Abstract
A family of peptides inducing rapid pH alkalinization in hybrid poplar (Populus trichocarpa × Populus deltoides) cell culture medium was isolated from hybrid poplar leaves. Five related approximately 5-kD peptides were purified by high-performance liquid chromatography and analyzed by matrix-assisted laser desorption ionization-mass spectrometry. The N-terminal sequence of one of the isolated peptides was very similar to a previously characterized peptide from tobacco (Nicotiana tabacum), rapid alkalinization factor (RALF), which causes a rapid increase in culture medium pH when added to tobacco cell cultures (G. Pearce, D.S. Moura, J. Stratmann, C.A. Ryan [2001] Proc Natl Acad Sci USA 98: 12843–12847). Two unique poplar RALF cDNAs (PtdRALF1 and PtdRALF2) were isolated from a poplar cDNA library and used to study RALF expression in poplar saplings and cultured poplar cells. Both genes were found to be expressed constitutively in poplar saplings and cultured cells. However, PtdRALF2 was expressed in leaves at very low levels, and its expression in suspension culture cells was transiently suppressed by methyl jasmonate (MeJa). Although the function of these novel peptides remains enigmatic, our experiments suggest their role may be developmental rather than stress related. Overall, our study confirms the presence of active RALF peptides in other plants, and provides new data on the complexity of the RALF gene family in poplar.
An essential feature of all plant cells is the electrochemical proton gradient across the plasma membrane, generated by the plasma membrane H+-ATPase, which uses ATP to pump H+ outside the cell. This H+ gradient is important for many physiological processes including ion uptake, solute transport, and cell wall growth (Sanders and Bethke, 2000). Moreover, transient changes in extracellular or intracellular concentrations of H+, and the accompanying plasma membrane depolarization or hyperpolarization, are implicated in the rapid responses of cells to environmental stimuli; for example, changes in turgor, gravity, and pathogen attack (Blumwald et al., 1998; Felix et al., 1999, 2000; Johannes et al., 2001). A common observation is the rapid alkalinization of the extracellular solution, which can be conveniently observed in suspension cell cultures; as a consequence, stress-induced culture medium alkalinization has become a useful tool for monitoring the rapid events that accompany stress signal transduction (Felix et al., 1993; Blumwald et al., 1998; Schaller and Oecking, 1999). Depending on the system, these rapid increases in medium pH could potentially be caused by several mechanisms, including activation of K+/H+ antiporters, H+/solute cotransporters, and other ion channels, as well as an inhibition of the plasma membrane H+-ATPase (Mathieu et al., 1993).
The response of cells to plant pathogens and pathogen-derived signal molecules called elicitors has been extensively studied using changes in culture medium pH. For example, glycoproteins, peptides, sterols, lipo-chitooligosaccharides, and oligosaccharide elicitors can all induce alkalinization of culture media (Boller, 1995, and refs. therein). A well-characterized elicitor is bacterial flagellin, which induces medium alkalinization of tomato (Lycopersicon peruvianum) cell cultures within minutes, and active oxygen species several hours later (Felix et al., 1999). Active oxygen and the oxidative burst are known to be important components of plant defense against pathogens and are induced in many plant-pathogen interactions (Bolwell, 1999). In parsley (Petroselinum crispum) cells, a 13-amino acid peptide called pep-13, derived from a glycoprotein of Phytophthora megasperma, also induces rapid medium pH alkalinization. This is later followed by the induction of Phe ammonia lyase (PAL) and the formation of defensive phytoalexins (Nürnberger et al., 1994), suggesting a link between alkalinization and the defense response.
In addition to pathogen elicitors, plant-derived signals can cause culture medium alkalinization. Felix and Boller (1995) found that a tomato cell culture homogenate wound induced alkalinization in tomato cultures, as did the peptide wound hormone systemin. Alkalinization is associated with the specific binding of systemin to a receptor (Scheer and Ryan, 1999), and is followed by the induction of ethylene production and PAL activity (Felix and Boller, 1995). By using the fungal toxin fusicoccin to activate the H+-ATPase and block extracellular alkalinization in tomato seedlings, Schaller and Oecking (1999) showed that the alkalinization is required for the induction of tomato defense genes. Therefore, the rapid pH changes observed in cell culture are essential components of signal transduction pathways. Furthermore, other plant peptides have been shown to trigger cell culture alkalinization: A recent report described the purification of a novel peptide from tobacco (Nicotiana tabacum) that causes rapid culture medium alkalinization (Pearce et al., 2001). However, this peptide, named the rapid alkalinization factor (RALF), does not induce defense-signaling pathways, but inhibits root growth when present in the surrounding medium. Homologs to RALF-encoding genes are found in many plant expressed sequence tag (EST) databases. Although the functions of these peptides and genes is as yet unknown, their high degree of sequence conservation and wide representation in plant sequence databases suggests a fundamental function in plants (Pearce et al., 2001).
Populus as a genus is very amenable to tissue culturing, and cell culture lines derived from hybrid poplar (Populus trichocarpa × Populus deltoides) have been maintained for many generations (de Sá et al., 1992). Due to its rapid growth, ease of vegetative propagation, and tractability to Agrobacterium tumefaciens-mediated genetic transformation, Populus has become a model organism for tree biotechnology, genomics, and proteomics (Sterky et al., 1998; Mijnsbrugge et al., 2000). These features also make poplar an economically important tree species, and poplar is grown in large-scale plantations for pulp production. Based on our interest in poplar and its responses to environmental stress, we began an investigation of pH alkalinization activities in hybrid poplar leaf extracts. In this report, we describe the isolation and characterization of several culture medium-alkalinizing peptides from poplar leaves, the cloning of two corresponding cDNAs and analysis of homologous genes in the databases, and their expression in poplar saplings and cell culture. While the work was in progress, we learned that a peptide alkalinizing factor, RALF, had been identified and characterized in tobacco (Pearce et al., 2001), and that our peptides were closely related to it. Thus, our work on RALF from poplar extends the characterization of these biologically active peptides to another plant family. It also provides additional data on the expression of the poplar RALF genes, which should help in elucidating in planta function of these novel peptides.
