-Microbe Interactions. Vol. 1. New York: Chapman and Hall; 1996. pp. 81–106. [Google Scholar]
- Parker JE, Coleman MJ, Szabò V, Frost LN, Schmidt R, van der Biezen EA, Moores T, Dean C, Daniels MJ, Jones JDG. The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Cook G, Feys BJ, Parker JE, Baulcombe DC. An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J. 2002a;29:569–579. doi: 10.1046/j.1365-313x.2002.029005569.x. [DOI] [PubMed] [Google Scholar]
- Peart JR, Liu R, Sadanandom A, Malcuit I, Moffett P, Brice DC, Schauser L, Jagard DAW, Xiao S, Coleman MJ et al. Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc Natl Acad Sci USA. 2002b;99:10865–10869. doi: 10.1073/pnas.152330599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhänsel C, Freialdenhoven A, Kurth J, Kolsch R, Schulze-Lefert P. Interaction analyses of genes required for resistance responses to powdery mildew in barely reveal distinct pathways leading to leaf cell death. Plant Cell. 1997;9:1397–1409. doi: 10.1105/tpc.9.8.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, Collins NC, Panstruga R, Schulze-Lefert P. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129:1076–1085. doi: 10.1104/pp.010954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procissi A, Piazza P, Tonelli C. A maize r1 gene is regulated post-transcriptionally by differential splicing of its leader. Plant Mol Biol. 2002;49:239–248. doi: 10.1023/a:1014959230492. [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P, Vogel J. Closing the ranks to attack by powdery mildew. Trends Plant Sci. 2000;5:343–348. doi: 10.1016/s1360-1385(00)01683-6. [DOI] [PubMed] [Google Scholar]
- Shen KA, Chin DB, Arroyo-Garcia R, Ochoa OE, Lavelle DO, Wroblewski T, Meyers BC, Michelmore RW. Dm3 is one member of a large constitutively expressed family of nucleotide binding site-leucine-rich repeat encoding genes. Mol Plant-Microbe Interact. 2002;15:251–261. doi: 10.1094/MPMI.2002.15.3.251. [DOI] [PubMed] [Google Scholar]
- Shirasu K, Lahaye T, Tan M, Zhou F, Azevedo C, Schulze-Lefert P. A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell. 1999a;99:355–366. doi: 10.1016/s0092-8674(00)81522-6. [DOI] [PubMed] [Google Scholar]
- Shirasu K, Nielsen K, Piffanelli P, Oliver R, Schulze-Lefert P. Cell-autonomous complementation of mlo resistance using a biolistic transient expression system. Plant J. 1999b;17:293–299. [Google Scholar]
- Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JD. Molecular genetics of plant disease resistance. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tör M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert-Turk F, Can C, Dangl JL, Holub EB. Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell. 2002;14:993–1003. doi: 10.1105/tpc.001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Merritt P, Sadanandom A, Shirasu K, Innes RW, Dangl JL. RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell. 2002;14:1005–1015. doi: 10.1105/tpc.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen EA, Jones JDG. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- Varet A, Parker J, Tornero P, Nass N, Nürnberger T, Dangl JL, Scheel D, Lee J. NHL25 and NHL3, two NDR1/HIN1-like genes in Arabidopsis thaliana with potential role(s) in plant defense. Mol Plant-Microbe Interact. 2002;15:608–616. doi: 10.1094/MPMI.2002.15.6.608. [DOI] [PubMed] [Google Scholar]
- Wang G-L, Ruan D-L, Song W-Y, Sideris S, Chen L, Pi LY, Zhang S, Zhang Z, Fauquet C, Gaut BS et al. Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell. 1998;10:765–780. doi: 10.1105/tpc.10.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZX, Yamanouchi U, Katayose Y, Sasaki T, Yano M. Expression of the Pib rice-blast-resistance gene family is up-regulated by environmental conditions favouring infection and by chemical signals that trigger secondary plant defences. Plant Mol Biol. 2001;47:653–661. doi: 10.1023/a:1012457113700. [DOI] [PubMed] [Google Scholar]
- Wang ZX, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 1999;19:55–64. doi: 10.1046/j.1365-313x.1999.00498.x. [DOI] [PubMed] [Google Scholar]
- Wei F, Gobelman-Werner K, Morroll SM, Kurth J, Mao L, Wing R, Leister D, Schulze-Lefert P, Wise RP. The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics. 1999;153:1929–1948. doi: 10.1093/genetics/153.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Wing R, Wise RP. Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell. 2002;14:1903–1917. doi: 10.1105/tpc.002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B. The product of the tobacco mosaic virus gene N: similarity to toll and interleukin-1 receptor. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- Willis AE. Translational control of growth factor and proto-oncogene expression. Int J Biochem Cell Biol. 1999;31:73–86. doi: 10.1016/s1357-2725(98)00133-2. [DOI] [PubMed] [Google Scholar]
- Wise RP. Disease resistance: What's brewing in barley genomics? Plant Dis. 2000;84:1160–1170. doi: 10.1094/PDIS.2000.84.11.1160. [DOI] [PubMed] [Google Scholar]
- Wise RP, Ellingboe AH. Infection kinetics of Erysiphe graminis f. sp. hordei on barley with different alleles at the Mla locus. Phytopathology. 1983;73:1220–1222. [Google Scholar]
- Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang ZX, Kono I, Kurata N, Yano M, Iwata N, Sasaki T. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc Natl Acad Sci USA. 1998;95:1663–1668. doi: 10.1073/pnas.95.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Kurth J, Wei F, Elliott C, Valé G, Yahiaoui N, Keller B, Somerville S, Wise R, Schulze-Lefert P. Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1 independent signaling pathway. Plant Cell. 2001;13:337–350. doi: 10.1105/tpc.13.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]