Abstract
Herpes simplex virus type I (HSV) typically enters peripheral nerve terminals and then travels back along the nerve to reach the neuronal cell body, where it replicates or enters latency. To monitor axoplasmic transport of HSV, we used the giant axon of the squid, Loligo pealei, a well known system for the study of axoplasmic transport. To deliver HSV into the axoplasm, viral particles stripped of their envelopes by detergent were injected into the giant axon, thereby bypassing the infective process. Labeling the viral tegument protein, VP16, with green fluorescent protein allowed viral particles moving inside the axon to be imaged by confocal microscopy. Viral particles moved 2.2 ± 0.26 μm/sec in the retrograde direction, a rate comparable to that of the transport of endogenous organelles and of virus in mammalian neurons in culture. Electron microscopy confirmed that 96% of motile (stripped) viral particles had lost their envelope but retained tegument, and Western blot analysis revealed that these particles had retained protein from capsid but not envelope. We conclude that (i) HSV recruits the squid retrograde transport machinery; (ii) viral tegument and capsid but not envelope are sufficient for this recruitment; and (iii) the giant axon of the squid provides a unique system to dissect the viral components required for transport and to identify the cellular transport mechanisms they recruit.
Herpes simplex virus (HSV) is a major cause of infectious corneal blindness, as well as a host of other diseases, ranging in severity from the common cold sore to life-threatening encephalitis (1, 2). After infecting sensory nerve endings in mucous membranes, the virus travels back within the neuron to its cell body, either in a dorsal root ganglion or in the eye, from the nerve terminals in the cornea along the trigeminal nerve to the trigeminal ganglion (3–7). This retrograde transport at 3–5 mm/h (7) is a crucial step in the viral life cycle because without transport to the cell nucleus, no latency or neuronal replication can occur. Neither the viral proteins required for this transport nor the cellular motors that mediate it are known.
Various viral components have been implicated as mediators of transport. The virus is composed of an inner DNA core, a capsid, the tegument, and an outer envelope, which is a lipid membrane containing glycoproteins (6, 8, 15). During infection, the viral envelope fuses with the cell membrane, liberating the nucleocapsid and associated tegument proteins into the cytoplasm of the cell, where they are transported to the nucleus (10–14). The viral proteins most likely to recruit the cellular transport machinery are the ≈11 tegument proteins, although these are the least well characterized viral proteins. Some tegument proteins play a role in the nucleus, such as transactivation (VP16 and ICP4), shut-off of host protein synthesis (vhs), and phosphorylation (UL13 kinase) (3, 8). These proteins are therefore believed to be transported along with the capsid to the nucleus, where they mediate these early events in viral replication. Biochemical interactions between capsid and tegument proteins and between different tegument proteins themselves argue that these components remain together as a structural unit during retrograde transport (15–18).
The mechanism of this viral transport might be discovered if transport could be directly observed in a living axon. However, direct observations in real time of intracytoplasmic movements of HSV particles in living axons has been difficult, although some analyses in fixed cultured neurons or living Vero cells have been done (7, 19–22).** What was needed was a living axon with intact viable transport that was long enough to image the rapid movement of viral particles over long distances. It was also important that viral particles of a known composition be delivered into this axon at a specific time and at a discrete, identifiable, location—none of which are features of viral delivery through the normal infection process.
The giant axon of the squid provides an opportunity to meet these conditions. The giant axon is up to 7 cm long and 0.6 mm in diameter, making it possible to inject labeled viral components whose transport can then be directly observed over long distances (23–25). Transport within the axon persists for up to 6 h at room temperature, even in axons isolated from cell bodies and synapses. The highly conserved nature of this transport machinery has allowed the discovery in squid of the molecular mechanism of axoplasmic transport common to many other systems. For example, microtubule-based transport of axoplasmic organelles was first observed in squid axons (26), and purification of the squid protein responsible for this activity, kinesin (27), enabled the identification of homologous proteins in many other species (28, 29). We thus reasoned that, even though HSV is a human virus that would not infect squid, it was possible that, once inside the squid axon, this human virus would be capable of recruiting conserved transport machinery. Direct observation of viral transport would allow us to dissect the viral components required for transport and the cellular machinery they recruit. A virus-based bioassay could also identify universal principles governing the normal recruitment of specific retrograde motors by endogenous organelles.
