Abstract
Expansin is a family of proteins that catalyze long-term expansion of cell walls and has been considered a principal protein that affects cell expansion in plants. We have identified the first root-specific expansin gene in soybean (Glycine max), GmEXP1, which may be responsible for root elongation. Expression levels of GmEXP1 were very high in the roots of 1- to 5-d-old seedlings, in which rapid root elongation takes place. Furthermore, GmEXP1 mRNA was most abundant in the root tip region, where cell elongation occurs, but scarce in the region of maturation, where cell elongation ceases, implying that its expression is closely related to root development processes. In situ hybridization showed that GmEXP1 transcripts were preferentially present in the epidermal cells and underlying cell layers in the root tip of the primary and secondary roots. Ectopic expression of GmEXP1 accelerated the root growth of transgenic tobacco (Nicotiana tabacum) seedlings, and the roots showed insensitivity to obstacle-touching stress. These results imply that the GmEXP1 gene plays an important role in root development in soybean, especially in the elongation and/or initiation of the primary and secondary roots.
The root is a plant organ that has adapted to acquire water and nutrients from the environment (Schiefelbein et al., 1997). The root system has recently been the focus of interest as a useful system for understanding organ development because it is a relatively simple organ, its growth pattern is uniform, and it has a small number of differentiated cell types (Aeschbacher et al., 1994). Furthermore, the development of new roots (secondary or lateral roots) from an existing root (primary root) provides a novel opportunity to investigate cellular differentiation and development in plants.
Although the primary and secondary roots share many basic structural features, they are different in their origin (Scheres et al., 1996). A basic feature of the root is its radial pattern, which is made up of concentric layers of tissues. Three fundamental types of the tissues are the epidermis, the cortex, and the vascular tissues (Esau, 1977; Dolan et al., 1993; Raven et al., 1999). In the longitudinal section, the root can be divided into three different regions: those of cell division, elongation, and maturation (specialization) (Dolan et al., 1993; Baluska et al., 1996; Howell, 1998; Raven et al., 1999). The region of cell division contains the root apical meristem, which carries out new cell divisions but does not elongate newly divided cells immediately. The cells derived from the region of cell division expand and elongate mostly in the region of elongation. After they have elongated, the cells begin to differentiate in the region of maturation, where root hairs and the secondary roots are initiated. The events of cell elongation and maturation occurring in the root have been suggested to be controlled by the extensibility of the cell wall and the turgor pressure inside the cell (Cosgrove, 1996).
It has been proposed that development of the secondary root is different from that of the primary root in many aspects (Scheres et al., 1996). Whereas the primary root is formed as a radicle at the base of the hypocotyl during embryogenesis, the secondary root is developed from the primary root but not by the root apical meristem at the root tip. The secondary root formation is initiated by periclinal and anticlinal divisions in pericycle and/or endodermal cells of the primary root depending on the species (Dolan et al., 1993; Malamy and Benfey, 1997).
It is known that the extent of plant cell elongation is confined by cell walls. The cell wall is composed of polysaccharides, proteins, phenolic compounds, and other materials (Varner and Lin, 1989). The plant cell wall plays a determinative role in establishing the size and shape of the plant cell. For the elongation or maturation event, however, the plant cell needs to selectively modify its cell wall. The agents for cell wall modification in the plant cell include various cell wall components, such as expansins, endoglucanases, xyloglucan endotransglycosylases, and hydroxyl radicals (Cosgrove, 1999, 2000a, 2000b).
Expansins are mainly considered as a primary agent for cell wall elongation, whereas the other substances could also modify wall structure and aid the primary agent for wall elongation (Vissenberg et al., 2000). However, the functional roles of expansins in the cell elongation have been mostly studied in vitro rather than in vivo (Cosgrove, 1996; Fleming et al., 1997). Although the molecular basis for expansin action on wall rheology is still uncertain, however, most evidence indicates that expansins cause wall creep by loosening hydrogen bonds between cellulose microfibrils and matrix polymer (McQueen-Mason and Cosgrove, 1994; Cosgrove, 1997). Since the first cloning of an expansin gene by Shcherban et al. (1995), many expansin genes have been identified from a variety of plant species. The expansin genes fall into three subfamilies, called the α-, β-, and γ-expansin subfamilies (Li et al., 2002). The α-expansins compose a major portion of the expansins, including the ones from tomato (Lycopersicon esculentum; Keller and Cosgrove, 1995), rice (Oryza sativa; Cho and Kende, 1997a), oat (Avena sativa; Li et al., 1993), and Arabidopsis (Cosgrove, 1998; Li et al., 2002). The α-expansin subfamily can be further divided into A, B, C, and D groups, as suggested by Link and Cosgrove (1998).