RESULTS
Isolation and Characterization of RALF Peptides from Poplar Leaves
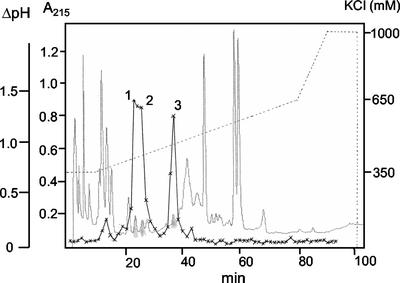
To investigate alkalinization factors in poplar, we obtained a hybrid poplar suspension cell culture (de Sá et al., 1992). Preliminary tests showed that the cells respond to the addition of fungal elicitors with culture medium alkalinization. The addition of crude poplar leaf extracts to the culture also induced a rapid alkalinization in the medium pH. Using C18 open column chromatography, we separated this alkalinization activity from leaf extracts into two active fractions, eluting with 60% and 20% (v/v) methanol, respectively. The 60% (v/v) MeOH fraction was further fractionated using Sephadex G-25 chromatography, strong cation exchange (SCX) HPLC, and C18 HPLC. The SCX HPLC elution profile showed the presence of three activity peaks inducing a rapid alkalinization response (Fig. 1). These three activity peaks all showed identical alkalinization kinetics, suggesting they all contained similar alkalinizing factors (data not shown). Incubation of the active fractions with the proteolytic enzyme proteinase K caused a loss of the alkalinizing activity of greater than 90%, suggesting that the active compounds were peptides. The three SCX column peaks were each further purified by C18 HPLC, from which they all eluted with identical retention times (data not shown). Peak 1 yielded the most active compound, which we estimated at 200 pmol by comparison with chromatographic peaks of known peptides, and which corresponded to about 94.8 g of leaf tissue. The yield was later confirmed by the peaks obtained during the Edman sequencing.
Figure 1.
SCX HPLC of RALFs from poplar leaf extracts. Active compounds were separated using a 350 to 650 mm KCl gradient (dashed line). Medium pH alkalinization activity was assayed by adding 2 μL of each fraction to 2 mL of cells and measuring the pH every 5 min for 35 min. The maximum pH increase (ΔpH) was calculated and plotted with the elution profile monitored at 215 nm. HPLC peaks 1 through 3 (small solid peaks in the elution profile) that corresponded to the highest alkalinization activity were recovered and further purified by C18 HPLC.
The three active peaks were subjected to matrix-assisted laser desorption ionization (MALDI)-mass spectrometry (MS) analysis. This analysis indicated that HPLC peaks 1 and 2 were pure compounds, with molecular masses of 5,488.7 and 5,402.7 mass units, respectively (Table I). MS analysis of peak 3 revealed that it was a mixture of three different molecules of 5,514.4, 4,979.8, and 4,966.0 mass units each. The compound in peak 1 was subjected to N-terminal Edman sequencing, which yielded ATTKYVSYGALQ(W)NXVPXSSXGASY(Y)N (X = unreadable residue) as a sequence. Querying GenBank databases with this amino acid sequence using BLAST revealed that many plant EST databases contained similar predicted peptides. The highest BLAST score was obtained with an EST of unknown function from hybrid aspen (Populus tremula × Populus tremuloides; EST clone AI163551; Sterky et al., 1998). Furthermore, we learned that our peak 1 peptide was very similar (14 of 17 amino acids identical) to a peptide that had been purified and characterized from tobacco (RALF; Pearce et al., 2001). Therefore, we named the peptide that we had identified poplar RALF 1. We inferred that peaks 2 and 3 likely contain similar RALF peptides also, based on their similar masses, susceptibility to proteolytic inactivation, similar chromatographic behavior on reverse phase and cation exchange HPLC, and identical kinetics of alkalinization. We named these other peptides poplar RALF 2, RALF 3-1, RALF 3-2, and RALF 3-3, respectively.
Table I.
Characteristics of RALF peptides identified in hybrid poplar
| Peptide | Retention Time | Mass | cDNA |
|---|---|---|---|
| min | |||
| RALF1 | 23.5 | 5,488.7 | PtdRALF1 |
| RALF2 | 26.1 | 5,402.7 | PtdRALF2 |
| RALF3-1 | 36.8 | 5,514.4 | – |
| RALF3-2 | 36.8 | 4,979.8 | – |
| RALF3-3 | 36.8 | 4,966.0 | – |
Three RALF peaks were separated by SCX HPLC and further purified using C18 HPLC as described in “Materials and Methods.” MALDI-MS analysis indicated that RALF1 and RALF2 are pure compounds, whereas RALF3 is a mixture of three related peptides. cDNAs encoding RALF3-1, RALF3-2, and RALF3-3 were not obtained.
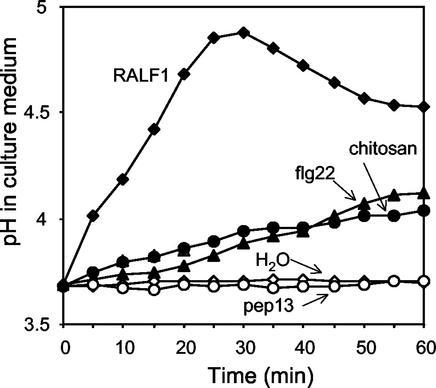
To gain more insight into the nature and function of these alkalinization factors, we compared the alkalinizing activity of poplar RALF1 with that of known defense response elicitors. We tested chitosan, a component of fungal cell walls (Felix and Boller, 1995); flg22, a 22-amino acid conserved peptide found in flagellin from bacteria (Felix et al., 1999); and pep-13, a 13-amino acid peptide derived from a larger fungal elicitor protein (Nürnberger et al., 1994). Alkalinization activity was performed by measuring culture medium pH every 5 min after elicitor addition. The alkalinization by poplar RALF1 we observed was clearly stronger and more rapid than any of the other elicitors tested, reaching its maximum within 30 min of addition and then declining (Fig. 2). In contrast, the pH change caused by chitosan and flg22 was slower and less dramatic, despite higher concentrations applied, and both induced increases of approximately 0.5 pH units over a 60-min period (Fig. 2). This pH increase is comparable with the one reported in tomato cells (Felix and Boller, 1995; Felix et al., 1999). RALF-induced alkalinization was consistently faster and dropped sooner than alkalinization triggered by the other elicitors, where it continued to rise until at least 60 min before declining. RALF2 and RALF3 fractions showed induction kinetics identical to that of RALF1 (data not shown). Therefore, we suspected that the RALF-induced response is distinct from the elicitor-induced responses, and speculate that RALF does not trigger a defense-related reaction (see “Discussion”). Pep-13, although active in parsley cells as an elicitor of alkalinization and phytoalexin synthesis (Nürnberger et al., 1994), did not induce alkalinization in the poplar cell culture.
Figure 2.
Medium alkalinization in poplar suspension culture in response to RALF and elicitors. Cell culture medium was monitored every 5 min. RALF1, flg22, and pep13 peptides were assayed at concentrations of 1 nm, 1 μm, and 5 μm, respectively. Chitosan was assayed at a final concentration of 1 μg mL−1.