Materials and Methods
Generation of Green Fluorescent Protein (GFP)-Labeled HSV.
To image HSV in the squid axon, a fusion construct was generated encoding the N terminus of VP16 and GFP in the pEGFP plasmid. The plasmid (pNFT, American Type Culture Collection) containing the HSV type 1 VP16 gene was digested with PstI and SacII, thereby retaining the 5′ start codon and removing 86 amino acids from the carboxyl terminus of the encoded protein (30). This was ligated into the N-terminal fusion vector, pEGFP/neoR (CLONTECH), after digestion with the same restriction enzymes, yielding the in-frame fusion transcript, VP16-GFP, under the control of the CMV IE promoter, based on the findings of McKnight et al. (31) that VP16 fusion proteins of this type can participate in virion assembly and targeting. Vero cells were transfected with pVP16-GFP and were cloned under G418 selection. A stable transfected clonal line chosen for high fluorescence was infected with HSV1 hrR3 (32) at a multiplicity of infection of 2. Forty-eight hours later, progeny virus were harvested by pooling cells and medium, freeze thawing three times, and centrifuging at 72,000 × g for three hours through a 25% sucrose cushion in PBS. Virus was suspended in PBS, was titered as plaque forming units on Vero cells, and was stored at −80°C in aliquots. Resultant virions contained both normal and GFP-labeled VP16. Typically, there are 10 virions per plaque forming unit.
Injection into the Giant Axon.
VP16-GFP-labeled virus diluted 1:1 was stripped by treatment for 30–40 min on ice by diluting virus 1:1 with 1.2 M KCl and 0.2% Triton X-100. Microscopic examination of the viral stock before and after stripping revealed wide variation in fluorescence intensity between different viral particles within the same preparation. Viral suspension (≈10 pl of 108 plaque forming units/ml) was loaded into a micropipette, the pipette tip was sealed with mineral oil, and both solutions were injected into a freshly dissected giant axon of known orientation whose synaptic and neurokaryon ends were color-coded with string during dissection (24, 25). The axon was transferred to the stage of a Bio-Rad confocal microscope, and the oil droplet marking the injection site was identified with Nomarski optics. The region of axoplasm adjacent to the oil droplet on the side toward the cell body was then examined by laser scanning confocal microscopy using a fluorescein filter set with a 40× oil immersion lens on a Zeiss upright microscope. Images were captured by using the Bio-Rad confocal software program (33).
Electron Microscopy.
Virus (10 μl) treated in parallel for electron microscopy were mounted on a Formvar-coated, deionized copper grid, were stained with 1% aqueous uranyl acetate for 1 min, were dried, and were imaged in a JEOL 200CX electron microscope.
Western Blots.
Viral particles either from stock preparations or after treatment with detergent were sedimented at 15,000 × g for 15 min, and the resultant supernatants and pellets were separated and subjected to SDS/gel electrophoresis. Blots were probed with either anti-GFP antibody (CLONTECH), a polyclonal anti-gD antibody (Goodwin Institute, Plantation, FL), or anti-capsid (VP5) monoclonal antibody (BioDesign, Saco, ME).
Results
Retrograde Movement of Viral Particles.
GFP-labeled viral particles moving in the axon were readily identified by confocal microscopy (Fig. 1). All particles moved in the retrograde direction (toward the cell body). For each experiment, 100 digital images were collected at 3- or 4-sec intervals from a single microscopic field in an axon. Typically, a sequence of 100 frames captured the movements of ≈20 or more different particles. Individual particles varied in brightness, possibly because they were at different depths within the axoplasm, because they had accumulated different amounts of the labeled VP16, or because they were aggregates containing different numbers of viral particles. In some microscopic fields, all particles disappeared at the same point, suggesting that the tracks on which they moved had left the plane of focus. The linear continuity of the transport pathways was apparent in instances in which particles moved within the plane of focus throughout the field. Very little lateral movement was observed—particles deviated from frame to frame less than 4 μm from a straight trajectory in any given field. No particles reversed to move in the anterograde direction. Rarely, a particle that had moved away from the injection site was no longer moving, presumably because it had lost contact with its motor or transport tracks, or because it became entangled in cytoskeletal elements. These stationary particles served as useful internal controls demonstrating that the movement of the other particles was not attributable to movement of the whole axon.