Expression patterns of the α-expansin genes have been characterized extensively in deepwater rice (Cho and Kende, 1997a, 1997b, 1998) and tomato (Reinhardt et al., 1998; Brummell et al., 1999a, 1999b). For example, the transcript of an expansin gene in deepwater rice, OsEXP4, increases in abundance before onset of increased wall extensibility and faster growth, supporting the role of expansins in cell elongation (Cho and Kende, 1998). In tomato, expression of the LeEXP18 gene was localized in a group of cells in the shoot apical meristem, where incipient leaf primordium initiation takes place (Reinhardt et al., 1998). Thus, the LeEXP18 expression is considered to be a molecular marker for leaf initiation, predicting a site of primordium formation before histological changes can be detected in the shoot apical meristem (Reinhardt et al., 1998). A recent study showed that the LeEXP18 expression does not correlate with elongation growth and that some of the tomato expansin genes are differentially expressed during fruit development, suggesting that not all the expansins are involved in the expansion process (Brummell et al., 1999a, 1999b; Caderas et al., 2000; Rose et al., 2000). Therefore, the complexity of the expansin family and the tissue-specific expression patterns of some of its members suggest that different expansins may play different roles in various cell types during organ development in plants (Rose et al., 1997; Reinhardt et al., 1998).
Our previous studies have shown that expression of an extensin gene encoding a soybean (Glycine max) Hyp-rich glycoprotein, SbHRGP3, was up-regulated by the maturation of the primary root (Ahn et al., 1996). Its expression was also observed in the mature region of the secondary root, suggesting its role in the maturation of soybean roots by cessation of cell elongation in the region of maturation (Ahn et al., 1998). On the other hand, although it has long been speculated that the cell elongation in roots is caused by expansins, the root-specific expansin gene has not yet been characterized in sufficient detail. Therefore, we have carried out an investigation to understand root development by comparing the expression patterns of extensin and expansin genes in soybean roots. Here, we report that in contrast to the SbHRGP3 gene, a soybean root-specific expansin gene, GmEXP1 (accession no. AF516879), is expressed in the region of elongation, particularly in the epidermal cells and underlying cell layers, but not in the region of maturation in the primary root. We also show that expression of the GmEXP1 gene was re-up-regulated at the secondary root initials. Furthermore, overexpression of GmEXP1 in transgenic tobacco (Nicotiana tabacum) plants caused accelerated growth and reduced sensitivity to the obstacle-touching stress. These facts suggest that the GmEXP1 gene induces cell elongation in the region of elongation in the soybean roots, whereas the SbHRGP3 gene plays an opposite role in the region of maturation.
RESULTS
Isolation of Expansin cDNAs from a Root cDNA Library of Soybean
To isolate expansin cDNAs in soybean, we designed two degenerate primers targeted to the conserved regions of expansin genes and carried out PCR with soybean genomic DNA as a template. The PCR-amplified product was used as a probe to screen a root cDNA library of soybean, which led to isolation of two cDNA clones, designated as GmEXP1 and GmEXP2 (accession no. AF516880). The GmEXP1 cDNA consists of 1,089 bp with an open reading frame encoding a polypeptide of 255 amino acids, which contains a putative signal sequence of 16 amino acids at the N terminus. In contrast, the GmEXP2 cDNA is composed of 1,312 bp with an open reading frame encoding 258 amino acids including a putative signal sequence of 21 amino acids (data not shown).
For homology analysis, the deduced amino acid sequences of GmEXP1 and GmEXP2 were compared with those of other expansins. GmEXP1 showed strong sequence similarities to a group of α-expansins including NtEXP3 of tobacco (86%; Link and Cosgrove, 1998), CsEXP2 of cucumber (Cucumis sativus; 92%; Shcherban et al., 1995), OsEXP1 of rice (85%; Cho and Kende, 1997b), and AtEXP1 of Arabidopsis (82%; Shcherban et al., 1995). GmEXP2 also exhibited strong similarity to other α-expansins, such as PsEXP1 of pea (Pisum sativum; 82%; Michael, 1996), AtEXP6 of Arabidopsis (77%; Shcherban et al., 1995), and OsEXP4 of rice (71%; Cho and Kende, 1997b). To investigate the phylogenetic status of the GmEXP1 and GmEXP2 genes among α-expansin genes, a phylogenetic tree was constructed with 20 α-expansins by using the maximum likelihood method (HKY 85 model). The results indicated that the GmEXP1 and GmEXP2 genes belong to the D and A groups of the α-expansin subfamily, respectively. An alignment of the amino acid sequences of six α-expansins and the phylogenetic tree are available as supplemental data at http://www.plantphysiol.org.
The GmEXP1 and GmEXP2 Genes Are Members of the Expansin Multigene Family in Soybean
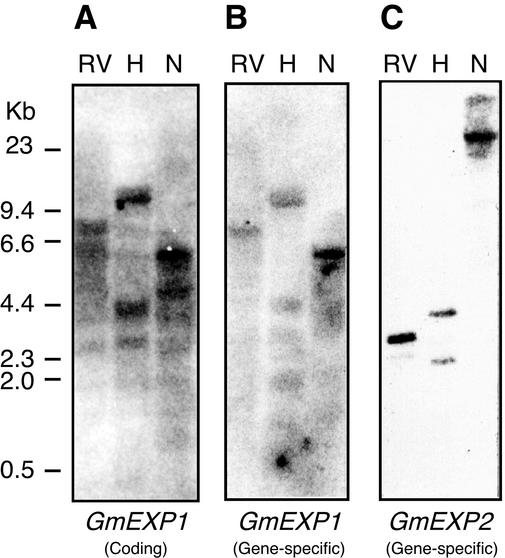
Expansin genes generally consist of a multigene family in various plant species (Shcherban et al., 1995; Cho and Kende, 1997b; Link and Cosgrove, 1998). Hence we performed genomic DNA gel-blot analyses to estimate the number of expansin genes in the soybean genome. As shown in Figure 1A, hybridization with the coding region of the GmEXP1 cDNA detected many bands in each lane, suggesting that various expansin genes are present in the soybean genome, including its α-expansin homologs and probably those encoding pollen allergens (β-expansins). However, when the gene-specific probes for the GmEXP1 and GmEXP2 were used, a strong single band was apparent along with occasional weaker bands in each blot (Fig. 1, B and C). The band patterns detected by the gene-specific probes differ from each other, suggesting that the GmEXP1 and GmEXP2 genes exist as single-copy genes in the soybean genome.