Cloning and Characterization of RALF cDNAs from Poplar
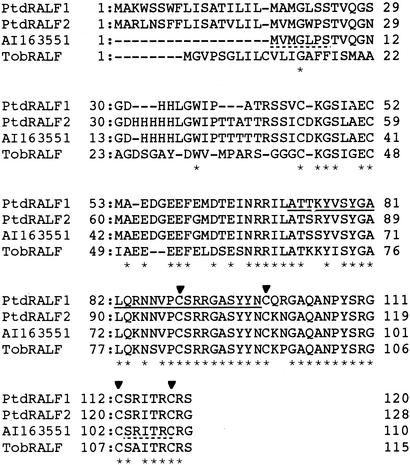
To further characterize poplar RALFs at the molecular level, we isolated poplar RALF cDNAs. The high DNA sequence similarity between poplars and aspens (Constabel et al., 2000; Haruta et al., 2001b) allowed us to make use of the aspen EST clone, identified as highly homologous to the RALF peptide, to design PCR primers. Using a poplar cDNA library as a template, we amplified a 327-bp fragment, which was sequenced and found to contain a partial RALF sequence. After screening the cDNA library (5 × 105 plaques) with this fragment, we isolated 10 plaques with positive signals. Seven clones were sequenced, and classified into two different groups based on the nucleotide sequences. Among these, five had identical coding sequences encoding a protein of 120 amino acids. These cDNAs all contained the exact N-terminal sequence of the purified RALF1 peptide obtained by Edman degradation (Fig. 3); thus, we conclude that these cDNAs encode RALF1. The N-terminal sequence of the RALF peptide was found at position 72 in the predicted protein sequence, indicating that, like tobacco RALF, the poplar RALF1 peptide is synthesized as a 120-amino acid precursor protein and processed into a biologically active mature form of 49 amino acid residues (Fig. 3). The Mr of the poplar RALF1 peptide as predicted from the nucleotide sequence is 5,493.1. Active tobacco RALF contains two disulfide bridges (Pearce et al., 2001); accounting for the loss of the four protons from the Cys during disulfide bridge formation, the expected Mr of active RALF1 would be 5,489.1. This Mr is very close to the mass of 5,488.7 we obtained from the MS analysis, further confirming that these cDNAs encode the RALF1 peptide. We chose an 838-bp cDNA as a representative of these cDNAs and named it PtdRALF1, and used this for subsequent experiments. Surprisingly, despite identical nucleotide sequences in the coding region, we observed that all five of these cDNA clones had variable 3′-untranslated region (UTR) sequences and sizes from each other (data not shown). This is suggestive of posttranscriptional modifications or alternative polyadenylation sites. In other eukaryotic organisms, transcript stability or translatability can be regulated by signals in the 3′-UTR (Hunt and Messing, 1998).
Figure 3.
Deduced amino acid sequences of two PtdRALF cDNAs compared with the aspen EST sequence (accession no. AI163551) and the tobacco RALF sequence (accession no. AF407278). The N-terminal sequence determined by Edman sequencing of RALF1 is underlined. The triangles indicate conserved Cys residues. Regions underscored with a dashed line correspond to sequences used for PCR primers. The GenBank accession numbers are AY172330 and AY172331 for PtdRALF1 and PtdRALF2, respectively.
Two additional cDNA clones obtained were distinct from PtdRALF1. They both contained the identical nucleotide sequence as the PCR product used to screen the cDNA library. We designated one clone as PtdRALF2, and used this cDNA for the further experiments. PtdRALF1 and PtdRALF2 showed a nucleotide identity of 88.4% over the coding sequences, and 84.2% similarity at the amino acid sequence levels, suggesting they represent different genes (Fig. 3; Table II). PtdRALF2 encodes a predicted protein of 128 amino acid residues, which by analogy to PtdRALF1 is predicted to be processed into a mature peptide of 49 amino acid residues. The Mr of this predicted peptide is 5,406.99. Again, if the active form of the mature Group 2 RALF peptide contains two disulfide bridges as expected, the measurable mass should be 5,402.99, very close to the mass of RALF 2 peptide we isolated (Table I). Therefore, we concluded that PtdRALF2 likely encodes the RALF2 peptide. Again, both PtdRALF2 cDNA had variable 3′-UTRs; interestingly, there are several possible polyadenylation sites in this region (data not shown).
Table II.
Comparison of PtdRALF1 and PtdRALF2 with P. tremula and P. tremuloides EST sequences
| PtdRALF1 | UB62BP.E02 | PtdRALF2 | UB31BPC05 | A044P29U | UB33PBE02 | G058P65Y | |
|---|---|---|---|---|---|---|---|
| PtdRALF1 | – | 95.9 | 88.4 | 87.9 | 84.1 | 65.3 | 57.8 |
| UB62BP.E02 | 94.7 | – | 86.6 | 87.5 | 77.8 | 65.3 | 53.4 |
| PtdRALF2 | 84.2 | 84.2 | – | 97.1 | 97.9 | 62.8 | 61.7 |
| UB31BPC05 | 85.8 | 86.0 | 95.3 | – | 99.1 | 63.7 | 61.0 |
| A044P29U | 81.8 | 76.4 | 97.3 | 99.1 | – | 64.0 | 62.1 |
| UB33PBE02 | 40.8 | 41.2 | 38.3 | 38.6 | 41.8 | – | 69.7 |
| G058P65Y | 45.1 | 39.6 | 47.3 | 47.3 | 47.3 | 75.8 | – |
Nucleotide sequences of EST clones homologous to PtdRALF were obtained from PopulusDB (http://poppel.fysbot.umu.se/blastsearch.html). Sequence similarities were analyzed by GeneTool and PepTool Analysis Software (Biotools, Inc., Edmonton, AB, Canada). Percent amino acid similarity is shown in bold, and nucleotide identity is shown in normal type.
The PtdRALF1 and PtdRALF2 coding sequences both have 62% overall amino acid similarity with the previously described tobacco RALF cDNA (Pearce et al., 2001). Over the 49 amino acids of both predicted mature peptides, however, the amino acid similarities with tobacco RALF are 86%, demonstrating that the mature RALF peptide sequence is highly conserved. As noted by Pearce et al. (2001), the sequence database searches contain numerous unannotated RALF-like ESTs from a diversity of species. Outside of the poplar ESTs (see below), the highest score was with an EST from Medicago trunculata (accession no. AJ501009), with amino acid similarities of 62% and 57% with PtdRALF1 and PtdRALF2, respectively.