Figure 1.
Time lapse sequence of GFP-labeled HSV transported in a living axon. Individual frames of a single microscopic field in a squid axon injected with stripped HSV labeled with VP16-GFP are shown in sequence from left to right (5.3 sec between frames). The stationary particle at the lower right of each frame (arrowhead) serves as reference point for the changes in position of two other particles as they rapidly cross the field on the left of each frame (one of these is indicated by a diagonal arrow). The two moving particles move approximately the same distance between frames, thereby remaining separated from each other by a similar distance. The cell body was toward the top of the page. A live video of this sequence can be accessed at E.L.B.'s web site, http://biomed.brown.edu/faculty/B/Bearer.html. (Bar = 40 μm.)
No particulate fluorescence was observed in un-injected axons, demonstrating that all detectable particles contained GFP. The maximum number of moving particles observed in a single axon injected with stripped virus was 113. Because we can only image a small area of the axon at any one time, observing so many particles moving in the retrograde direction suggests that a large number of stripped virus are picked up by the retrograde transport system.
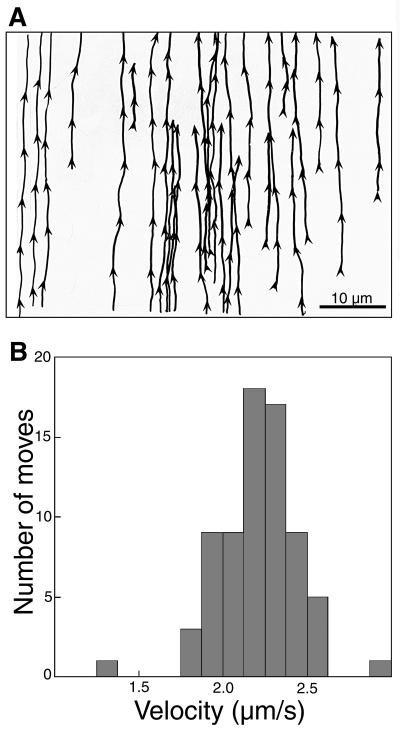
Tracings from a series of 100 frames illustrate the consistency of speeds and tracks (Fig. 2). At any one time, only two to four particles appear in the field, and each particle crosses the field in five to six frames. All particles move at similar rates, with the distance moved between each frame almost identical. By tracing the movements of 26 different particles as they moved across the same microscopic field, the tracks they each followed through the axon can be superimposed. The distribution of the groups of tracks closely resembles the actin-microtubule transport network we have recently described (34). Individual tracks in the example overlap but cannot be superimposed, suggesting that there are numerous different tracks on which a viral particle can move. Interweaving of the trajectories of individual particles could be a result of particles switching tracks.
Figure 2.
Herpes viral particles are transported with uniform directionality and velocity along multiple, interweaving tracks in the living axon. (A) Tracings of pathways of 26 particles in a single field of an axon: Tracings were taken from a series of 100 frames captured at 4-sec intervals with a 40× objective and a 5× zoom in a single field from an axon after injection of VP16-GFP-labeled, stripped virus. The cell body was toward the top of the page. Arrowheads indicate the site where an image of each particle was captured in a frame as well as its direction of movement as determined by its location in the subsequent frame. (B) Viral particles move with an average velocity of 2.2 μm/sec. Measurements of the distances moved between frames of 71 particles, all going in the retrograde direction, are summarized in a histogram.
Measurements of 71 different position changes between frames demonstrate that the particles move at a consistent rate of 2.2 ± 0.26 μm/sec. Only 2 of the 71 moves varied from this rate by more than 0.4 μm. This consistency in the distance moved between 4-sec frames demonstrates that very little pausing occurs during transport. This is not a saltatory type movement, such as described for mitochondria, which also move in the retrograde direction in squid, but at velocities 1/10 those of the viral particles (35). Untreated virus typically remained at the injection site, but very occasionally individual particles made moves in either the anterograde or retrograde direction.
