Abstract
The biochemical and molecular properties of the β-oxidation enzymes from algae have not been investigated yet. The present study provides such data for the phylogenetically old alga Euglena (Euglena gracilis). A novel multifunctional β-oxidation complex was purified to homogeneity by ammonium sulfate precipitation, density gradient centrifugation, and ion-exchange chromatography. Monospecific antibodies used in immunocytochemical experiments revealed that the enzyme is located in mitochondria. The enzyme complex is composed of 3-hydroxyacyl-coenzyme A (-CoA) dehydrogenase, 2-enoyl-CoA hydratase, thiolase, and epimerase activities. The purified enzyme exhibits a native molecular mass of about 460 kD, consisting of 45.5-, 44.5-, 34-, and 32-kD subunits. Subunits dissociated from the complete complex revealed that the hydratase and the thiolase functions are located on the large subunits, whereas two dehydrogenase functions are located on the two smaller subunits. Epimerase activity was only measurable in the complete enzyme complex. From the use of stereoisomers and sequence data, it was concluded that the 2-enoyl-CoA hydratase catalyzes the formation of l-hydroxyacyl CoA isomers and that both of the different 3-hydroxyacyl-CoA dehydrogenase functions on the 32- and 34-kD subunits are specific to l-isomers as substrates, respectively. All of these data suggest that the Euglena enzyme belongs to the family of β-oxidation enzymes that degrade acyl-CoAs via l-isomers and that it is composed of subunits comparable with subunits of monofunctional β-oxidation enzymes. It is concluded that the Euglena enzyme phylogenetically developed from monospecific enzymes in archeons by non-covalent combination of subunits and presents an additional line for the evolutionary development of multifunctional β-oxidation enzymes.
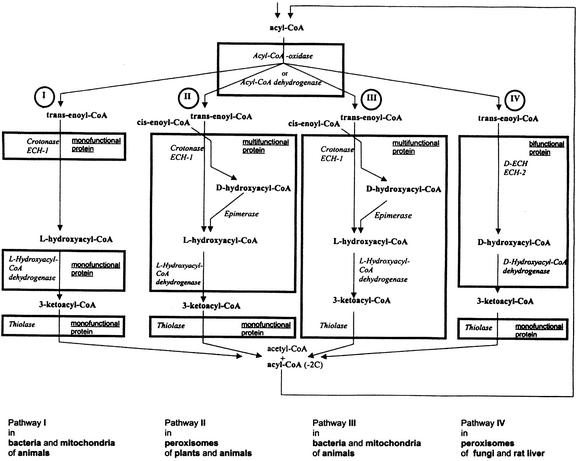
The degradation of activated fatty acids by β-oxidation principally requires four different enzyme-catalyzed reactions. The acyl-CoA esters are desaturated in an initial reaction. The resulting 2-enoyl-CoA esters are then hydrated to 3-hydroxyacyl-CoA esters, which in a third step are dehydrogenated concomitant with the reduction of NAD. The fourth reaction is the acetyl-CoA yielding thiolytic cleavage of the 3-ketoacyl-CoA esters formed in the dehydrogenation process. Because the fatty acid chains are reduced by only two carbon atoms in the course of these successive reactions, the reaction sequence has to be repeated until the fatty acids have been completely degraded (Fig. 1).
Figure 1.
Most common pathways for the degradation of fatty acid CoA esters by β-oxidation enzymes in the different organisms. In the first reaction of the β-oxidation cycle, acyl-CoA esters are desaturated to Δ2-trans enoyl-CoA esters by acyl CoA oxidases or acyl CoA dehydrogenases. Oxidases are located in microbodies of higher plants and animal tissue. Dehydrogenases are found in bacteria, animal mitochondria, and microbodies of some fungi and in algae. For the following reaction of the β-oxidation cycle, different pathways are used. Pathway I, The Δ2-trans enoyl-CoA esters are metabolized by monofunctional crotonases (also named ECH-1), l-hydroxyacyl-CoA dehydrogenases, and thiolases. This pathway is located in bacteria and animal mitochondria. Pathway II, Crotonase, and l-hydroxyacyl-CoA dehydrogenase are domains of a common polypeptide that also metabolizes Δ2-cis-enoyl-CoA esters occurring in unsaturated fatty acids. These Δ2-cis-enoyl-CoA esters are also suitable substrates for the crotonase, but the resulting product is d-hydroxyacyl-CoA, which is transformed to the l-form by an epimerase function of the polypeptide. This MFP is specifically found in microbodies of higher plants and animals. Pathway III, The polypeptide that carries functions for crotonase, l-hydroxyacyl-CoA dehydrogenase, and epimerase is combined with a second polypeptide that displays thiolase activity. This type of MFP is found in bacteria and animal mitochondria. Pathway IV, The Δ2-trans enoyl-CoA esters are metabolized by a d-specific 2-enoyl-CoA hydratase (d-ECH; ECH-2) that forms d-specific products and a d-hydroxyacyl-CoA dehydrogenase that is specific to d-hydroxyacyl-CoA esters as substrates. Both enzyme functions are located at a common polypeptide. This bifunctional protein is located in microbodies of fungi and in rat liver tissue.
The first reaction, in which acyl-CoA esters are desaturated can be catalyzed by different types of enzymes, either an acyl-CoA oxidase transferring electrons to molecular oxygen and thereby producing H2O2 or an acyl-CoA dehydrogenase coupled—via an electron transferring protein—with an electron transport chain reducing oxygen to H2O, but incapable of forming H2O2. So far, the acyl CoA oxidase has been detected in microbodies of higher plants (Kindl, 1993), animals (Hashimoto, 1987), and some fungi (Kunau et al., 1987). Acyl-CoA dehydrogenase, however, is localized in bacteria (Klein, 1973), the mitochondria of animals (Hall, 1978), and primitive algae (Stabenau, 1992), but it was also found in the microbodies of developed algae (Winkler et al., 1988) and in the fungus Neurospora crassa (Kionka and Kunau, 1985). The existence of biochemical pathways without an H2O2 metabolism may be characteristic of microbodies in phylogenetically old organisms (Stabenau, 1992; Kunau et al., 1995).
In the following steps, as in the first reaction, there are different enzyme types catalyzing the hydration and dehydrogenation reactions. In particular, they differ in stereospecificity of substrates required and products formed. All in all, the different enzymes of both reaction steps are assigned to two different metabolic routes. The first and most widespread route consists of a 2-enoyl-CoA hydratase, also termed enoyl-CoA hydratase 1 or crotonase. It hydrates 2-trans-enoyl-CoA esters to l-3-hydroxyacyl-CoA esters, but also 3-cis-enoyl-CoA esters to d-3-hydroxyacyl-CoA esters. Because only l-dependent 3-hydroxyacyl-CoA dehydrogenases are available for the subsequent reaction step in this first metabolic route, d-3-hydroxyacyl-CoA esters must be transformed into the corresponding l-form in case they arise, for example, during metabolism of unsaturated fatty acids. This transformation takes place by means of an epimerase reaction.
The second metabolic route is characterized by a 2-enoyl-CoA hydratase that hydrates 2-trans-enoyl-CoA to d-3-hydroxyacyl-CoA esters. It is called d-trans-enoyl-CoA hydratase, enoyl-CoA hydratase 2, or novel enoyl-CoA hydratase. The resulting d-3-hydroxyacyl-CoA esters are further metabolized by d-3-hydroxyacyl-CoA dehydrogenases.