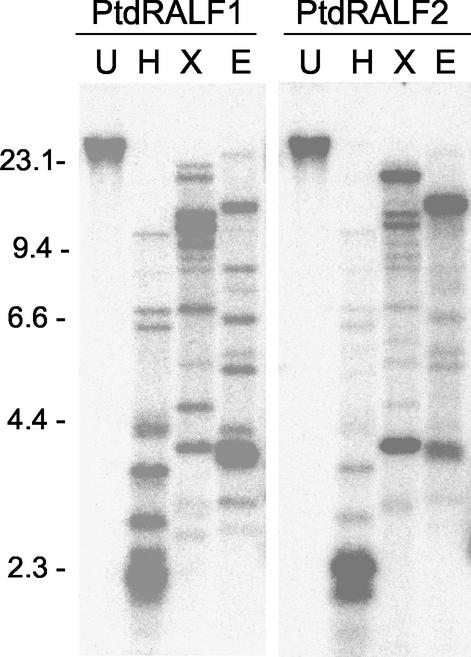
Using both PtdRALF1 and PtdRALF2 cDNAs as probes, we performed Southern analysis on hybrid poplar genomic DNA to estimate the size of the RALF gene family in poplar. We first hybridized Southern-blotted membrane with the full-length PtdRALF1 cDNA, and subsequently stripped the membrane and rehybridized with PtdRALF2. With either probe, the same set of approximately 10 to 12 bands was detected in the restricted genomic DNAs (Fig. 4). However, for each enzyme digest, the most strongly hybridizing bands were different for PtdRALF1 and PtdRALF2; that is, the relative strengths of the signals differed between the probes. Therefore, we conclude that both probes are recognizing essentially the same members of the PtdRALF gene family, and that the strongest one or two bands in each case represent the specific PtdRALF1 and PtdRALF2 genes. Because our material is an interspecific hybrid, we are likely detecting both alleles of each PtdRALF gene as distinct bands. Thus, it appears that the poplar genome contains a small family of RALF-like genes. This is consistent with our observation of five potential RALF peptides in poplar extracts.
Figure 4.
Southern analysis of the RALF gene family in poplar. Ten micrograms of restricted genomic DNA was probed with the full-length PtdRALF1 cDNA (left). The same membrane was stripped and rehybridized with the PtdRALF2 cDNA (right). U, Undigested; H, HindIII; X, XbaI; E, EcoRV.
Additional Homologs of RALF Genes in Populus
Although we have evidence for five distinct RALF peptides from the biochemical purification and detected a number of RALF-like genes in the H11-11 hybrid poplar genome by Southern analysis, we could only isolate two types of cDNAs by library screening. The PopulusDB database (http://poppel.fysbot.umu.se/blastsearch.html), which contains P. tremula and P. tremuloides ESTs, provided us with an additional tool to investigate RALF homologs in poplar. Using poplar RALF cDNA sequences for homology searches, we identified five unique ESTs (UB62BP.E02, UB31BPC05, A044P29U, UB33BPE02, and G058P65Y) in the PopulusDB. EST A044P29U is the previously identified RALF EST (AI163551; see above). Multiple sequence alignments indicated that four Cys residues are conserved in all sequences (not shown). As noted by Pearce et al. (2001), the C-terminal portions of the predicted proteins (corresponding to the mature RALF peptides) were most conserved, whereas the N-terminal regions were more divergent. Pair-wise analyses of the sequences showed that these RALF homologs were 38.3% to 99.1% similar at the amino acid sequence level (Table II). Based on their high scores, it appears that the aspen EST UB62BP.E02 is an ortholog of PtdRALF1 (95.9% nucleotide identity) and that aspen EST clones UB31BPC05 and A044P29U are orthologs of PtdRALF2 (97.1 and 97.9% nucleotide identity, respectively). Therefore, all sequences identified to date appear to represent a total of four distinct poplar RALF genes, represented by PtdRALF1, PtdRALF2, UB33PBE02, and G058P65Y (Table II).
We also considered if the most divergent RALF ESTs (UB33BPE02 and G058P65Y) might encode the peptides corresponding to the RALF3 MS peaks we found in poplar leaf extracts. Calculation of Mrs of their predicted RALF peptides, however, gave Mrs significantly different from the measured masses of RALF3 peaks (Table I). Therefore, ESTs UB33BPE02 and G058P65Y likely represent distinct members of the poplar RALF gene family, corresponding to peptides not yet identified in poplar leaf extracts. Analysis of the complete set of poplar RALF peptides and genes will require the availability of additional ESTs or genomic sequences.
Expression Analysis of PtdRALF mRNA in Poplar Saplings and Cell Culture
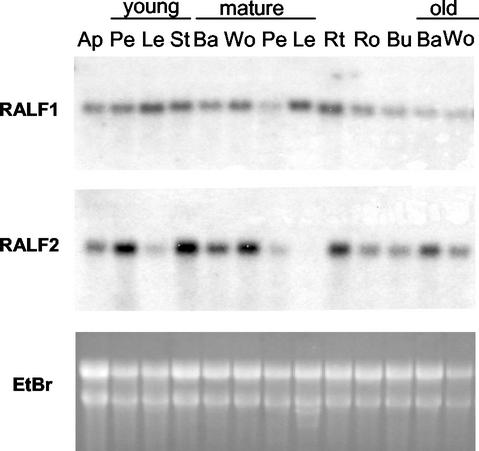
To obtain clues for understanding the biological roles of RALF peptides in planta, we conducted northern analysis to study the expression of PtdRALF. The availability of two different RALF cDNA sequences allowed us to investigate the differential expression of PtdRALF mRNAs in plants and cell cultures. For this purpose, gene-specific probes for PtdRALF1 and PtdRALF2 were generated from the sequence of 5′-UTRs of both cDNAs, where the sequence was least conserved. The specific probes shared 60.2% nucleotide sequence identity and their cross hybridization was negligible (data not shown). We first analyzed expression of PtdRALF in a variety of poplar sapling tissues. PtdRALF1 was expressed in all tissues tested, including shoot apex, petiole, leaf, stem, root, bud, bark, and wood (Fig. 5A). In contrast, expression of PtdRALF2 was observed in most tissues but absent or expressed at a very at low level in young and mature leaves (Fig. 5B). A general comparison of Figure 5, A with B, shows that the expression of PtdRALF2 varied more between tissues than did PtdRALF1. We also detected constitutive expression of both PtdRALF genes in poplar suspension culture (Fig. 6, control).
Figure 5.
Northern analysis of PtdRALF expression. RNA was extracted from tissues of different ages from a 3-month-old sapling. Total RNA (20 μg lane−1) was blotted onto membranes and hybridized with gene-specific probes for PtdRALF1 or PtdRALF2 (see “Materials and Methods”). EtBr, Ethidium bromide-stained gel; Ap, apical tissue; Pe, petiole; Le, leaf; St, stem; Ba, bark; Wo, wood; Rt, root tip, Ro, root; Bu, bud.
Figure 6.