Moving particles must contain at least VP16 because the GFP we imaged is fused to that tegument protein. These fluorescent particles are likely to contain several copies of VP16-GFP [our microscope is unlikely to be able to detect single GFP molecules (36)], as well as the other viral proteins to which VP16 binds, probably including the entire capsid together with its complement of tegument proteins. GFP alone would not be expected to be packaged into the virus because it lacks viral targeting sequences, and we detected no 27-kDa GFP in Western blots of labeled virions (data not shown).
Electron Microscopy of Stripped Particles.
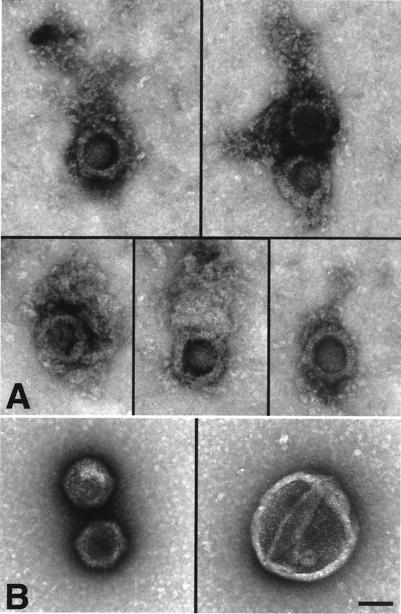
To determine the structure of the moving particles, aliquots of virus treated like those used for injections were examined by negative stain electron microscopy. Treated virions were compared with untreated virus from the same viral stock (Fig. 3). Preparations of virus that moved in the axon primarily contained particles with the typical hexagonal shape of the capsid that were uniformly decorated with a net-like matrix, often attached to one apex of the hexagon (Fig. 3A). This net-like shell closely resembles the tegument as imaged in vitreous ice (37). In contrast, enveloped virus (Fig. 3B Right) and naked capsids without associated material (Fig. 3C Left) predominated in the untreated viral stock. Morphometric analysis demonstrated that 0.1% Triton treatment increases the proportion of de-enveloped particles from 55% to 96% (Table 1). Control treatment with salt (0.667 KCl), intended to mimic the intracellular osmolarity of the squid axon (23–27, 38, 39), only increased the number of de-enveloped virus by 11%. More importantly, after 0.1% Triton, the majority of de-enveloped particles (77%) were associated with net-like tegument whereas all de-enveloped particles observed in the untreated viral stock were naked capsids with no identifiable tegument. The small number of tegument-associated capsids in the salt-treated preparation could correspond to those few moving particles seen in axons injected with untreated virus.
Figure 3.
Motile particles are composed of capsid plus a net-like tegument but no envelope. (A) Five images demonstrating the typical structure of stripped virus: a hexagonal capsid with a net-like tegument attached to it, either at one apex or surrounding it. (B and C) Two types of particles predominate in untreated viral preparations: enveloped virus (B) and naked capsid (C). A–C are at the same magnification. (Bar = 100 nm.)
Table 1.
Morphometric analysis of the effects of Triton treatment on tegument
| Treatment | Virus without envelope, % (n1) | De-enveloped capsids retaining tegument, % (n2) |
|---|---|---|
| No treatment | 55 (23) | 0 (26) |
| Salt | 66 (32) | 8 (13) |
| 0.1% Triton + salt | 96 (58) | 77 (30) |
| 1% Triton + salt | 100 (34) | 60 (10) |
Method: For the first series, comparison of virus with and without envelopes, all viral particles in a field were included, and the presence of envelope was determined by size and morphological characteristics matching the typical envelope (Fig. 3B). Those particles with envelope measured 200–230 nm in diameter whereas capsids measured ≈130 nm. For the second series, capsids retaining tegument were determined structurally only in those micrographs in which all particles were sufficiently well preserved and well stained to distinguish the presence or absence of this delicate structure. Micrographs were photographed by one observer (E.L.B.) and were counted blind on prints by two different second observers. n1, Total number of viral particles counted in each group; n2, Total number of capsids counted in each group.
Western Blots of Viral Particles.