Figure 1.
Genomic DNA gel-blot analyses of the GmEXP1 and GmEXP2 genes. Ten micrograms of soybean genomic DNA was digested with EcoRV (RV), HindIII (H), or NcoI (N) and hybridized with the coding and gene-specific probes of the GmEXP1 and GmEXP2 cDNAs after labeling. A, Filter hybridized with the GmEXP1 coding sequence as a probe. B, Filter hybridized with the GmEXP1 gene-specific probe. C, Filter hybridized with the GmEXP2 gene-specific probe. Numbers on the left side indicate size markers.
GmEXP1 Is a Root-Specific Expansin Gene in Soybean
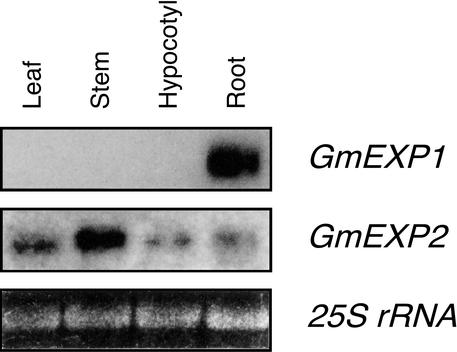
To investigate the expression patterns of the GmEXP1 and GmEXP2 genes in soybean, RNA gel-blot analyses were performed with the gene-specific probes using total RNAs isolated from various tissues in 20-d-old soybean seedlings. In the RNA gel-blot analysis with the GmEXP1-specific probe, hybridization signals were detected only with RNA isolated from roots (Fig. 2). The size of the GmEXP1 mRNA was about 1.1 kb, which was similar to that predicted from its cDNA sequence. However, the GmEXP2-specific probe hybridized with RNAs from all the tissues, although its transcripts were most abundant in the stems. These indicated that the GmEXP1 gene is root specific and may be involved in root development in soybean, whereas the GmEXP2 gene is not expressed in a tissue-specific manner, suggesting that the GmEXP1 and GmEXP2 genes are differentially regulated in soybean. Therefore, we decided to further dissect the role(s) of the GmEXP1 gene in soybean root development.
Figure 2.
Organ-specific expression of the GmEXP1 and GmEXP2 genes in soybean. Each lane contains 10 μg of total RNA extracted from leaves, stems, hypocotyls, and roots of 20-d-old soybean seedlings grown in the Hoagland solution. For comparison of both the GmEXP1 and GmEXP2 genes, the membrane was stripped and reprobed with the other probe. The bottom panel shows the ethidium bromide (EtBr)-stained 25S rRNA in the gel, indicating that an equal amount of RNA was loaded in each lane.
GmEXP1 Expression Is Up-Regulated When Soybean Roots Elongate Rapidly
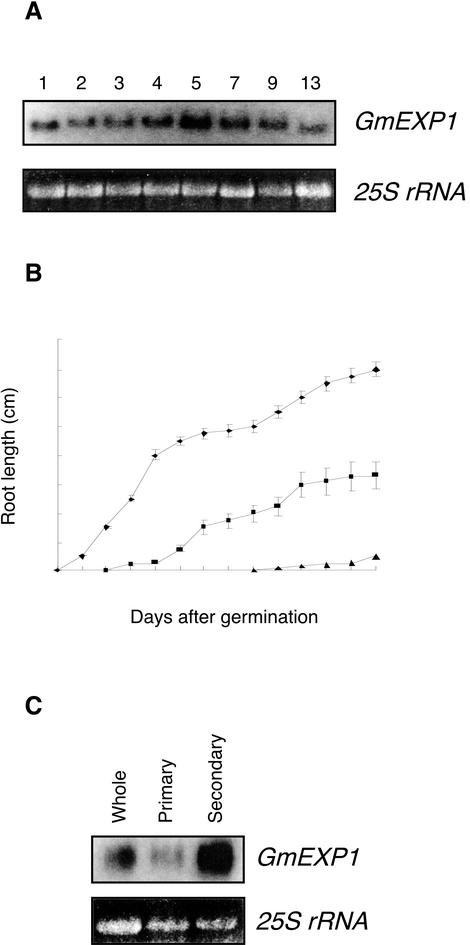
Because the GmEXP1 gene exhibited root-specific expression in soybean, we evaluated the expression levels of the GmEXP1 gene in roots at different developmental stages. Total RNAs were extracted from roots of 1-, 2-, 3-, 4-, 5-, 7-, 9-, and 13-d-old seedlings and were subjected to RNA gel-blot analysis. As shown in Figure 3A, although the GmEXP1 gene was expressed at all the developmental stages, the expression level was very high at 1 d after germination and reached a maximum level at 5 d after germination.