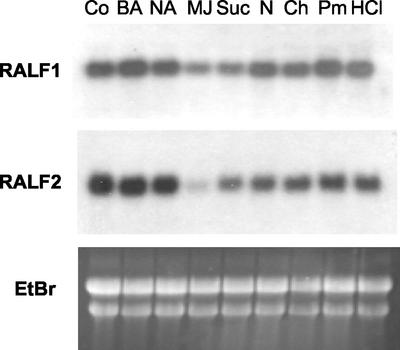
Northern analysis of PtdRALF in poplar cells after various treatments. Three-day-old cultures were treated for 5 h with compounds at concentrations as described in “Materials and Methods,” and analyzed for PtdRALF1 and PtdRALF2 gene expression. EtBr, Ethidium bromide-stained gel; Co, control; BA, benzyl adenine; NA, naphthalene acetic acid; MJ, MeJa; Suc, Suc; N, ammonium nitrate; Ch, chitosan; Pm, P. megasperma elicitor; HCl; hydrochloric acid.
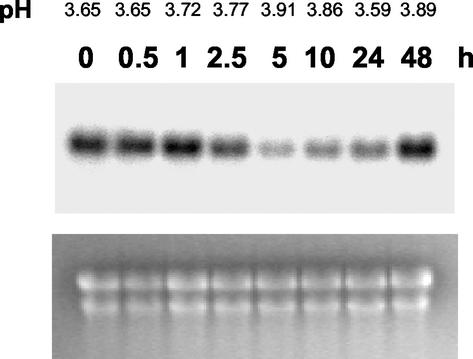
This almost ubiquitous pattern of expression led us to hypothesize that RALF gene products are likely involved in fundamental cellular processes, such as in hormone responses or primary metabolism. Therefore, we tested the effects of manipulating culture conditions on PtdRALF gene expression. Specifically, we asked whether the level of PtdRALF transcripts in the cells could be altered by the application of phytohormones (cytokinin, auxin, or methyl jasmonate [MeJa]), by modulation of the nutrient concentration (increasing Suc 3-fold and increasing nitrogen 10-fold), or by treatment with pathogen elicitors. We also tested the effect of culture medium acidification on PtdRALF expression by reducing the pH to 2.5 using HCl. After the addition of the test substances, the cultures were incubated for 5 h, and then harvested for northern analysis. Relative to control cells, the expression of PtdRALF1 and PtdRALF2 did not change significantly in the cells after most of the treatments. However, MeJa almost completely suppressed PtdRALF2 mRNA, and greatly reduced the abundance of PtdRALF1 mRNA (Fig. 6). We investigated the effect of MeJa on PtdRALF2 expression in more detail in time course experiments. The level of PtdRALF2 transcript decreased dramatically by 5 h after MeJa treatment, and then recovered again by 48 h (Fig. 7). In parallel, the pH of the medium increased moderately, peaking 5 h after MeJa addition and then decreasing by 24 h. At 48 h, the medium pH increased again, but control experiments indicated that this was due to the normal rise in medium pH due to aging of the cells through the culture cycle (data not shown). The inverse correlation of medium pH with PtdRALF2 transcript was observed consistently in repeat experiments; we note, however, that the kinetics of this extracellular pH increase by MeJa were much slower than the alkalinization observed with RALF itself (compare with Fig. 2). In additional experiments, we treated cells with KOH and NaOH to artificially increase the pH of the culture medium. However, these treatments did not result in any change in the level of PtdRALF2 transcripts (data not shown), suggesting that the reduction of PtdRALF2 transcript level by MeJa is not simply a consequence of MeJa-induced pH alkalinization.
Figure 7.
Northern analysis showing the effect of MeJa and PtdRALF2 expression in poplar cell cultures. Cultures (40 mL) were treated with 50 μm MeJa, and at the times indicated the cells were harvested for RNA extraction and the medium pH was measured. The ethidium-stained gel is shown on the lower panel as a loading control.
DISCUSSION
A Family of Poplar RALF Peptides and Genes
The first peptide with hormone-like properties in plants was reported more than 10 years ago with the discovery of the tomato wound signal systemin (Pearce et al., 1991), yet the widespread importance of peptide hormones in the plant kingdom is only beginning to be recognized (Ryan et al., 2002). RALF, a novel peptide causing rapid alkalinization of cell culture media, was very recently discovered in tobacco, alfalfa (Medicago sativa), and tomato, and similar ESTs from a number of plant species were identified in sequence databases (Pearce et al., 2001). In this study, we isolated a series of RALF peptides from poplar, cloned two distinct poplar RALF cDNAs, and identified additional genes from poplar ESTs. Using MS analysis, we identified five individual compounds from three HPLC peaks of poplar leaf extracts (Fig. 1; Table I). All three peaks induced the identical alkalinization response and exhibited very similar chromatographic profiles on both reverse phase and SCX HPLC, indicating they all contain similar active constituents. Direct peptide sequencing of the purified compound in peak 1 identified it as a RALF peptide, which we later named RALF1 (encoded by the PtdRALF1 gene). The mass of the alkalinizing compound in HPLC peak 2 corresponded exactly with the predicted Mr of the PtdRALF2-encoded peptide, strongly suggesting that this compound is the RALF2 peptide. Although we have no sequence information on the alkalinizing compounds in HPLC peak 3, based on the MS analysis, chromatographic behavior, and alkalinization kinetics, these constituents are very likely to be additional RALF peptides. Thus, our analysis extends the work of Pearce et al. (2001) by demonstrating the presence of multiple distinct RALF peptides within the same leaf extract. This could represent functional redundancy or specialization based on differential expression.
We isolated two distinct cDNAs representing two RALF genes (PtdRALF1 and PtdRALF2) from a poplar cDNA library, encoding the RALF1 and RALF2 peptides. Given the number of RALF peptides that appear to be present in leaf extracts, additional RALF genes are likely expressed; presumably, the sequence similarity of our probe with other RALF genes was insufficient for these to hybridize during our cDNA library screening. Southern analysis of hybrid poplar genomic DNA identified 10 to 12 bands, which given the hybrid nature of our plant material, could represent up to five or six individual genes. In the PopulusDB database, which contains ESTs from a number of different libraries, we identified five RALF homologs with nucleotide identities ranging from 53.4% to 99.1% (Table II). It should be noted that the C-terminal portions of the predicted proteins, which contain the mature RALF peptide, are more conserved than the N-terminal portions (not shown; Pearce et al., 2001). Our analysis of these sequences indicates there are at least four genes within the poplar genome (Table II); therefore, the ESTs provide direct evidence for at least two additional RALF genes in poplar. Together, these data provide evidence for the presence of a small gene family in poplar. In addition, there are likely to be other, more distantly related genes in poplar; a recent search for small peptides in Arabidopsis identified 34 genes with similarity to RALF (Olsen et al., 2002).