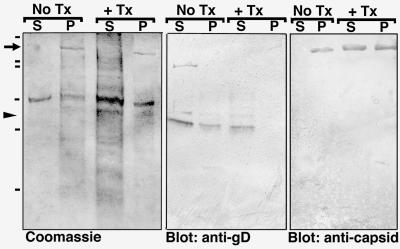
To determine whether the loss of the viral membrane corresponded to loss of envelope proteins, we separated viral particles from soluble proteins before and after detergent treatment by centrifugation and analyzed the fractions by Coomassie blue gel and Western blot analysis (Fig. 4). Coomassie staining revealed that the majority of viral proteins in untreated samples collect in the particulate (insoluble) fraction (Fig. 4, lanes labeled P) whereas Triton treatment solubilizes much of the total protein, shifting it from the particulate to the soluble fraction. Triton-solubilized proteins include a set of bands between ≈45 and 60 kDa, representing the various forms of gD, a major envelope glycoprotein (6, 7). In contrast, most of the major capsid protein, VP5 (6, 7), remains in the particulate fraction after treatment, appearing as a prominent ≈155-kDa band in the Coomassie gels, as confirmed by Western blot analysis. VP16-GFP was detected only in the particulate fraction of both preparations by Western blotting, and no fluorescence was detected by microscopy in the supernatant with or without Triton treatment. These results confirm the electron microscopic observations and identify specific proteins that have been removed from the motile particles by Triton treatment.
Figure 4.
Biochemical analysis of stripped, motile, viral particles. Shown are Coomassie-stained gels and parallel Western blots showing soluble (S) and particulate (P) fractions of viral preparations either untreated (No Tx) or treated (+ Tx) with 0.1% Triton and high salt (see Fig. 1).
Discussion
De-enveloped human HSV particles are transported in the retrograde direction after injection into the squid axon. The absence of the envelope in preparations of motile viral particles rules out the envelope as a component required for retrograde transport. Because untreated whole virus was not seen to move in these experiments, some internal component must be exposed to the axoplasm for successful recruitment into the retrograde transport system.
That the retrograde transport of HSV reported here depends on axonal transport motors is supported by (i) the uniformity of instantaneous velocities of viral particles; (ii) the similarity in rate of transport with that of minus-end directed motility (2–4 μm/sec) of isolated squid organelles on microtubules in vitro (23–27); and (iii) the similarity of rate of viral transport with the rate of transport of squid organelles (0.1–4 μm/sec) in the axon (23, 26). Furthermore, the speed and direction of HSV transport in squid axons mimic those observed in mammalian neurons in culture (7), thus supporting the relevance of our observations in squid to the normal pathway of Herpes transport in its natural hosts. Finally, the retrograde transport in the squid axon is likely to be specific for injected virus because beads similarly injected into the squid axon move only in the anterograde direction (24, 25).
The consistency of rate and uniformity of direction argue for a single transport mechanism. The characteristics of this transport are similar to the motility observed with cytoplasmic dynein, a microtubule-based motor that moves cargo toward the minus-end (retrograde) in vitro.
There has been considerable speculation about the identity of the motor that mediates retrograde transport of HSV (2–4, 7, 12, 19–22).** In addition to microtubules, actin filaments are now known to act as tracks for axoplasmic organelle motility (38–43). Thus, motors for either system could be co-opted by the virus. Currently, there is only one known microtubule-based retrograde motor in axons, dynein (44, 45), although kinesins with minus-end directed motility have recently been identified in other systems (28, 29). Little is known so far about actin-based motors in axons, and much remains to be understood about how axonal motors interact with cargo. Studies on dynein are complicated by the fact that the dynein heavy chain responsible for the motor activity is very large (>400 kDa) and exists in a huge macromolecular complex containing at least eight other subunits. In addition, dynein requires the presence and activity of a cofactor, dynactin, another large, multimolecular complex, to bind to organelles (46, 47). Which molecule(s) within these complexes directly contact the cargo remains unknown. Identification of the viral proteins and their specific amino acid sequences that are required for retrograde transport should provide a powerful tool for the discovery of endogenous sequences on cellular components that recruit retrograde motor(s) for transport.