Enzymes of the first route are monospecific proteins, which are characteristic of animal mitochondria (Osumi and Hashimoto, 1980; Fong and Schulz, 1981) but have also been detected in some bacteria (O‘Connell et al., 1990; Klenk et al., 1997). Hydratase and dehydrogenase functions, however, may also be localized on a common polypeptide that most likely resulted from a fusion process between the genes of both monospecific enzymes during evolution (Kamijo et al., 1993). Corresponding multifunctional proteins (MFPs) are the multifunctional enzyme 1 in animal peroxisomes (Furuta et al., 1980), the trifunctional and tetrafunctional proteins from plant glyoxysomes (Kindl, 1993), the α-subunit of the trifunctional protein from rat liver mitochondria (Uchida et al., 1992), and the α-subunit of the multifunctional fatty acid oxidation complex from Escherichia coli (Binstock et al., 1977) and other eubacteria (Kunau et al., 1995). The MFPs from rat liver mitochondria and eubacteria additionally contain β-subunits with thiolase function.
Hydratases and dehydrogenases of the second metabolic route are always domains of a common polypeptide. So far, such bifunctional enzymes have only been detected in peroxisomes, like the multifunctional enzyme 2 from rat (Dieuaide-Noubhani et al., 1997) and the β-oxidation enzymes found in different fungi (Thieringer and Kunau, 1991). All d-enoyl-CoA hydratase and all d-3-hydroxyacyl-CoA dehydrogenase domains from the different organisms exhibit pronounced amino acid sequence similarities among themselves (Qin et al., 1997). The same is true for monospecific enzymes or domains displaying 2-enoyl-CoA hydratase 1 or l-3-hydroxyacyl-CoA dehydrogenases activity, respectively (Baldwin, 1993; Kamijo et al., 1993). Because there are no sequence relations between the d-specific bifunctional enzymes of the second route and the l-specific β-oxidation enzymes of the first one (Hiltunen et al., 1992), the enzymes of both routes obviously belong to different evolutionary families.
Enzymes of all four reactions of the β-oxidation pathway have also been detected in algae. In the group of Heterokontophyta, corresponding enzyme activities have only been found in the mitochondria (Gross et al., 1985; Winkler and Stabenau, 1994). In the green alga Chara spp., however, they are exclusively localized in peroxisomes (H. Stabenau, U. Winkler, and W. Säftel, unpublished data). A localization in mitochondria as well as in peroxisomes was found in the green algae Mougeotia spp. and Eremosphaera spp. (Stabenau et al., 1984; Winkler et al., 1988) and in heterotrophically grown Euglena (Euglena gracilis; Graves and Becker, 1974).
The biochemical and molecular properties of the β-oxidation enzymes from algae have not been investigated yet. Hence, the present study provides such data for the first time, to our knowledge. We chose the alga Euglena for our investigations because it is one of the few algal species that can be cultured heterotrophically on fatty acids (Hosotani et al., 1988). Moreover, Euglena is one of the oldest eukaryotes (Tessier et al., 1997) and may therefore be a useful organism to enhance our knowledge about the evolution of β-oxidation enzymes.
RESULTS
Enzymes of the Fatty Acid β-Oxidation Pathway in Euglena
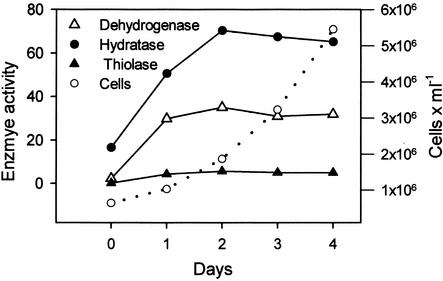
In Euglena, an increase in the activity of the enzymes involved in the β-oxidation pathway was measured after the conditions were changed from autotrophic to heterotrophic growth with hexanoic acid as the sole source of carbon and energy (Fig. 2). Enzyme activity reached a maximum after 2 d of heterotrophic growth. Cells were harvested at this time and used for the experiments described here.
Figure 2.
Increase in β-oxidation enzyme activity in Euglena. Autotrophically grown cells were transferred to a growth medium containing 5 mmol L−1 hexanoic acid as sole carbon source and cultured in the dark for 6 d. Enzyme activities for 3-hydroxyacyl-CoA dehydrogenase (Dehydrogenase), 2-enoyl-CoA hydratase (Hydratase), and thiolase were measured in the crude homogenates and are expressed as nanomoles per minute per 106 cells. Growth is indicated by the cell number on the right ordinate.
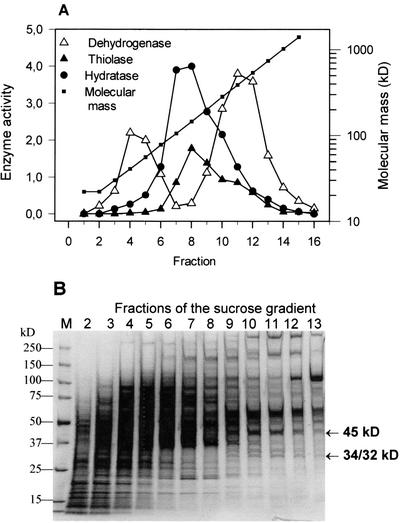
Enzymes of β-oxidation in the crude homogenate of hexanoic acid-grown algae were separated according to their molecular mass by Suc gradient centrifugation. As shown in Figure 3A, the distribution patterns of 2-enoyl-CoA hydratase and thiolase each show one peak in fraction eight, indicating a molecular mass of around 146 kD. Thiolase and 2-enoyl-CoA hydratase having similar molecular masses have already been described (Staak et al., 1978; Fong and Schulz, 1981). The distribution of the 3-hydroxyacyl-CoA dehydrogenase reveals two peaks. In fraction four, the enzyme protein should possess a molecular mass of around 41 kD, which makes it comparable with monospecific l-3-hydroxyacyl-CoA dehydrogenase from animal mitochondria (Osumi and Hashimoto, 1980). The molecular mass of the enzyme in fractions 11 to 12 is expected to be somewhere between 390 and 540 kD, yet no comparable 3-hy-droxyacyl-CoA dehydrogenase has been found in other organisms. That is why we have directed our attention to this enzyme. The biochemical and molecular properties of the protein associated with the 3-hydroxyacyl-CoA dehydrogenase activity will be detailed in the following section.
Figure 3.
Distribution of β-oxidation enzymes from hexanoic acid-grown Euglena in a Suc gradient. Proteins of a crude homogenate were separated according to their molecular masses in a linear gradient from 5% to 20% (w/w) Suc. Centrifugation was performed at 140,000g for 18 h and at 20°C. A, Distribution of enzyme activities in the gradient. Enzyme activity is expressed as micromoles per minute per milliliter fraction for 3-hydroxyacyl-CoA dehydrogenase (Dehydrogenase) and 2-enoyl-CoA hydratase (Hydratase). Full-scale activity for the thiolase reaction corresponds to 50 nmol min−1 mL−1 fraction. The distribution of molecular masses in the fractions was obtained from the distribution of standard proteins. B, Analysis of the proteins distributed in the fractions from the gradient by SDS-PAGE and Coomassie staining. Proteins were separated in a 5% to 15% (w/v) gradient gel. Lane M, Molecular mass markers as indicated on left. The arrows indicate the protein bands that correlate with the main peak of activity for 3-hydroxyacyl-CoA dehydrogenase in fractions 10 to 11.