Figure 3.
Expression patterns of the GmEXP1 gene in soybean roots during development. A, Total RNA was extracted from roots of 1-, 2-, 3-, 4-, 5-, 7-, 9-, and 13-d-old soybean seedlings. For comparison of expression levels of the GmEXP1 gene, 10 μg was used on each lane. The bottom panel shows the EtBr-stained 25S rRNA in the gel. B, The growth patterns of the primary, secondary, and tertiary roots in soybean seedlings were analyzed during seedling development. ♦, Primary root; ▪, secondary root; ▴, tertiary root. C, The whole, primary, and secondary roots were dissected separately from 5-d-old seedlings. Ten micrograms of total RNA isolated from the roots were used in the RNA gel-blot analysis. The bottom panel shows the EtBr-stained 25S rRNA in the gel.
We hence examined root development to determine the physiological relevance of the high expression of the GmEXP1 gene. We monitored development of the primary root and the secondary and tertiary roots of six young seedlings growing in a liquid medium by measuring their root length. As shown in Figure 3B, the primary roots grew rapidly until 4 d after germination, and then their growth gradually decelerated. The secondary roots were initiated from the primary root at 2 d after germination and grew rapidly from 4 to 6 d after germination. The tertiary roots emerged at 8 d after germination.
We subsequently compared the expression pattern of the GmEXP1 gene with the growth pattern of soybean roots. The comparison showed that rapid emergence of the primary root is correlated with the high level of GmEXP1 expression at 1 d after germination. In addition, it revealed that the primary and secondary roots contributed the maximum expression level of the GmEXP1 gene at 5 d after germination. Therefore, we examined relative levels of GmEXP1 expression in the primary and secondary roots at the developmental stage. As shown in Figure 3C, although both the primary and secondary roots expressed the GmEXP1 gene, its level was much higher in the secondary roots in 5-d-old seedlings. This is probably because in the 5-d-old seedlings, the growth of the primary root decelerated, whereas the secondary root accelerated its growth, indicating that the expression level of the GmEXP1 gene is closely related with the growth rate of the roots. Furthermore, the expression levels of the GmEXP1 gene decreased gradually as the growth of the secondary root concurrently decelerated. Taken together, our data suggested that expression of the GmEXP1 gene is up-regulated when rapid root elongation takes place during root development in soybean.
Up-Regulation of the GmEXP1 Expression Is Closely Linked to Cell Elongation in Soybean Roots
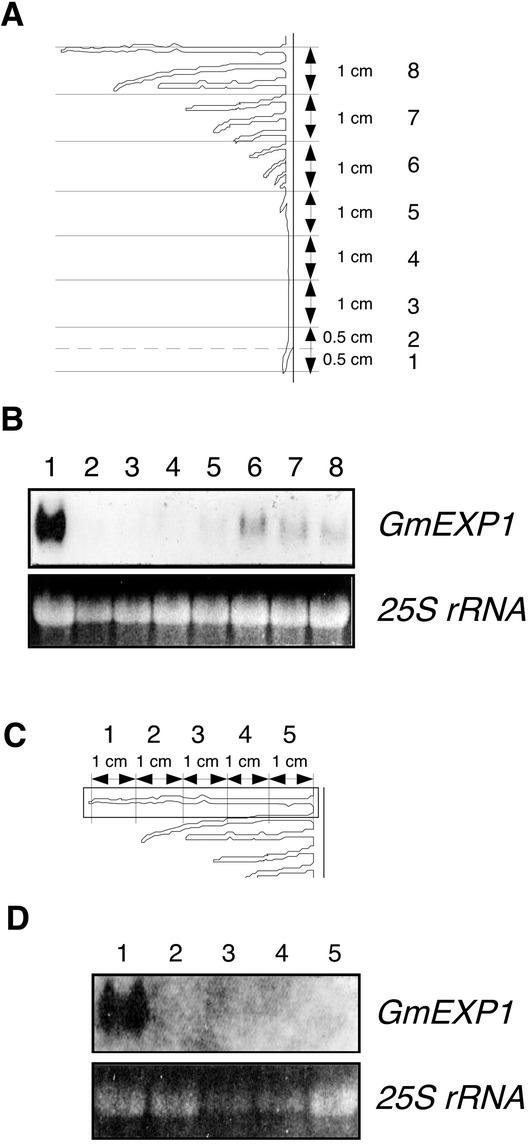
We investigated spatial the distribution of the GmEXP1 mRNA in the primary root of the 5-d-old seedlings that showed the maximum level of GmEXP1 expression. We serially dissected the primary roots that reached a length of 9 cm in 5-d-old seedlings as depicted in Figure 4A. Total RNAs were extracted from each section and subjected to an RNA gel-blot analysis. As shown in Figure 4B, the GmEXP1 transcripts were predominantly detected in section 1, representing the root tip region, which mostly comprises the region of cell division and elongation (Ahn et al., 1998). In contrast, hybridization signals were very weak in the region of maturation (sections 2–8). Interestingly, section 6 also revealed elevated levels of GmEXP1 expression, which was most likely contributed by the emerging secondary roots as observed in Figure 3C.
Figure 4.