Acknowledgments
We thank Dr. B. Sodeik for helpful discussion, Dr. Cornel Fraefel for advice and help in producing the GFP-VP16 virus, Michelle Schlief for performing injections and helping with the microscopy, and John Chludzinski for computer support. This work was supported in part by National Institutes of Health Grants GM47638 (E.L.B.) and EYO8773 (J.H.L.), the Frederik Bang Fellowship, and the Evelyn and Melvin Spiegel Fellowship Fund from the Marine Biological Laboratory (Woods Hole, MA).
Abbreviations
- HSV
herpes simplex virus
- GFP
green fluorescent protein
Footnotes
Sodeik, B., 24th International Herpesvirus Workshop, July 21, 1999, Massachusetts Institute of Technology, Cambridge, MA.
References
- 1.Whitley R J. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippencott; 1996. pp. 2297–2336. [Google Scholar]
- 2.Whitley R J, Kimberlin D W, Roizman B. Clin Infect Dis. 1998;26:541–555. doi: 10.1086/514600. [DOI] [PubMed] [Google Scholar]
- 3.Cook M L, Stevens J G. Infect Immun. 1973;7:272–288. doi: 10.1128/iai.7.2.272-288.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margolis T, Togni B, LaVail J, Dawson C R. Curr Eye Res. 1987;6:119–126. doi: 10.3109/02713688709020078. [DOI] [PubMed] [Google Scholar]
- 5.Topp K S, Meade L B, LaVail J H. J Neurosci. 1994;14:318–325. doi: 10.1523/JNEUROSCI.14-01-00318.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roizman B, Sears P. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippencott; 1996. pp. 2231–2295. [Google Scholar]
- 7.Lycke E, Kristensson K, Svennerholm B, Vahlne A, Ziegler R. J Gen Virol. 1984;65:55–64. doi: 10.1099/0022-1317-65-1-55. [DOI] [PubMed] [Google Scholar]
- 8.Haarr L, Skulstad S. APMIS. 1994;102:321–346. doi: 10.1111/j.1699-0463.1994.tb04882.x. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs A, Breakefield X O, Fraefel C. Neoplasia. 1999;1(4):1–15. [Google Scholar]
- 10.Wittels M, Spear P G. Virus Res. 1990;18:271–290. doi: 10.1016/0168-1702(91)90024-p. [DOI] [PubMed] [Google Scholar]
- 11.Elliott G, Mouzakitis G, O'Hare P. J Virol. 1995;69:7932–7941. doi: 10.1128/jvi.69.12.7932-7941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sodeik B, Ebersold M W, Helenius A. J Cell Biol. 1998;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajcani J, Kudelova M. Virus Genes. 1999;18:81–90. doi: 10.1023/a:1008025520655. [DOI] [PubMed] [Google Scholar]
- 14.Spear P. Semin Virol. 1993;4:167–180. [Google Scholar]
- 15.McNabb D S, Courtney R J. J Virol. 1991;190:221–232. doi: 10.1016/0042-6822(92)91208-c. [DOI] [PubMed] [Google Scholar]
- 16.Elliott G, Mouzakitis G, O'Hare P. J Virol. 1995;69:7932–7941. doi: 10.1128/jvi.69.12.7932-7941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smibert C A, Popova B, Xiao P, Capone J P, Smiley J R. J Virol. 1994;68:2339–2346. doi: 10.1128/jvi.68.4.2339-2346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLaughlin J, Rixon F J. J Gen Virol. 1992;73:269–276. doi: 10.1099/0022-1317-73-2-269. [DOI] [PubMed] [Google Scholar]
- 19.Elliott G, O'Hare P. J Virol. 1999;73:4110–4119. doi: 10.1128/jvi.73.5.4110-4119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suomalainen M, Nakano M Y, Keller S, Boucke K, Stidwill R P, Greber U F. J Cell Biol. 1999;114:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penfold M E, Armati P, Cunningham A L. Proc Natl Acad Sci USA. 1994;91:6529–6533. doi: 10.1073/pnas.91.14.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristensson K, Lycke E, Roytta M, Svennerholm B, Vahlne A. J Gen Virol. 1986;67:2023–2028. doi: 10.1099/0022-1317-67-9-2023. [DOI] [PubMed] [Google Scholar]