Purification of 3-Hydroxyacyl-CoA Dehydrogenase from Euglena
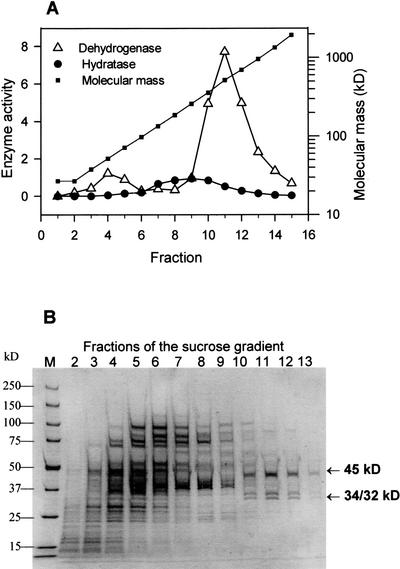
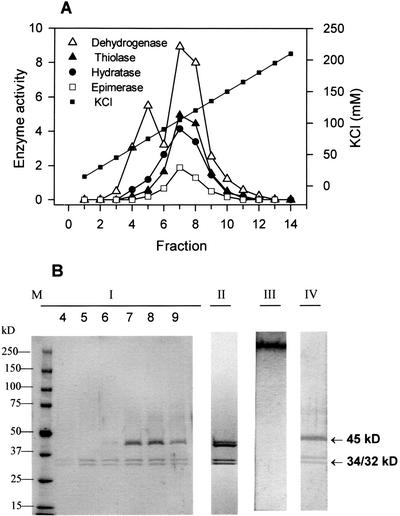
Enzyme purification was performed by successive ammonium sulfate (AS) precipitation, Suc gradient centrifugation, and ion-exchange chromatography on sulfopropyl- and dimethylaminoethyl columns (Table I). The highest 3-hydroxyacyl-CoA dehydrogenase activity was obtained in the fraction of 52.5% to 62.5% AS. Most of the proteins precipitating together with this activity were removed by Suc density centrifugation. The main peak of 3-hydroxyacyl-CoA dehydrogenase appeared in fraction 11 of the gradient (Fig. 4A). In accordance with the enzyme activity profile in the SDS gel of fractions 10 to 12, three bands of 45, 34, and 32 kD were found (Fig. 4B). Further purification was obtained using cation-exchange chromatography on sulfopropyl columns. The 3-hydroxyacyl-CoA dehydrogenase was not bound to this column, but contaminating proteins with β-oxidation activity were removed. Final purification of the enzyme was achieved by means of dimethylaminoethyl anion-exchange chromatography (Fig. 5). The 3-hydroxyacyl-CoA dehydrogenase coeluated with activity of 2-enoyl-CoA hydratase, thiolase, and epimerase. The activity profile of all enzymatic reactions agreed with the abundance of three proteins bands of 45, 34, and 32 kD in an SDS gel (Fig. 5B-I). As demonstrated in Figure 5B-III, fraction 7 of the eluate only contained one native protein with activity for all four enzymatic reactions, which are summarized in Table II. In an SDS gel it could be shown that the native protein is composed of subunits with 45, 34, and 32 kD (Fig. 5B-IV). Using an improved gel system, however, we could separate the protein in the 45-kD band into two peptides with only minor differences in their molecular masses (Fig. 5B-II). Thus, the MFP actually consists of four different subunits. In Table II, some data are presented characterizing the enzyme, which was apparently purified to homogeneity after dimethylaminoethyl anion-exchange chromatography.
Table I.
Purification of 3-hydroxyacyl-CoA dehydrogenase from Euglena
| Purification Step | Activity of 3-Hydroxyacyl-CoA Dehydrogenasea
|
||||
|---|---|---|---|---|---|
| Total protein | Total activity | Specific activity | Purification | Recovery | |
| mg | μmol min−1 | μmol min−1 mg−1 protein | -fold | % | |
| Crude homogenate | 177 | 105 | 0.6 | 1.0 | 100 |
| AS (52.5 to 62.5%) | 14.5 | 24 | 2.0 | 3 | 27 |
| Suc gradient | 2.2 | 20 | 9 | 15 | 18 |
| Sulfopropyl column; void volumeb | 0.8 | 16 | 20 | 33 | 14 |
| Dimethylaminoethyl column | 0.3 | 12 | 40 | 67 | 11 |
Enzyme activity was measured at the standard substrate concentration of 15 μmol L−1. The specific activity for the 3-hydroxyacyl-CoA dehydrogenase reaction of 40 μmol min−1 mg−1 protein of the purified enzyme corresponds to a specific activity of 260 μmol min−1 mg−1 protein at substrate saturation.
Most of the activity was not bound to this column, but some contaminating proteins were removed.
Figure 4.
Distribution of proteins obtained by AS precipitation (52.5%–62.5%) in a Suc gradient. A, Distribution of 3-hydroxyacyl-CoA dehydrogenase (Dehydrogenase) and 2-enoyl-CoA hydratase (Hydratase) in the gradient. Enzyme activity is expressed as micromoles per minute per milliliter fraction. The distribution of molecular masses for proteins separated in the gradient was obtained from the distribution of standard proteins. B, Analysis of the proteins in the different fractions of the gradient by SDS-PAGE and Coomassie staining. Proteins were separated in 5% to 15% (w/v) gradient gels. Lane M, Molecular mass markers as indicated on left. The arrows indicate the proteins which correlate with the main peak of activity for 3-hydroxyacyl-CoA dehydrogenase in fraction 11 of the gradient.
Figure 5.
Enzyme purification on a dimethylaminoethyl anion-exchange column. The proteins not bound to the sulfopropyl anion-exchange column were injected on the column and eluted in a gradient of 0 to 500 mm KCl prepared in 10 mm HEPES, pH 7.5, containing 1 mm dithiothreitol (DTT) and 0.1% (v/v) Tween 20. The KCl concentration increased by 10 mm per minute. Fractions of 1.5 mL were collected. A, Distribution of enzyme activities in the fractions eluted from the column. Enzyme activity is expressed as micromoles per minute per milliliter fraction for 3-hydroxyacyl-CoA dehydrogenase (Dehydrogenase). Full-scale activity corresponds to 50 nmol min−1 mL−1 fraction for 2-enoyl-CoA hydratase (Hydratase), thiolase, and epimerase. B, Monitoring the purification by PAGE. Lanes under I, 5% to 15% (w/v) SDS gradient gel of the fractions eluted from the column. In fractions 4 to 6, only two 32- and 34-kD bands are present. In fractions 7 to 9, an additional 45-kD band appears. Lanes under II, 5% to 12% (w/v) SDS gradient gel from fraction 7. Two separated bands are visible at 45 kD. Lanes under III, Preparative native gel from fraction 7 containing only one single protein. Lanes under IV, 5% to 15% (w/v) SDS gradient gel of the protein extracted from the native protein in lane III: All three bands that are present in the SDS gel from fraction 7 (Lanes under I) are seen. Lane M, Molecular mass markers as indicated on the left. The right arrows indicate the molecular masses of the subunits from the purified protein.