Spatial distribution of the GmEXP1 mRNA in the primary roots of 5-d-old seedlings and in the 5-cm-long secondary roots of 9-d-old seedlings. A, The primary root was serially dissected along the root axis as shown. B, Total RNA was extracted from each section in A and analyzed on a gel blot. Sections 5 to 8 also included the secondary roots initiated from the primary root. C, The 5-cm-long secondary roots were serially dissected. D, Total RNA was prepared from each section of the secondary roots in C and analyzed on a gel blot. The bottom panels in B and D show the EtBr-stained 25S rRNA in the gel.
We also carried out RNA gel-blot analysis to examine the spatial distribution of the GmEXP1 mRNA in the secondary roots. The secondary roots of 5-cm length in 9-d-old seedlings were serially dissected along the root axis as depicted in Figure 4C. Total RNAs extracted from each section were subjected to RNA gel-blot analysis. As shown in Figure 4D, hybridization signals were also detected mainly in section 1 that included the tip region of the secondary root as observed in the primary root (Fig. 4B). These results indicated that the GmEXP1 gene is highly expressed in a specific zone that includes the regions of cell division and elongation in the primary and the secondary roots, suggesting its role in cell elongation during root development.
GmEXP1 mRNA Is Preferentially Localized to the Epidermal and Underlying Cell Layers in the Region of Cell Elongation of the Primary Root in Soybean
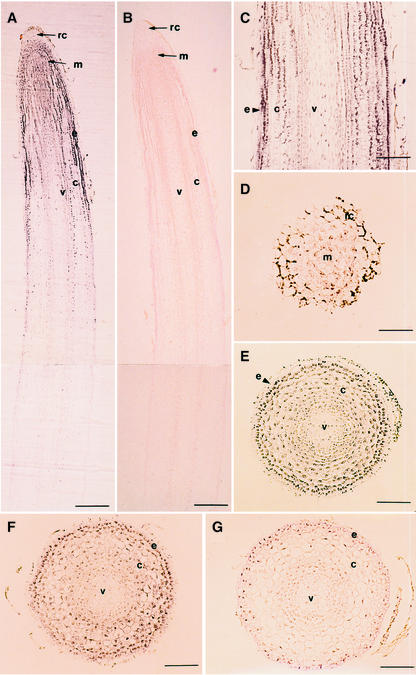
In situ hybridization was conducted with 5-d-old seedlings to localize GmEXP1 expression in soybean roots. When longitudinal sections of the distal part of the primary root were hybridized with an antisense probe, hybridization signals were highly detected in the root tip region (Fig. 5A), which is in good agreement with our previous results obtained from the RNA gel-blot analysis (Fig. 4B). On the basis of the anatomical features including the vascular bundle development (Ahn et al., 1998; data not shown), levels of the GmEXP1 transcripts gradually increased from the region of cell division to the region of elongation and then decreased in the region of maturation (Fig. 5A). The GmEXP1 transcripts were detected in most cells in the region of cell division, where little cell differentiation takes place. As the cells differentiated, the GmEXP1 transcripts were localized in the epidermis, some underlying cell layers, and the vascular cylinder in the region of elongation (Fig. 5C). However, the hybridization signals were hardly detectable in the epidermis of the region of maturation (Fig. 5G).
Figure 5.
Localization of GmEXP1 mRNA in the primary root of 5-d-old soybean seedlings by in situ hybridization. A to C, Longitudinal sections from the root tip region. A was hybridized with the antisense probe for the GmEXP1 gene. B was hybridized with the sense probe for the GmEXP1 gene. C is the magnification image of the region of cell elongation in A. D to G, Cross sections from the root were hybridized with the antisense probe for the GmEXP1 gene. D is the region that is 80 μm apart from the root tip, which is the region of the root cap. E is the region that is 1.0 mm apart from the root tip. F is the region that is 1.3 mm apart from the root tip. G is the region that is 4.0 mm apart from the root tip. rc, Root cap; e, epidermis; v, vascular cylinder; m, meristem; c, cortex. In A and B, bars = 250 μm; in D, bar = 50 μm; in C, E, F, and G, bars = 100 μm.
Because the longitudinal sections revealed that the GmEXP1 gene was highly expressed in the epidermal and underlying cell layers in the region of elongation (Fig. 5C), we carried out in situ hybridization with the cross sections of the primary roots to identify cell layers in which GmEXP1 is expressed. On the basis of anatomical features of the root regions (Ahn et al., 1998), we chose sections representing the regions of cell division, elongation, and maturation and hybridized with the antisense probe. As observed in Figure 5A, the cells in the root cap were weakly stained, whereas the cells in other layers in the region of cell division were evenly stained (Fig. 5D). In the region of elongation, the GmEXP1 transcripts were strongly detected in cells of the epidermis and vascular cylinder (Fig. 5, E and F). However, the epidermal cells in the region of maturation were barely stained (Fig. 5G). These results suggested that the GmEXP1 gene is expressed in the regions of cell division and elongation, preferentially in their epidermal cells and some underlying cell layers.