Expression of PtdRALF Genes
Our northern blots indicated that RALF genes were expressed in all tissues tested (Fig. 5). This widespread pattern of expression is consistent with the presence of RALF in plant EST databases constructed from a wide variety of tissues and organs, including roots, flowers, conducting tissues, and fruit (Pearce et al., 2001). PtdRALF2 transcript levels appeared to be more variable than PtdRALF1; specifically, PtdRALF2 mRNA was barely detectable in old leaves, and expressed at lower levels in younger leaves than any other tissue (Fig. 5). Nonetheless, we were able to isolate the RALF2 peptide during the original peptide purification; it was likely present in petioles or other tissue types that were included in the large-scale extraction.
Both PtdRALF genes were highly expressed in suspension cells (Fig. 6). Their expression levels showed no detectable variation after several environmental and hormonal stimuli, including changing the medium pH, addition of auxin, cytokinin, fungal elicitors, or nitrogen. We did, however, observe a strong reduction in RALF expression after MeJa treatment, especially for PtdRALF2 (Figs. 6 and 7). The decrease in PtdRALF2 expression over time was tightly correlated with a very slow increase in cell medium pH, which may suggest a link between these phenomena. This alkalinization was measured in hours rather than minutes; therefore, it appeared to be unrelated to the previous rapid alkalinization responses we had observed. Simply increasing the pH of the medium with NaOH or KOH did not influence PtdRALF2 expression (data not shown), which suggests that it is not alkalinization itself that represses PtdRALF2 expression in cells. Therefore, there appears to be no causal connection between the shift in pH and the expression of RALF gene expression, but rather both are manifestations of the cells' response to MeJa. MeJa is known to repress a number of housekeeping genes, such as Rubisco, chlorophyll a/b-binding protein, carbonic anhydrase, and α-tubulin (Wasternack et al., 1998; Schenk et al., 2000). Thus, MeJa may be inducing developmental changes and cellular differentiation (see below). We note that because we hybridized our northern blots with only the PtdRALF1 and PtdRALF2 probes, we have little information on the expression of other members of the PtdRALF gene family. It is possible that other RALF genes show a much greater degree of tissue-specific gene expression, or that they vary more dramatically in response to environmental stimuli in suspension cells.
Sequence analysis using PSORT software (http://psort.nibb.ac.jp) predicts that RALF is most likely to be secreted to the extracellular space. Because in normally dividing cultures RALF genes are highly expressed, it is surprising that these cells still respond to additional, exogenous RALF peptide. This may indicate that RALF in the medium is rapidly degraded. Other posttranscriptional regulatory mechanisms, such as processing or sequestration, could also account for this. Elucidating these aspects of RALF will require more in-depth studies.
Toward Possible Functions for RALF
Alkalinization of culture medium has been most commonly reported in response to microbially or plant-derived defense signals, for example flagellin, chitin, xylanase, pep-13, and systemin, where it appears to be a necessary part of the signal transduction pathway (Felix et al., 1993, 1999; Nürnberger et al., 1994; Felix and Boller, 1995; Enkerli et al., 1999). In contrast, RALF appears not to be a defense signal because it does not induce the antiherbivore proteinase inhibitors in tomato plantlets, but rather causes inhibition of root growth (Pearce et al., 2001). Our experiments with poplar cell cultures support the idea that RALF is unlikely to be involved in plant defense for the following reasons: (a) there was no induction of either RALF gene by these elicitor treatments (Fig. 6); (b) unlike flg22 or chitosan, RALF did not induce expression of the defense marker PAL in our cell cultures (data not shown); and (c) alkalinization triggered by RALF is much more rapid than that induced by flg22 or chitosan, elicitors of the defense response (Fig. 2).
A non-defensive role for RALF could imply a housekeeping or developmental function, consistent with the broad pattern of expression of both poplar RALF genes (Fig. 5). Although we tested a series of treatments, the only significant change in RALF gene expression that we observed was after the addition of MeJa to the cells. Although MeJa plays a key role in inducing pest and pathogen defense responses, it can also stimulate or inhibit developmental processes such as senescence, flower formation, and pollen maturation, and is important for tendril curling and internode elongation (Sembdner and Parthier, 1993; Weiler, 1997). In poplar cell culture, we speculate that the MeJa-induced changes are more of a developmental rather than a stress-related nature because in these cultures MeJa induces a developmentally regulated isoform of polyphenol oxidase, but not the wound-induced form (data not shown). In other systems, extracellular alkalinization has also been linked to processes other than defense. For example, Felle et al. (2000) observed extracellular alkalinization of roots in response to nod factors, and Felix et al. (2000) documented the alkalinization response in cell culture following changes in osmotic pressure. Other groups have demonstrated that extracellular alkalinization correlates with gravitropic responses (Johannes et al., 2001), and medium pH was found to be important for tracheary element differentiation in cultures of Zinnia elegans (Roberts and Haigler, 1994). Ultimately, the search for functions of RALF in growth and development will have to rely on more direct tests using transgenic plants or mutants.
In summary, we have identified a family of RALF genes and peptides in poplar. Our analysis of RALF gene expression in poplar indicates that they are expressed in most plant tissues and organs, as well as suspension cell cultures. Significantly, we observed reduced RALF transcript abundance in cells after MeJa treatment, which may be a first clue in working toward a function for these novel peptides.
MATERIALS AND METHODS
Plant Material
Hybrid poplar (Populus trichocarpa × Populus deltoides) clone H11-11 was propagated from green cuttings in peat (Terra-Lite Redi-Earth, W.R. Grace, Ajax, ON, Canada) in 15-cm-diameter pots as described (Constabel et al., 2000). Plants were maintained in environmental chambers under 16-h days at 18°C and 75% relative humidity. Light intensity was 300 mE m−2 s−1 at pot height, composed of approximately 20% incandescent (2,700 W) and 80% cool-white (11,880 W) light. Plants were watered daily with solution containing 1 g L−1 20-20-20 Plant-Prod complete fertilizer (Plant Products, Brampton, ON, Canada).
Cell Cultures, Alkalinization Assays, and Elicitor Treatments
Hybrid poplar suspension cells were obtained from Dr. Carl Douglas (University of British Columbia, Vancouver, Canada; de Sá et al., 1992) and maintained in Murashige and Skoog medium (Sigma, St. Louis) adjusted to pH 5.5 to 5.6 with KOH. For routine maintenance, 5 mL of a 1-week-old culture was transferred into 40 mL of medium in 200-mL flasks and maintained on an orbital shaker at 100 rpm in the dark at room temperature. A 2-mL aliquot of cells was transferred into each well of 12-well tissue culture plates (Corning, Corning, NY) and allowed to equilibrate on an orbital shaker at 120 rpm for 50 min. Fractions (2 μL) were added to the cells and the change in pH of the medium was measured every 5 min for 35 min using an Accumet pH meter with an Accuphast pH electrode (Fisher Scientific, Nepean, ON, Canada).