Table II.
Enzymatic and molecular properties of the MFP from Euglena
| Molecular Mass | Enzymatic Properties
|
||||
|---|---|---|---|---|---|
| Optimum reaction pH (K-phosphate buffer) | Km value | Ratio of enzyme functions
|
|||
| Dimethylaminoethyl column | Native gel | ||||
| kD | μmol L−1 | % | |||
| 3-Hydroxyacyl-CoA dehydrogenase | 6.5 | 50a | 100 | 100 | |
| 2-Enoyl-CoA hydratase | 6.25 | 70b | 1.9 | 1.4 | |
| Thiolase | 2.2 | 4.2 | |||
| Epimerase | 0.9 | 1.2 | |||
| Native Protein | 460 | ||||
| Subunits | 45.5, 44.5, 34, and 32 | ||||
Substrate was acetoacetyl CoA.
Substrate was crotonoyl CoA.
Distribution of Enzyme Functions on the Subunits
The molecular structure of the Euglena MFP proved to be unstable under certain experimental conditions. Complete dissociation into protomers was obtained by applying low temperatures, high concentration of some salts and homogenization procedures like sonification, suggesting that the subunits are combined through non-covalent interactions. Graduated dissociation, by which the smaller 32- and 34-kD subunits were obtained, was observed after eluting the MFP from the dimethylaminoethyl column (Fig. 5B-I). In the fractions containing dissociated 32- and 34-kD subunits only 3-hydroxyacyl-CoA dehydrogenase activity could be measured. From analogous experiments with anion-exchange columns resulting in partial dissociation of the 45-kD subunits, it was concluded that the 2-enoyl-CoA hydratase and thiolase functions are located on 45-kD subunits (data not shown). Activity for epimerase was only measured in the native MFP.
Stereospecificity of the 2-Enoyl-CoA Hydratase Reaction
Due to its dehydrating activity, the 2-enoyl-CoA hydratase from Euglena is capable of catalyzing both the hydratase reaction and the reverse reaction. To determine the stereospecificity of the enzyme, we have measured the dehydratase activity of the purified MFP by using either the R(l) or S(d) isomers of 3-hydroxy-3-phenylpropionyl-CoA as substrates (Baes et al., 2000). Significant activity was only determined to take place with the R-isomer (Table III). In this regard, the 2-enoyl-CoA hydratase from Euglena is similar to the mitochondrial monospecific 2-enoyl-CoA hydratase and the 2-enoyl-CoA hydratase function of the peroxisomal multifunctional enzyme 1 but dissimilar to peroxisomal multifunctional enzyme 2, which is exclusively specific to S(d)-isomers (Mao et al., 1994).
Table III.
Stereospecificity of the 2-enoyl-CoA dehydratase function of the MFP purified from Euglena
| Enzyme Reaction | Substrate | Substrate Concentration | Activity |
|---|---|---|---|
| nmol mL−1 | nmol min−1 mL−1 fraction | ||
| Hydratase | Crotonoyl-CoA | 15 | 85.0 |
| Dehydratase | S-3-Phenyl-2-propenoyl-CoA | 25 | 0.4 |
| Dehydratase | R-3-Phenyl-2-propenoyl-CoA | 25 | 15.4 |
Protein Microsequencing and Search for Homologies
Upon analyzing the amino acid sequence, only the 32- and 34-kD subunits yielded significant data. A search for proteins related to the 32-kD subunit revealed that its amino acid sequence is 51% to 31% homologous to monospecific 3-hydroxyacyl-CoA dehydrogenases and to domains for 3-hydroxyacyl-CoA dehydrogenases of MFPs that are known to possess l-specific dehydrogenases (Fig. 6). In addition, the monospecific dehydrogenase from Mus musculus, producing sequence homology to the Euglena 32-kD subunit, is also an l-specific enzyme (the stereospecificity for the other monospecific enzymes was not indexed in the databases). No similarities to domains of d-specific 3-hydroxyacyl-CoA dehydrogenases in MFPs were detected. Altogether, the sequence analysis and the activity measurements indicate that an l-3-hydroxyacyl-CoA dehydrogenase function is located on the 32-kD subunit of the Euglena MFP. The amino-terminal sequence of the 34-kD subunit shows only limited similarities to the 32-kD subunit, but is homologous to monospecific 3-hydroxyacyl-CoA dehydrogenases from bacteria. Therefore the 34 kD should also carry a 3-hydr-oxyacyl-CoA dehydrogenase function. Many dehydrogenases possess a highly conserved GxGxxG sequence that is located around the NAD-binding site (Kamijo et al., 1993). Such GxGxxG motives are also present in the sequences of both subunits from the Euglena MFP (Fig. 6), providing additional evidence that they carry dehydrogenase functions.
Figure 6.
Alignment of the amino-terminal sequences of 32- and 34-kD subunits of the Euglena MFP with several β-oxidation enzymes. Individual homologies were obtained with the BLAST program (see “Materials and Methods”). For the 32-kD subunit from Euglena, the identity with A. fulgidus is 51%, the similarity is 62%; the identity with Rattus sp. peroxisomal MFP I is 51%, the similarity is 67%; the identity with P. aerophilum is 47%, the similarity is 65%; the identity with M. musculus is 49%, the similarity is 59%; the identity with Caenorhabditis sp. is 38%, the similarity is 62%; the identity with Arabidopsis is 46%, the similarity is 59%; the identity with Myobacterium sp. is 38%, the similarity is 54%; the identity with Cucumis sp. is 31%, the similarity is 44%; the identity with Rattus sp. mitochondrial MFP is 31%, the similarity is 49%; the identity with Euglena, 34-kD subunit is 51%, the similarity is 61%. For the 34-kD subunit from Euglena, the identity with Acinetobacter sp. is 39%, the similarity is 50%; the identity with Pseudomonas sp. is 36%, the similarity is 47%. Amino acids identical with the 32- and 34-kD subunits are shaded. Aligned sequences are indicated by the first amino acid. The GxGxxG motive representing a conserved sequence around the NAD-binding side of dehydrogenases is boxed in. 3-HOADH, 3-Hydroxyacyl-CoA dehydrogenase; ECH, 2-enoyl-CoA hydratase; TFP, tetrafunctional protein. The numbers in parentheses are the GenBank accession numbers from the National Center of Biotechnology Information.
Cellular Localization of the Euglena MFP
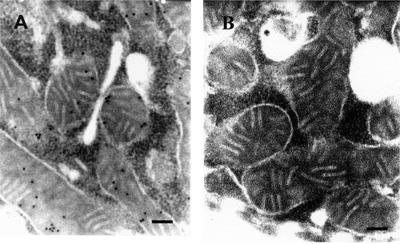
Polyclonal antibodies directed against the Euglena MFP were raised in rabbits. After immunoblot analysis, the antibodies proved to be monospecifically directed against all subunits of the MFP (Fig. 8). On ultrathin sections from Euglena cells grown on hexanoic acid, antibodies against the MFP were significantly bound to mitochondria exclusively when gold particle-conjugated secondary antibodies were used for visualization (Fig. 7). No significant binding of the conjugated gold particles was observed when using preimmune serum in control experiments. Thus the mitochondria are the only compartment in Euglena containing the MFP.
Figure 8.