GmEXP1 mRNA Is Also Localized in the Secondary Root Initials in Soybean
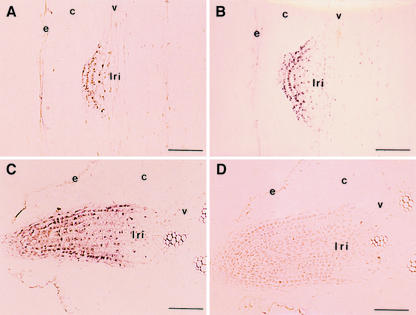
It has been suggested that formation of organ initials involves oriented cell expansion and cell wall modification, coupled with controlled cell division (Lyndon, 1990; Fleming et al., 1997; Jacobs, 1997; Meyerowitz, 1997). Therefore, we investigated expression patterns of the GmEXP1 gene in secondary root initials by in situ hybridization analysis to test whether the gene is also involved in formation of the root initials. The secondary root-initiating region in the primary root of 5-d-old seedlings (about 3.5 cm from the root tip) was longitudinally sectioned and subjected to in situ hybridization analysis. As shown in Figure 6, A and B, the GmEXP1 gene was expressed in the secondary root initials, preferentially in their epidermal cells. Cross sections of the primary root containing the secondary root initials also revealed that the GmEXP1 gene was mainly expressed in the tip region of the emerging secondary root (Fig. 6C). In contrast, the tissue hybridized with a sense probe did not show any detectable signal at the same developmental stage (Fig. 6D).
Figure 6.
Localization of GmEXP1 mRNA in the secondary root of 5-d-old soybean seedlings by in situ hybridization. A and B, Longitudinal sections from the secondary root initiation zone (3.5 cm from the primary root tip) that shows a secondary root primordium at the early developmental stage. C and D, Cross sections from the secondary root initiation zone in the primary root. A to C were hybridized with the antisense probe for the GmEXP1 gene. D was hybridized with the sense probe of the GmEXP1 gene. lri, Lateral root initial; e, epidermis; v; vascular cylinder; c, cortex. Bars in A to D = 100 μm.
It is known that the secondary roots are initiated from the pericycle in the region of maturation of the primary root and then penetrate through cell layers of cortex and epidermis to emerge from the primary root (Malamy and Benfey, 1997). The fact that the primary and secondary roots employ the same GmEXP1 gene in cell expansion suggest that although the origin of the secondary root is different from that of the primary root, they are not significantly different in their morphology, organization, and gene expression in some aspects (Dolan et al., 1993).
Overexpression of GmEXP1 in Tobacco Plants Reduces Sensitivity to the Obstacle-Touching Stress
To investigate in vivo functions of the expansin gene during organ formation in plants, we carried out ectopic expression of the GmEXP1 gene in tobacco plants. The entire coding region of the GmEXP1 cDNA was amplified by PCR, and the product was fused with the cauliflower mosaic virus 35S promoter in the pGA643 vector (An et al., 1988). The resulting transgene, 35S::GmEXP1, was introduced into tobacco plants by Agrobacterium tumefaciens-mediated transformation. After identifying transgenic plants on selection media, we divided the transgenic plants into two arbitrary groups based on the expression levels of the transgene (see supplemental data available at http://www.plantphysiol.org), namely strong and weak lines.
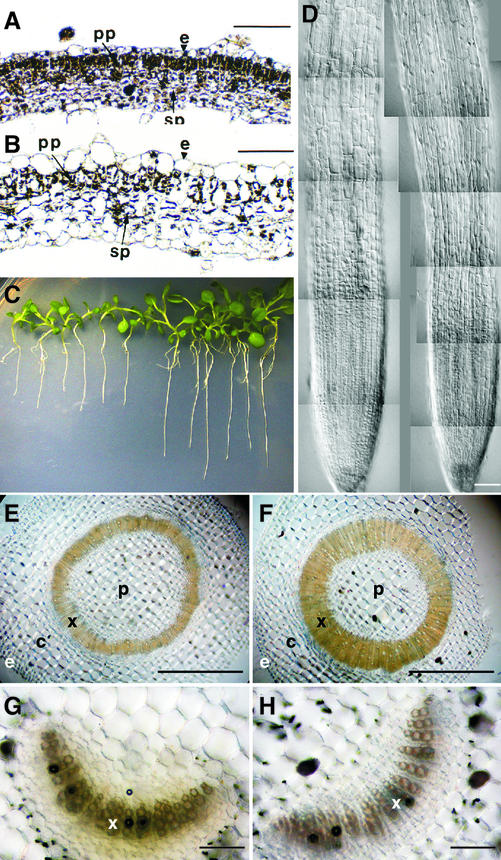
As shown in Figure 7C, the transgenic plants overexpressing the GmEXP1 gene were relatively bigger than the control plants, suggesting that GmEXP1 overexpression caused accelerated growth. In addition, they showed abnormal phenotypes in leaves, stems, and flowers, depending on the lines. The strong lines showed a bushy phenotype because multiple leaves developed almost simultaneously from the shoot apex, whereas the weak lines generated axillary shoots, probably due to weak apical dominance (data not shown). The cells in the leaves of the strong lines were enlarged and layered irregularly (Fig. 7, A and B), probably due to the cell elongation driven by the GmEXP1 overexpression. Interestingly, the transgenic plants showed thickened xylem cell layers in the stems (Fig. 7, E and F), although there was no significant difference in xylem tissues of the petioles of the transgenic plants (Fig. 7, G and H), as compared with those of wild-type plants. These data suggest that the GmEXP1 overexpression selectively affected developmental processes of the transgenic plants. Although the transgenic plants showed additional phenotypes, however, we focused on the root phenotype of the transgenic plants because GmEXP1 expression is root-specific in soybean.