Flg22 peptide and Pep-13 were kindly provided by Dr. Georg Felix, (Friedrich Miescher-Institut, Basel) and Dr. Thorsten Nürnberger (Institut für Pflanzenbiochemie, Halle, Germany), respectively. Partially acid-hydrolyzed chitosan was obtained from Dr. Armand Seguin (Canadian Forest Service, Quebec City). Stock solutions for flg22, Pep-13, and chitosan were prepared at concentrations of 1 mm, 5 mm, and 1 mg mL−1, respectively. Two microliters of elicitor stock solution was added into 2 mL of poplar cell for the pH alkalinization assay.
For northern analysis, 3-d-old cultures were treated with the elicitors for a 5-h period. Treatments consisted of final concentrations of 2.5 mg L−1 benzyl adenine, 2.5 mg L−1 naphthalene acetic acid, 50 μm MeJa (Bedoukian Research, Danbury, CT), 9% (w/v) Suc, 16.5 mg mL−1 NH4NO3, 0.5 μg mL−1 chitosan, 0.1% (v/v) Phytophthora megasperma elicitor (Lisker and Kuc, 1977), or 5 × 10−5 n HCl. For MeJa time course experiments, eight flasks of 40 mL of culture were individually treated with 50 μm MeJa and cells were harvested at different time points. Treated cells were harvested by centrifugation at 1,600g for 15 min, frozen in liquid nitrogen, and stored at −80°C until analyzed.
Peptide Isolation
Hybrid poplar leaf and petiole tissue (800 g) was collected from 2-month-old poplar saplings. The tissues were homogenized in 200-g batches in a blender with 1 L of 1% (v/v) trifluoroacetic acid (TFA), filtered through two layers of Miracloth (Calbiochem, La Jolla, CA), and the homogenate centrifuged at 10,000g for 15 min. The supernatant was separated on C18 media (J.T. Baker, Phillipsburg, NJ) using open column reverse-phase chromatography (2.5 × 20 cm). The 60% (v/v) methanol-eluting fraction showing rapid medium pH-alkalinizing activity was lyophilized, the active material was redissolved in 60% (v/v) methanol/0.1% (v/v) TFA, and separated on a Sephadex G-25 (Pharmacia, Uppsala) gel chromatography column (2.5 × 55.5 cm). A portion of the active fraction was lyophilized and further purified on a cation exchange Macro-spin tube (PolySulfoethyl A, The Nest Group, Southboro, MA). The 1 m KCl-eluting fraction was further separated using SCX HPLC (PolySulfoethyl A, 200 × 4.6 mm, The Nest Group) running a 350 to 650 mm KCl gradient in 25% (v/v) acetonitrile/5 mm potassium phosphate buffer (pH 3.0). Activity peaks 1, 2, and 3 were applied to a C18 HPLC column (Bondpack C18, 3.9 × 300 mm, Waters, Milford, MA) and separated with an acetonitrile gradient in 0.1% (v/v) TFA. The purified peptides were analyzed by MALDI-MS at the University of Alberta's MS Facility. N-terminal sequencing was carried out using Edman chemistry at Washington State University (Pullman).
Cloning and Sequence Analysis
All molecular cloning procedures were carried out following standard protocols (Sambrook et al., 1989). PCR primers for amplification of a RALF fragment for library screening were designed based on the sequence of an aspen EST in GenBank (accession AI163551), and consisted of sense primer ATGGTGATGGGCTTGCCAT and antisense primer GCACCTTGTAATGCGACTGCA. The RALF cDNA fragment was amplified by PCR using a hybrid poplar leaf cDNA library as template (Constabel et al., 2000). The PCR fragment was cloned, sequenced, and used for library screening (approximately 5 × 105 plaques). Ten positive clones isolated from the secondary screening were excised into the pBluescript phagemid, and seven clones were sequenced using a fluorescently labeled dideoxyterminator sequencing kit (Thermosequenase, Amersham, Buckinghamshire, UK) on an ABI Prism 377 DNA Sequencer (Applied Biosystems, Foster City, CA). Similarity searches (BLAST) were carried out at the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/). Multiple sequence alignment for RALF sequences from poplar and aspen was conducted using the Clustal program at http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html, and pair-wise similarities calculated using Peptool and Genetool Software (BioTools, Inc., Edmonton, Canada).
RNA and DNA Hybridization Analysis
For Southern-blot analysis, genomic DNA was isolated from poplar leaves as described (Haruta et al., 2001a). DNA (10 μg) was digested with HindIII, XbaI, and EcoRV (Life Technologies/Gibco-BRL, Gaithersburg, MD), electrophoresed through 0.8% (w/v) agarose, and blotted onto Zeta-Probe membranes (Bio-Rad, Hercules, CA) using standard protocols (Sambrook et al., 1989). The DNA was cross-linked to membranes using a GC gene Linker UV chamber (Bio-Rad) before prehybridization for 2 h at 65°C in 6× SSC, 5× Denhardt's solution, 0.5% (w/v) SDS, and 100 μg mL−1 denatured salmon sperm DNA. DNA probes of PtdRALF1 and PtdRALF2 were prepared using the Rediprime II DNA labeling kit (Amersham Biosciences, Piscataway, NJ), and hybridization carried out for 18 h. The membranes were rinsed twice with 2× SSC at room temperature, washed once with 1× SSC/0.1% (w/v) SDS at 65°C for 30 min, and washed once with 1× SSC/0.1% (w/v) SDS at 65°C for 10 min. Hybridizing bands were revealed by exposure to x-ray film or on a PhosphorImager screen (Molecular Dynamics, Sunnyvale, CA).
For northern analysis, total RNA was extracted from tissues or cell culture using the protocol described by Haruta et al. (2001a). Three-month-old poplar saplings were used as a source of different tissues for the gene expression study. Total RNA (20 μg per lane) was loaded onto 1.4% (w/v) agarose-formaldehyde gels in MOPS buffer (pH 7.0) and transferred onto Zeta-Probe membranes (Bio-Rad) using standard procedures (Sambrook et al., 1989). Hybridization and analysis was carried out as described above.
ACKNOWLEDGMENTS
The authors thank Gregory Pearce and Dr. Clarence Ryan (Washington State University, Pullman) for advice on peptide purification and for sharing data before publication. We also thank Dr. Randy Whittal (University of Alberta) for MALDI-MS analysis and useful discussions, Stefan Jansson, (Umeå University, Umeå, Sweden) for providing unpublished aspen EST sequence data, and Barry McCashin (University of Alberta, Edmonton, Canada) for help with HPLC. In addition, we acknowledge the gifts of flg22 from Dr. Georg Felix, pep-13 from Dr. Thorsten Nürnberger (Institut für Pflanzenbiochemie, Halle, Germany), and chitosan from Dr. Armand Seguin (Canadian Forest Service, Quebec City, Canada).