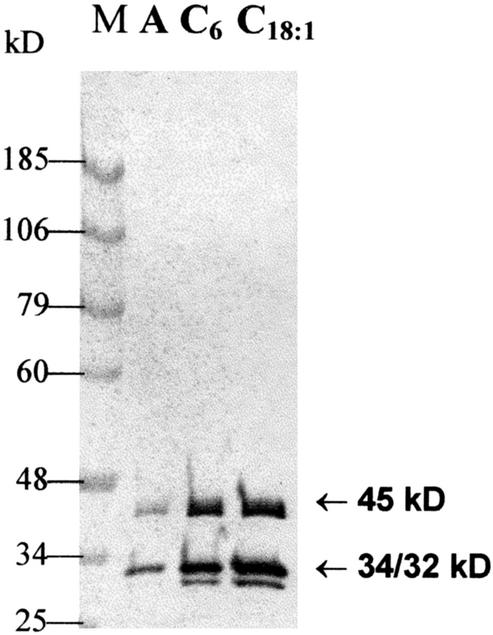
Immunoblot analysis of anti-MFP antibodies. Proteins of crude homogenates were separated in Suc gradients. Each crude homogenate contained the same amount of algal fresh weight peak fractions with activity for 3-hydroxyacyl-CoA dehydrogenase were electrophoresed on an SDS gel, and proteins were transferred to blotting membranes. The membrane was incubated with anti-MFP antibodies. The antibody-antigen reaction was detected by using alkaline phosphatase. Lane M, Molecular mass marker. Lane A, MFP from autotrophically grown algae. Lane C6, MFP from hexanoic acid-grown algae. Lane C18:1, MFP from oleic acids-grown algae.
Figure 7.
Thin sections of LR White resin embedded Euglena cells grown on hexanoic acid. A, Sections were labeled by the IgG gold method using antibodies against the Euglena MFP. The gold particles representing the sites of the antigen are localized exclusively over the mitochondria. B, Sections were labeled by the IgG gold method using the preimmune serum of the immunization experiment. Bars = 0.1 μm.
Physiological Significance of the MFP
Adding 0.1 m cycloheximide to algae growing with hexanoic acid stopped the increase of the l-3-hydroxyacyl-CoA dehydrogenase, whereas chloramphenicol and rifampicin were not effective. These results suggest that the MFP is neither coded nor synthesized in mitochondria, but is translated in the cytoplasm. To find whether the increase of enzyme activity was due to de novo synthesis or activation of pre-existing enzymes, we compared the enzyme levels in hexanoic acid, oleic acid, and photoautotrophically grown algae by western-blot analysis. Crude homogenates prepared from the same amount of algae were separated in Suc gradients. Fractions with main activity for the dehydrogenase reaction were separated in SDS gels, and proteins were electrotransferred to polyvinylidene difluoride membranes. The blotted proteins were probed using anti-MFP serum followed by anti-mouse IgG conjugated with alkaline phosphatase. In all cases, only the three bands of the MFP subunits appeared on the blots (Fig. 8). The increases of enzyme levels after the addition of hexanoic or oleic acid were immunochemically shown to be an increase in the amounts of enzyme proteins (Fig. 8). These results demonstrate that the MFP is induced by the short-chain saturated fatty acid but also by the long-chain unsaturated fatty acid, indicating that the Euglena enzyme complex is capable of metabolizing a broad range of substrates.
DISCUSSION
Algae are capable of metabolizing fatty acids via the β-oxidation pathway present in either mitochondria or microbodies or both of them (Stabenau, 1992). But little is known about the enzymes participating in this process. Especially lacking is information on biochemical and structural properties of the algal β-oxidation enzymes. In Euglena, as demonstrated in this paper, a tetrafunctional oligomeric protein is involved in the metabolism of fatty acid-CoA esters when the cells are grown on hexanoic or oleic acid. Immunocytological experiments revealed that this MFP is located in the mitochondria. The enzyme is composed of the two smaller 32- and 34-kD subunits and two larger subunits having molecular masses of 44.5 and 45.5 kD, respectively. The latter two subunits house the enzymes enoyl-CoA hydratase and thiolase, whereas two different hydroxylacyl-CoA dehydrogenases are present on the smaller subunits. Epimerase activity, also measurable in the Euglena MFP, could not be assigned to a special subunit, which may be due to the epimerization mechanism (Hiltunen et al., 1989). The data that we obtained indicate that acyl-CoA esters are degraded via l-isomers of hydroxyacyl-CoA. Therefore epimerization of d-isomers to the l-form seems to be necessary in MFPs, which are only capable of metabolizing the l-forms of 3-hydroxyacyl-CoA esters as in Euglena.
The degradation of solely acyl-CoA esters via l-isomers of substrates is a common characteristic of mitochondrial β-oxidation systems. Long-chain acyl-CoAs undergo one or more cycles of chain shortening catalyzed by long-chain-specific monofunctional enzymes that are bound to the inner mitochondrial membrane (Carpenter et al., 1992). The binding to membranes facilitates substrate channeling. It has been speculated that the organization of participating matrix enzymes in complexes would enable substrate channeling like the binding of enzymes to membranes and could also facilitate the tuning between the first and the last reactions of the β-oxidation (Kunau et al., 1995). Examples of these kinds of enzyme complexes are the trifunctional protein isolated from rat liver mitochondria (Uchida et al., 1992) and also the tetrafunctional MFP from Euglena mitochondria described in this paper. It seems that both of the MFPs serve the same purpose, although they are different with respect to their enzyme functions and their structural organization. For example, only the Euglena enzyme involves epimerase activity. Enoyl-CoA hydratase and the 3-hydroxyacyl-CoA dehydrogenase are usually domains of a common polypeptide. The Euglena enzyme is the only exception, because both of the enzyme functions are located on different subunits. The Euglena MFP subunits are comparable with subunits of monospecific enzymes with comparable functions. For example, the subunits with 32 to 34 kD of l-3-hydroxyacyl-CoA dehydrogenases from the archeon Archeoglobus fulgidus (Klenk et al., 1997) and from animal mitochondria (Osumi and Hashimoto, 1980) correspond in size and function to the small subunits of the Euglena MFP. The 43- to 45-kD subunits of the 2-enoyl-CoA hydratases from the archeon A. fulgidus (Klenk et al., 1997) and from Closterium acetobutylicum (Waterson et al., 1971) and the 42- to 45-kD subunits for monospecific thiolases from various organisms (Kunau et al., 1995) are comparable with respect to molecular sizes and enzyme functions to the large subunits of the Euglena MFP. Put together, these data suggest that the Euglena MFP is a combination of subunits from monospecific enzymes assembled in a multienzyme complex.