Figure 7.
Anatomical features of transgenic tobacco plants overexpressing the GmEXP1 gene. A and B, Cross sections of leaves of wild-type and transgenic plants, respectively. Leaf cells of the transgenic plants are enlarged and irregularly layered, reflecting ectopic expression of the GmEXP1 gene in the transgenic tobacco plants. e, Epidermis; pp, palisade parenchyma; sp, spongy parenchyma. C, Transgenic tobacco seedlings of line S1 in the T2 generation overexpressing the GmEXP1 cDNA under the obstacle-touching condition. After vernalization, the seedlings were incubated on 1.5% (w/v) agar plates in an inclined position (45°) for 17 d after germination. The seedlings on the left side are wild-type tobacco plants, and the ones on the right are transgenic. D, Comparison of the root apical region in a wild-type control tobacco plant (left) and a transgenic plant (right). The cells in the transgenic plant are more elongated than those of the wild-type control plant. Bar in D = 100 μm. E and F, Cross sections of stems of wild-type control plants (E) and the transgenic plants (F) at the same developmental stage. Bars in E and F = 1,000 μm. c, Cortex; e, epidermis; p, pith; x, xylem. G and H, Cross sections of petioles of wild-type control plants (G) and the transgenic plants (H) at the same developmental stage. Bars in G and H = 100 μm. x, Xylem.
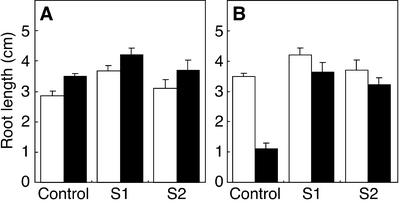
We self-pollinated the transgenic tobacco plants of the weak lines and obtained their homozygous seeds in the T2 generation because the strong lines were sterile. We grew the homozygous seedlings under the neutral condition (pH 7.0) to investigate the role of GmEXP1 during root development of the transgenic plants. We used two independent transgenic plants harboring one copy of the transgene, namely S1 and S2, respectively (data not shown). We subsequently examined whether the growth of the transgenic roots is affected under the acidic condition (pH 4.5), which has been known to cause acidic growth of the cell wall. Wild-type seedlings and the homozygous transgenic seedlings were incubated on 1.5% (w/v) agar plates (pH 7.0 and pH 4.5) in a vertical position for 17 d after vernalization in a dark chamber for 2 d and their root lengths were measured. The primary root of the seedlings was longer than that of the wild type under both neutral (pH 7.0) and acidic (pH 4.5) conditions (Fig. 8A). These data suggest that GmEXP1 is one of the major elements that induce acid growth in soybean.
Figure 8.
Comparison of root length under the acid growth condition (A) and under the obstacle-touching condition (B). A, White bars represent the roots grown under the neutral condition (pH 7.0), whereas black bars indicate the roots grown under acidic condition (pH 4.5). B, White bars represent the roots of the plants grown in vertical plates, whereas black bars indicate the ones in an inclined position (45°). S1 and S2 are the two independent transgenic plants containing 35S::GmEXP1.
We also investigated whether the GmEXP1 overexpression affected root growth under the obstacle-touching stress to see if an expansin activity is involved in a response to the stress (Okada and Shimura, 1990). Wild-type and homozygous transgenic seedlings were incubated on 1.5% (w/v) agar plates (pH 4.5) in a vertical position (90o) after cold treatment for 2 d. After 3 d, one-half of the plates containing both samples were inclined at 45o to give the obstacle-touching stress, and the rest were continually incubated in a vertical position. In the vertical plate, the primary roots of wild-type seedlings just grew straight downward on the surface of the agar (data not shown). As shown in Figure 7C, when in an inclined position, in which plants face the obstacle-touching stress, however, the growth of the primary roots of wild-type seedlings was severely affected. The roots were much shorter in the inclined position, with an average length being decreased from 3.5 to 1.1 cm (Fig. 8B). However, the primary roots of GmEXP1 overexpressing seedlings were barely affected under the obstacle-touching stress (Fig. 8B). This result showed that the GmEXP1 overexpression reduced sensitivity to the obstacle-touching stress, implying that GmEXP1 may play an important role in overcoming the stress, which the plant roots would face in soil.
Because the primary roots of the transgenic plants were longer than those of wild-type control plants in both the acidic and obstacle-touching conditions, we carefully examined anatomical features of the roots. No dramatic difference in overall anatomical features of the primary roots was found between the transgenic and control plants; however, the cell length was significantly different. Epidermal cells were longitudinally elongated along the root axis in the transgenic seedlings (Fig. 7D), and the cortex cells were also larger than those of the control plants (data not shown). We therefore measured the cell length by using the NIH image software (developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image) to analyze the composite images of the roots. As shown in Table I, the roots of the S1 plants showed significant elongation of epidermal cells along the root axis, which is likely to be a main reason for the rapid growth. However, there was no significant difference in vascular bundles of the transgenic roots in the cross sections (data not shown) compared with the ones in the stems (Fig. 7, E and F).
Table I.