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada (research grant to C.P.C.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.014597.
LITERATURE CITED
- Blumwald E, Aharon GS, Lam BC-H. Early signal transduction pathways in plant-pathogen interactions. Trends Plant Sci. 1998;3:342–346. [Google Scholar]
- Boller T. Chemoperception of microbial signals in plant cells. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:189–214. [Google Scholar]
- Bolwell GP. Role of active oxygen species and NO in plant defence responses. Curr Opin Plant Biol. 1999;2:287–294. doi: 10.1016/S1369-5266(99)80051-X. [DOI] [PubMed] [Google Scholar]
- Constabel CP, Yip L, Patton JJ, Christopher ME. Polyphenol oxidase from hybrid poplar. Cloning and expression in response to wounding and herbivory. Plant Physiol. 2000;124:285–295. doi: 10.1104/pp.124.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sá MM, Subramaniam R, Williams FE, Douglas CJ. Rapid activation of phenylpropanoid metabolism in elicitor-treated hybrid poplar (Populus trichocarpa Torr. & Gray X Populus deltoides Marsh) suspension-cultured cells. Plant Physiol. 1992;98:728–737. doi: 10.1104/pp.98.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkerli J, Felix G, Boller T. The enzymatic activity of fungal xylanase is not necessary for its elicitor activity. Plant Physiol. 1999;121:391–397. doi: 10.1104/pp.121.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Boller T. Systemin induces rapid ion fluxes and ethylene biosynthesis in Lycopersicon peruvianum cells. Plant J. 1995;7:381–389. [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- Felix G, Regenass M, Boller T. Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 1993;4:307–316. [Google Scholar]
- Felix G, Regenass M, Boller T. Sensing of osmotic pressure changes in tomato cells. Plant Physiol. 2000;124:1169–1179. doi: 10.1104/pp.124.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle HH, Kondorosi E, Kondorosi A, Schultze M. How alfalfa root hairs discriminate between Nod factors and oligochitin elicitors. Plant Physiol. 2000;124:1373–1380. doi: 10.1104/pp.124.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Major IT, Christopher ME, Patton JJ, Constabel CP. A Kunitz trypsin inhibitor gene family from trembling aspen (Populus tremuloides Michx.): cloning, functional expression, and induction by wound and herbivory. Plant Mol Biol. 2001a;46:347–359. doi: 10.1023/a:1010654711619. [DOI] [PubMed] [Google Scholar]
- Haruta M, Pedersen JA, Constabel CP. Polyphenol oxidase and herbivore defense in trembling aspen (Populus tremuloides): cDNA cloning, expression, and potential substrates. Physiol Plant. 2001b;112:552–558. doi: 10.1034/j.1399-3054.2001.1120413.x. [DOI] [PubMed] [Google Scholar]
- Hunt A, Messing J. mRNA polyadenylation in plants. In: Bailey-Serres J, Gallie DR, editors. A Look Beyond Transcription. Rockville, MD: American Society of Plant Biology; 1998. pp. 29–39. [Google Scholar]
- Johannes E, Collings DA, Rink JC, Allen NS. Cytoplasmic pH dynamics in maize pulvinal cells induced by gravity vector changes. Plant Physiol. 2001;127:119–130. doi: 10.1104/pp.127.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisker N, Kuc J. Elicitors of terpenoid accumulation in potato tuber slices. Phytopathology. 1977;67:1356–1359. [Google Scholar]
- Mathieu Y, Jouanneau J-P, Thomine S, Lapous D, Guern J. Cytosolic protons as secondary messengers in elicitor-induced defence responses. Biochem Soc Symp. 1993;60:113–130. [PubMed] [Google Scholar]
- Mijnsbrugge KV, Meyermans H, Van Montagu M, Bauw G, Boerjan W. Wood formation in poplar: identification, characterization, and seasonal variation of xylem proteins. Planta. 2000;210:589–598. doi: 10.1007/s004250050048. [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Nennstiel D, Jabs T, Sacks WR, Hahlbrock K, Scheel D. High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell. 1994;78:449–460. doi: 10.1016/0092-8674(94)90423-5. [DOI] [PubMed] [Google Scholar]
- Olsen AN, Mundy J, Skriver K. Peptomics, identification of novel cationic Arabidopsis peptides with conserved sequence motifs. In Silico Biol. 2002;2:0039. http://www.bioinfo-de/isb/2002/02/0039/ ( http://www.bioinfo-de/isb/2002/02/0039/) ) [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci USA. 2001;98:12843–12847. doi: 10.1073/pnas.201416998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science. 1991;253:895–898. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- Roberts AW, Haigler CH. Cell expansion and tracheary element differentiation are regulated by extracellular pH in mesophyll cultures of Zinnia elegans L. Plant Physiol. 1994;105:699–706. doi: 10.1104/pp.105.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CA, Pearce G, Scheer J, Moura DS (2002) Polypeptide hormones. Plant Cell Suppl S251–S264 [DOI] [PMC free article] [PubMed]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanders D, Bethke P. Membrane transport. In: Buchanan BB, Gruissem W, Jones RL, editors. Biochemistry and Molecular Biology of Plants. Rockville, MD: American Society of Plant Biologists; 2000. pp. 110–158. [Google Scholar]
- Schaller A, Oecking C. Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell. 1999;11:263–272. doi: 10.1105/tpc.11.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer JM, Ryan CA. A 160-kD systemin receptor on the surface of Lycopersicon peruvianum suspension-cultured cells. Plant Cell. 1999;11:1525–1535. doi: 10.1105/tpc.11.8.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sembdner G, Parthier B. The biochemistry and the physiological and molecular actions of jasmonates. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:569–589. [Google Scholar]
- Sterky F, Regan S, Karlsson J, Hertzberg M, Rohde A, Holmberg A, Amini B, Bhalerao R, Larsson M, Villarroel R et al. Gene discovery in the wood-forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proc Natl Acad Sci USA. 1998;95:13330–13335. doi: 10.1073/pnas.95.22.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Ortel B, Miersch O, Kramell R, Beale M, Greulich F, Feussner I, Hause B, Krumm T, Boland W et al. Diversity in octadecanoid-induced gene expression of tomato. J Plant Physiol. 1998;152:345–352. [Google Scholar]
- Weiler EW. Octadecanoid mediated signal transduction in higher plants. Naturwissenschaften. 1997;84:340–349. [Google Scholar]