The evolution of β-oxidation enzymes is characterized by two strategies for achieving the advantageous substrate channeling: integration of enzymes into membranes and formation of multienzyme complexes. Only in the archeon A. fulgidus that is the phylogenetically oldest organism known to possess the β-oxidation pathway, all functions are distributed among separate enzymes (Klenk et al., 1997). During evolution, multienzyme complexes were formed by combining monospecific enzymes through non-covalent interactions and/or gene fusion. The MFP from Euglena is the first example of a multienzyme complex formed exclusively by non-covalent interactions. The genome analysis of the archaeobacterium Pyrobaculum aerophilum (Fitz-Gibbon et al., 2002) led to the discovery of the fusion of the genes for 2-enoyl-CoA hydratase and 3-hydroxyacyl-CoA dehydrogenases. The products of this gene, however, are not known. The first common polypeptides having hydratase and dehydrogenase functions, probably forming a more stable gene product, occur in the α-subunits of multifunctional fatty acid oxidation complexes of eubacteria like E. coli or Mycobacterium leprae (Binstock et al., 1977; Cole et al., 2001). The α-subunits carrying the hydratase and dehydrogenase functions are combined by non-covalent interactions to a subunit housing a thiolase function in the bacterial enzymes and in the trifunctional protein from rat liver mitochondria (Uchida et al., 1992). The peroxisomal and glyoxysomal MFPs only consist of the product of the fused genes (Yang et al., 1991; Kindl, 1993).
Although information on the amino acid sequence of the Euglena MFP is limited to the N-terminal of the 32- and 34-kD subunits, significant alignments to other β-oxidation enzymes were produced, allowing a discussion on the role of Euglena MFP in the evolution of multifunctional β-oxidation enzymes. Euglena is believed to be one of the oldest known eukaryotic organisms (Tessier et al., 1997). Therefore the Euglena MFP should be more closely related to bacterial enzymes than to enzymes of higher developed eukaryotic organisms. In agreement with this assumption, it was found that the 32-kD subunit of the Euglena MFP shows a degree of similarity to the 3-hydroxyacyl-CoA dehydrogenases from A. fulgidus. These data, plus the similarities in the sizes of subunits between the Euglena MFP and the monospecific enzymes from A. fulgidus, provide evidence that the monospecific enzymes from the archeon could possibly represent the phylogenetic ancestors of Euglena MFP. In contrast to the archeon, there was only a low degree of similarity to the 3-hydroxyacyl-CoA dehydrogenases domain in the multienzyme complex of fatty acid oxidation from the eubacterium M. leprae, similar to the multifunctional fatty acid oxidation complex in E. coli. From the analysis of the 16S rRNA, it is also established that Euglena is phylogenetically more related to archaebacteria than to eubacteria (Giovanni et al., 1988). Therefore, one must consider that the Euglena enzyme could possibly represent an additional phylogenetic line of development for β-oxidation enzymes, different from the E. coli enzyme. This assumption would imply a polyphyletic development of β-oxidation systems, as was also proposed for plastids (Bhattacharya, 1995). The significance of Euglena MFP in the evolution of β-oxidation enzymes, however, still has to examined using more detailed sequence data.
MATERIALS AND METHODS
Algal Material and Growth Conditions
Euglena (Euglena gracilis), strain 1224-5/25, was obtained from the collection of algal cultures at the University of Göttingen (Germany). The nutrient medium contained 10 mm (NH4)2HPO4, 4 mm NaH2PO4, 5 mm K2HPO4, 1 mm MgSO4, and 0.1 mm CaCl2. To 1 L of medium were added: 1 mg of B12, 1 mg of B6, 0.05 mg of B12, 1 mL of micronutrient solution, and 1 mL of Fe-EDTA complex (Kuhl and Lorenzen, 1964). The algae were grown in glass tubes at 25°C under aeration with air plus 2% (v/v) CO2 either in continuous light of 40 μmol quanta m−2 s−1 or in the dark after the addition of 5 mm hexanoic acid or oleic acid.
Cell Homogenization and Enzyme Purification
Algae (5 g fresh weight) were washed and homogenized in a grinding buffer containing 10 mm HEPES, pH 7.5, 1 mm DTT, and 0.1 mm phenylmethylsulfonylfluoride. Cells were disrupted in a Virtis homogenizer in the presence of glass beads (0.5 mm). The homogenate was filtered (nylon sieve, 0.2-mm pore size) and centrifuged at 1,100g for 5 min. The resulting supernatant was used for measuring the activities of the β-oxidation enzymes and for protein separation in Suc gradients.
qEnzyme Purification
The crude homogenate was incubated with 500 mm KCl at 5°C for 30 min, followed by a treatment with 0.1% (v/v) Tween 20 for 30 min at 5°C. After centrifugation at 48,000g for 30 min, proteins in the supernatant were precipitated with 52.5% to 62.5% (of saturation) AS, collected by centrifugation, and suspended in 2 mL of grinding buffer containing 0.1% (v/v) Tween 20. The AS precipitate was layered onto a Suc gradient from 5% to 20% (w/w) Suc prepared in 30 mL of the same buffer and centrifuged at 27,000 rpm (140,000g) in a SW 28-rotor (Beckman Coulter, Fullerton, CA) at 20°C for 18 h. Fractions of 2 mL were collected.
The following protein chromatography was performed on an inert Merck-Hitachi system using the following columns: a Fractogel EMD-sulfopropyl-650 (S) cation-exchange column (Merck, Darmstadt, Germany) and a Fractogel EMD-dimethylaminoethyl-650 (S) anion-exchange column (Merck). Fractions 10 to 12 from the Suc gradient were first applied to the cation-exchange column and eluted in 10 mm HEPES, pH 7.5, containing 1 mm DTT and 0.1% (v/v) Tween 20. The first 10 mL of the non-bound proteins were collected and injected into the anion-exchange column. The bound proteins eluted in a KCl gradient of 0 to 500 mm prepared in 10 mm HEPES, pH 7.5, containing 1 mm DTT and 0.1% (v/v) Tween 20. The KCl concentration increased by 10 mm per minute. Fractions of 1.5 mL were collected.
Assays
Enoyl-CoA hydratase activities were measured as the hydration of crotonoyl-CoA with 15 μm crotonoyl-CoA as the substrate (Steinmann and Hill, 1975). The reverse reaction, dehydration of 3-hydroxyacyl-CoA esters, was observed by measuring the increase of A308 due to the formation of 3-phenyl-2-propenoyl-CoA in an assay containing enzyme fraction, 200 mm phosphate buffer, pH 8.0, 25 μm of the R- or S-isomers of 3-hydroxy-3-phenylpropionyl-CoA and 1 m EDTA (Baes et al., 2000). The activity of 3-hydroxyacyl-CoA dehydrogenase was monitored by using the oxidation of NADH in the presence of 15 μm acetoacetyl-CoA (Overath and Raufuss, 1967). Thiolase activity was observed by measuring the dissociation of the Mg-acetoacetyl-CoA complex at 303 nm (Middleton, 1975) using 15 μm acetoacetyl-CoA.
Epimerase activity was monitored by reducing NAD+ after complete oxidation of the l-isomer of dl-hydroxybutyryl-CoA by adding l-3-hydroxyacyl-CoA dehydrogenase (bovine liver) in a test medium 30 μm dl-hydroxybutyryl-CoA (Binstock and Schulz, 1981). The protein concentration was measured using the protein assay (Bio-Rad, Hercules, CA; modified Bradford method) according to the manufacturer’s instructions. Bovine serum albumin was used as the standard.
Gel Electrophoresis and Immunoblotting
SDS-PAGE was carried out in gradient gels using standard methods (Laemmli, 1970). Native gel separations were performed as described by Laemmli (1970) omitting SDS and 2-mercaptoethanol. For isoelectric focusing, the method described by Robertson et al. (1987) was applied. A Bio-Rad mini protean II cell was used for all separations. Gels were stained in 0.1% (w/v) Coomassie Brilliant Blue R 250 in a mixture of 40% (v/v) methanol and 10% (w/v) acetic acid and destained several times in a mixture of 40% (v/v) methanol and 10% (w/v) acetic acid. Immunoblot analysis was performed according to the method of Towbin et al. (1979). Antibodies were diluted 1:500 (v/v).