- 23.Allen R D, Metuzals J, Tasaki I, Brady S T, Gilbert S P. Science. 1982;218:1127–1129. doi: 10.1126/science.6183744. [DOI] [PubMed] [Google Scholar]
- 24.Terasaki M, Schmidek A, Galbraith J A, Gallant P E, Reese T S. Proc Natl Acad Sci USA. 1995;92:11500–11503. doi: 10.1073/pnas.92.25.11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galbraith J A, Reese T S, Schlief M L, Gallant P E. Proc Natl Acad Sci USA. 1999;96:11589–11594. doi: 10.1073/pnas.96.20.11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnapp B J, Vale R D, Sheetz M P, Reese T S. Cell. 1985;40:455–462. doi: 10.1016/0092-8674(85)90160-6. [DOI] [PubMed] [Google Scholar]
- 27.Vale R D, Reese T S, Sheetz M P. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstein L S, Philp A V. Annu Rev Cell Dev Biol. 1999;15:141–183. doi: 10.1146/annurev.cellbio.15.1.141. [DOI] [PubMed] [Google Scholar]
- 29.Kirchner J, Woehlke G, Schliwa M. J Biol Chem. 1999;380:915–921. doi: 10.1515/BC.1999.113. [DOI] [PubMed] [Google Scholar]
- 30.Byrne G W, Ruddle F H. Proc Natl Acad Sci USA. 1989;86:5473–5477. doi: 10.1073/pnas.86.14.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKnight J L, Doerr M, Zhang Y. J Virol. 1994;68:1750–1757. doi: 10.1128/jvi.68.3.1750-1757.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldstein D J, Weller S K. J Virol. 1988;62:196–205. doi: 10.1128/jvi.62.1.196-205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bearer E L, Schlief M L, Breakefield X O, Schuback D E, Reese T S, LaVail J H. Biol Bull (Woods Hole, Mass) 1999;197:257. doi: 10.2307/1542637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bearer E L, Reese T S. J Neurocytol. 1999;28:85–98. doi: 10.1023/a:1007025421849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martz D, Lasek R J, Brady S T, Allen R D. Cell Motil. 1984;4:89–101. doi: 10.1002/cm.970040203. [DOI] [PubMed] [Google Scholar]
- 36.Pierce D W, Vale R D. Methods Cell Biol. 1999;58:49–73. doi: 10.1016/s0091-679x(08)61948-2. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Z H, Chen D H, Jakana J, Rixon F J, Chiu W. J Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brady S T, Lasek R J, Allen R D. Science. 1982;218:1129–1131. doi: 10.1126/science.6183745. [DOI] [PubMed] [Google Scholar]
- 39.Morris J R, Lasek R J. J Cell Biol. 1982;92:192–198. doi: 10.1083/jcb.92.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuznetsov S A, Langford G M, Weiss D G. Nature (London) 1992;356:722–725. doi: 10.1038/356722a0. [DOI] [PubMed] [Google Scholar]
- 41.Bearer E L, DeGiorgis J A, Bodner R A, Kao A W, Reese T S. Proc Natl Acad Sci USA. 1993;90:11252–11256. doi: 10.1073/pnas.90.23.11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bearer E L, DeGiorgis J A, Medeiros N A, Jaffe H, Reese T S. Proc Natl Acad Sci USA. 1996;93:6064–6068. doi: 10.1073/pnas.93.12.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris R L, Hollenbeck P J. J Cell Biol. 1995;131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vallee R B, Gee M A. Trends Cell Biol. 1998;8:490–494. doi: 10.1016/s0962-8924(98)01379-8. [DOI] [PubMed] [Google Scholar]
- 45.Karki S, Holzbaur E L. Curr Opin Cell Biol. 1999;11:45–53. doi: 10.1016/s0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- 46.Gill S R, Schroer T A, Szilak I, Steuer E R, Sheetz M P, Cleveland D W. J Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holleran E A, Karki S, Holzbaur E L. Int Rev Cytol. 1998;182:69–109. doi: 10.1016/s0074-7696(08)62168-3. [DOI] [PubMed] [Google Scholar]