Molecular Mass Determination
The molecular masses of the proteins were determined after Suc gradient centrifugation as described by Martin and Ames (1961). Catalase, malate dehydrogenase, hexokinase, and peroxidase were used as Mr standards.
Production of Antibodies and Immunoelectron Microscopy
The MFP separated by Suc density centrifugation was separated from contaminating proteins in a preparative native gel. The gel slice containing the MFP was used as an antigen probe for the production of antibodies in rabbits according to the protocol of the manufacturer (Bioscience, Germany). The serum was partially purified on a cation-exchange column (Fractogel EMD-SO3-650 (S), Merck) according to the manufacturer's instructions. For electron-microscopic preparation, the algae were fixed with 1.0% (v/v) glutaraldehyde and 4% (v/v) depolymerized paraformaldehyde in a 100 mm cacodylate buffer, pH 7.4, at 20°C for 2 h. Cells were dehydrated in a graded ethanol series. Embedding in LR White resin was performed with a mixture of 33% and 66% (w/v) LR White resin in ethanol followed by three incubations with pure resin. All steps were carried out at 5°C for 6 h. Blocks were polymerized in gelatin capsules at 50°C for 30 h. Ultra-thin sections were mounted on uncoated nickel grids. The following steps were all performed at 30°C: The sections were treated with a blocking solution (2.5% [w/v] nonfat dry milk in phosphate buffered saline) for 30 min and then incubated for 2 h in a solution of the antibody diluted 1:100 (v/v) in blocking medium. After washing with phosphate buffered saline, the sections were incubated with goat anti-rabbit IgG conjugated to 15-nm gold particles (British Biocell, Cardiff, UK), diluted 1:20 (v/v) in a blocking medium for 1 h. The sections were washed with distilled water, and stained with 4% (w/v) uranyl acetate and lead citrate.
Protein Microsequencing and Search for Homologies
Protein N-terminal sequencing was performed by automated Edman degradation with electrophoretically purified enzyme subunits blotted on polyvinylidene difluoride membranes. Sequence homologies were determined using the BLAST program (Altschul et al., 1997) of the National Center for Biotechnology.
ACKNOWLEDGMENTS
We thank Dr. F. Buck (Institut für Zellbiologie und Klinische Neurobiologie, Universtität Hamburg) for the analysis of the amino acid sequences.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.013151.
LITERATURE CITED
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baes M, Huyghe S, Carmeliert P, Declercq E, Collen D, Mannaerts GP, Van Veldhoven PP. Inactivation of the peroxisomal multifunctional protein-2 in mice impedes the degradation of not only 2-methyl-branched fatty acids and bile acid intermediates but also of very long chain fatty acids. J Biol Chem. 2000;275:16329–16336. doi: 10.1074/jbc.M001994200. [DOI] [PubMed] [Google Scholar]
- Baldwin GS. Comparison of sequences of the 78 kDa gastrinbinding protein and some enzymes involved in fatty acid oxidation. Comp Biochem Physiol. 1993;104B:55–61. doi: 10.1016/0305-0491(93)90337-5. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D. The phylogeny of plastids: a review based on comparisons of small-subunit ribosomal RNA coding regions. J Phycol. 1995;31:489–498. [Google Scholar]
- Binstock J, Schulz H. Fatty acid oxidation complex from E. coli. Methods Enzymol. 1981;71:403–411. doi: 10.1016/0076-6879(81)71051-6. [DOI] [PubMed] [Google Scholar]
- Binstock JF, Pramalik A, Schulz H. Isolation of a multienzyme complex of fatty acid oxidation from Escherichia coli. Proc Natl Acad Sci USA. 1977;74:492–495. doi: 10.1073/pnas.74.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter K, Pollitt RJ, Middleton B. Human liver long-chain 3-hydroxyacyl coenzyme A dehydrogenase is a multifunctional membrane bound beta-oxidation enzyme of mitochondria. Biochem Biophys Res Commun. 1992;183:443–448. doi: 10.1016/0006-291x(92)90501-b. [DOI] [PubMed] [Google Scholar]
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honore N, Ganier T, Churcher C, Harris D et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- Dieuaide-Noubhani M, Novikov D, Vandekerkhove J, Van Veldhoven PP. Identification and characterization of the 2-enoyl-CoA hydratases involved in peroxisomal β-oxidation in rat liver. Biochem J. 1997;321:253–259. doi: 10.1042/bj3210253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz-Gibbon ST, Ladner H, Kim UJ, Stetter KO, Simon MI, Miller JH. Genome sequence of the hyperthermophillic crenarchaeon Pyrobaculum aerophilum. Proc Natl Acad Sci USA. 2002;99:984–989. doi: 10.1073/pnas.241636498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JC, Schulz H. Short chain and long chain enoyl-CoA hydratase from pig heart muscle. Methods Enzymol. 1981;71:390–398. doi: 10.1016/0076-6879(81)71049-8. [DOI] [PubMed] [Google Scholar]
- Furuta S, Miyazawa S, Osumi T, Hashimoto T, Ui N. Properties of mitochondrial and peroxisomal enoyl-CoA hydratases from rat liver. J Biochem. 1980;88:1059–1070. doi: 10.1093/oxfordjournals.jbchem.a133057. [DOI] [PubMed] [Google Scholar]
- Giovanni SJ, Turner S, Olsen GJ, Barns S, Lane DJ, Pace NR. Evolutionary relation ships among cyanobacteria and green chloroplasts. J Bacteriol. 1988;170:3584–3592. doi: 10.1128/jb.170.8.3584-3592.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves LB, Becker WM. Beta-oxidation in glyoxysomes from Euglena. J Protozool. 1974;21:771–774. doi: 10.1111/j.1550-7408.1974.tb03750.x. [DOI] [PubMed] [Google Scholar]
- Gross W, Winkler U, Stabenau H. Characterization of peroxisomes from the alga Bumilleriopsis. Plant Physiol. 1985;75:292–296. doi: 10.1104/pp.77.2.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CL. Acyl-CoA dehydrogenase and electron transferring flavoprotein. Methods Enzymol. 1978;53:502–518. doi: 10.1016/s0076-6879(78)53053-x. [DOI] [PubMed] [Google Scholar]
- Hashimoto T. Comparison of enzymes of lipid β-oxidation in peroxisomes and mitochondria. In: Fahimi HD, Sies H, editors. Peroxisomes in Biology and Medicine. Berlin: Springer-Verlag; 1987. pp. 97–104. [Google Scholar]
- Hiltunen JK, Palosaari M, Kunau H-D. Epimerization of 3-hydroxyacyl-CoA esters in rat liver. J Biol Chem. 1989;23:13356–13540. [PubMed] [Google Scholar]
- Hiltunen JK, Wenzel B, Beyer A, Erdmann R, Fossa A, Kunau W-H. Peroxisomal multifunctional β-oxidation protein of Saccharomyces cerevisiae. J Biol Chem. 1992;267:6646–6659. [PubMed] [Google Scholar]
- Hosotani K, Ohkochi T, Iniu H, Yokota A, Nakano Y, Kitaoka S. Photoassimilation of fatty acids, fatty alcohols and sugars by Euglena gracilis Z. J Gen Microbiol. 1988;134:61–66. [Google Scholar]
- Kamijo T, Aoynama T, Miyazaki J, Hashimoto T. Molecular cloning of the cDNAs for the subunits of rat mitochondrial fatty acid β-oxidation multienzyme complex. J Biol Chem. 1993;268:26452–26460. [PubMed] [Google Scholar]
- Kindl H. Fatty acid degradation in plant peroxisomes: function and biosynthesis of the enzymes involved. Biochemie. 1993;75:225–230. doi: 10.1016/0300-9084(93)90080-c. [DOI] [PubMed] [Google Scholar]
- Kionka C, Kunau H-D. Inducible β-oxidation pathway in Neurospora crassa. J Bacteriol. 1985;161:153–157. doi: 10.1128/jb.161.1.153-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein K. Acyl-CoA Dehydrogenasen und ETF in Escherichia coli. Studien zum Fettsäureabbau. Doctoral dissertation. Germany: University of Cologne; 1973. [Google Scholar]
- Klenk H-P, Clayton RA, Tomb J-F, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD et al. The complete genome sequence of the hyperthermophilic, sulfate-reducing archeon Archeaoglobus fuligdus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- Kuhl A, Lorenzen H. Handling and culturing of Chlorella. In: Prescott DM, editor. Methods in Cell Physiology. I. New York: Academic Press; 1964. pp. 152–187. [Google Scholar]
- Kunau W-H, Dommes V, Schulz H. β-Oxidation of fatty acids in mitochondria, peroxisomes, and bacteria: a century of continued progress. Prog Lipid Res. 1995;34:267–342. doi: 10.1016/0163-7827(95)00011-9. [DOI] [PubMed] [Google Scholar]
- Kunau W-H, Kionka C, Ledebur A, Kionka C, Mateblowski M, Moreno de la Garca M, Schultz-Borchard U, Thieringer R, Veenhuis M. β-Oxidation systems in eucaryotic microorganisms. In: Fahimi HD, Sies H, editors. Peroxisomes in Biology and Medicine. Berlin: Springer-Verlag; 1987. pp. 128–140. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mao L-F, Chu C, Schulz H. Hepatic β-oxidation of 3-phenylpropionic acid and the stereospecific dehydration of (R)- and (S)-3-hydroxy-3-phenylpropionyl-CoA by different enoyl-CoA hydratases. Biochemistry. 1994;33:3320–3326. doi: 10.1021/bi00177a024. [DOI] [PubMed] [Google Scholar]
- Martin RG, Ames BN. A method of determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961;236:1372–1379. [PubMed] [Google Scholar]
- Middleton B. 3-ketoacyl-CoA thiolases of mammalian tissue. Methods Enzymol. 1975;35:128–136. doi: 10.1016/0076-6879(75)35148-3. [DOI] [PubMed] [Google Scholar]
- O'Connell MA, Orr G, Shapiro L. Purification and characterization of fatty acid β-oxidation enzymes from Caulobacter crescentus. J Bacteriol. 1990;172:997–1004. doi: 10.1128/jb.172.2.997-1004.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi T, Hashimoto T. Purification and properties of mitochondrial and peroxisomal 3-hydroxyacyl-CoA dehydrogenase from rat liver. Arch Biochem Biophys. 1980;203:372–383. doi: 10.1016/0003-9861(80)90189-7. [DOI] [PubMed] [Google Scholar]
- Overath P, Raufuss EM. The induction of the enzymes of fatty acid degradation in Escherichia coli. Biochem Biophys Res Commun. 1967;29:28–33. doi: 10.1016/0006-291x(67)90535-9. [DOI] [PubMed] [Google Scholar]
- Qin Y-M, Poultanen MH, Helander HM, Kvist A-P, Sivari KM, Schmitz W, Conzelmann E, Hellmann U, Hiltunen JK. Peroxisomal multifunctional enzyme of β-oxidation metabolizing d-3-hydroxyacyl-CoA esters in rat liver: molecular cloning, expression and characterization. Biochem J. 1997;321:21–28. doi: 10.1042/bj3210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson EF, Dannelly HK, Malloy PJ, Reeves HC. Rapid isoelectric focusing in a vertical minigel system. Anal Biochem. 1987;167:290–294. doi: 10.1016/0003-2697(87)90166-7. [DOI] [PubMed] [Google Scholar]
- Staak H, Binstock JF, Schulz H. Purification and properties of a pig heart thiolase with broad chain length specificity and comparison of thiolases from pig heart and Escherichia coli. J Biol Chem. 1978;253:1827–1831. [PubMed] [Google Scholar]
- Stabenau H. Evolutionary changes of enzymes in peroxisomes and mitochondria of green algae. In: Stabenau H, editor. Phylogenetic Changes in Peroxisomes of Algae: Phylogeny of Plant Peroxisomes. Oldenburg, Germany: University Press; 1992. pp. 112–129. [Google Scholar]
- Stabenau H, Winkler U, Säftel H. Enzymes of β-oxidation in different types of algal microbodies. Plant Physiol. 1984;75:531–533. doi: 10.1104/pp.75.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann HM, Hill RL. Bovine liver crotonase (ECH) Methods Enzymol. 1975;35:136–151. doi: 10.1016/0076-6879(75)35149-5. [DOI] [PubMed] [Google Scholar]
- Tessier LH, van der Speck H, Gualberto JM, Grieneberger JM. The cox 1 gene modifications. Curr Genet. 1997;31:208–213. doi: 10.1007/s002940050197. [DOI] [PubMed] [Google Scholar]
- Thieringer R, Kunau W-D. The β-oxidation system in catalase-free microbodies of the filamentous fungus Neurospora crassa. J Biol Chem. 1991;266:13110–13117. [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from acrylamide gels to nitrocellulose sheets: procedure and some applications. J Proc Natl Acad Sci USA. 1979;76:4350–4353. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y, Izai K, Hashimoto T. Novel fatty acid β-oxidation enzymes in rat liver mitochondria. Biol Chem. 1992;267:1034–1041. [PubMed] [Google Scholar]
- Waterson RM, Castellino FJ, Hass GM, Hille RL. Purification and characterization of crotonase from clostridium acetobutylicum. J Biol Chem. 1971;247:5266–5271. [PubMed] [Google Scholar]
- Winkler U, Säftel W, Stabenau H. β-Oxidation of fatty acids in algae: localization of thiolase and acyl-CoA oxidizing enzymes in three different organisms. Planta. 1988;175:91–98. doi: 10.1007/BF00402885. [DOI] [PubMed] [Google Scholar]
- Winkler U, Stabenau H. Isolation and characterization of peroxisomes from diatoms. Planta. 1994;195:403–407. [Google Scholar]
- Yang S-Y, Schulz H, Elzinga M, Yang S-Y. Nucleotide sequence of the promoter and fadB gene of the fad BA operon and primary structure of the multifunctional fatty acid oxidation protein from Escherichia coli. Biochemistry. 1991;30:6788–6795. doi: 10.1021/bi00241a023. [DOI] [PubMed] [Google Scholar]