Comparison of longitudinal length of root epidermal cells
| Distance from the Root Tip | Cell Length
|
|
|---|---|---|
| C | S1 | |
| μm | ||
| 400 | 17.6 ± 3.9 | 30.2 ± 6.9 |
| 800 | 32.0 ± 5.0 | 71.9 ± 15.7 |
| 1,200 | 140.5 ± 21.6 | 216.0 ± 25.5 |
| 1,600 | 188.7 ± 33.7 | 207.9 ± 30.7 |
Wild-type seedlings and transgenic seedlings harboring 35S::GmEXP1 were grown on 1.5% (w/v) agar plates in an inclined position, and the length of epidermal cells was measured along the root axis at 17 d after germination. The NIH image software (v1.62) was used to analyze the composite digital camera images of the roots. At least 10 cells were measured for each measurement. C, Wild-type control plant; S1, a transgenic plant line overexpressing the GmEXP1 gene.
DISCUSSION
Our previous studies have shown that the SbHRGP3 gene encoding an extensin in soybean is expressed mainly in the region undergoing maturation in the primary and secondary roots and may be involved in the cessation of cell elongation in the region of maturation of the soybean roots (Ahn et al., 1996, 1998). In this study, we have identified a root-specific expansin gene from soybean and examined its expression patterns and its role during root development in soybean and transgenic tobacco plants. The results have shown that the GmEXP1 gene was expressed differentially in roots at different developmental stages, as previously observed with the SbHRGP3 gene. However, the GmEXP1 expression was confined mainly to the regions undergoing cell division and elongation in the primary and secondary roots, suggesting its roles in root elongation in soybean. Taken together, we suggest that GmEXP1 and SbHRGP3 play important roles in the elongation and maturation processes of soybean root development.
The GmEXP1 and GmEXP2 Genes Are New Members of the α-Expansin Subfamily in Soybean
Because the cell elongation induced by the acidic condition occurs in algae, mosses, ferns, gymnosperms, and angiosperms and because expansins mediate expansion of isolated plant cell walls under such conditions, it has been considered that expansins may play a role in cell enlargement in both lower and higher plants (Cosgrove, 1996). To understand the mechanisms governing root development by comparing the expression patterns between extensin and expansin genes in soybean roots, we isolated two expansin cDNA clones, GmEXP1 and GmEXP2, from soybean roots. Sequence alignment of GmEXP1 and GmEXP2 with other expansins indicated that they show high homology to other expansins at the amino acid levels (supplemental data available at http://www.plantphysiol.org).
Many expansin genes have been isolated from a variety of plant species and it has been shown that they form a multigene family (Cosgrove, 1998). Li et al. (2002) have recently classified the expansins into the α-, β-, and γ-expansin subfamilies based on their phylogenetic relationship. Link and Cosgrove (1998) have also previously classified the α-expansin subfamily into four groups. Our phylogenetic analysis indicated that GmEXP1 belongs to the D group, which includes NtEXP3 (Link and Cosgrove, 1998), OsEXP1 (Cho and Kende, 1997b), and CsEXP2 (Shcherban et al., 1995), whereas GmEXP2 belongs to the A group, which includes AtEXP6 (Shcherban et al., 1995), PsEXP1 (Michael, 1996), and LeEXP18 (Reinhardt et al., 1998; see supplement data available at http://www.plantphysiol.org). The high sequence conservation among various expansins from monocotyledonous and dicotyledonous plants, such as rice, cucumber, Arabidopsis, pea, and soybean, may support several important points: (a) The expansin gene family has arisen by gene duplication before the evolutionary divergence of monocotyledonous and dicotyledonous plants some 150 million years ago; (b) expansin proteins have strict functional constraints that limit the structural modifications while possibly maintaining functions; and (c) their functions are important to the normal development and physiology of angiosperms (Shcherban et al., 1995).
Temporal and Spatial Expression of the GmEXP1 Gene during Root Development in Soybean
Although expansins constitute a multigene family in a variety of plant species, each expansin gene is differentially expressed in various parts of plants at different developmental stages. Nevertheless, expansin transcripts are most abundant in actively growing organs, such as leaf primordia in tomato (Fleming et al., 1997, 1999; Reinhardt et al., 1998), internodes in rice (Cho and Kende, 1997b), pollen in maize (Zea mays; Cosgrove et al., 1997) and soybean (Crowell, 1994), pistil in tobacco (Pezzotti et al., 2002), fruits in strawberry (Fragaria spp.; Civello et al., 1999) and tomato (Brummell et al., 1999a, 1999b), and coleoptiles in oat (Cosgrove and Li, 1993), indicating that they are involved in critical developmental processes. To date, however, most experiments with expansin genes have not clarified the roles of each expansin gene in plant organ development in sufficient detail.
Our results from RNA gel-blot analyses showed that the GmEXP1 gene is root-specific, whereas the GmEXP2 gene is expressed in various tissues in soybean (Fig. 2). The GmEXP1 gene was highly expressed in roots, especially when the primary roots were rapidly emerging and when they were forming the secondary roots (Fig. 3, A and B). Furthermore, in 5-d-old seedlings, in which the growth of the primary roots decelerated and the secondary roots were just emerging, its expression was predominant in the secondary roots at the developmental stage (Fig. 3C). These results strongly suggested that the GmEXP1 gene is involved in root elongation in soybean.
Further experiments have indicated that the GmEXP1 gene was mainly expressed in the regions of cell division and elongation of the primary and secondary roots (Fig. 4). In situ hybridization revealed that GmEXP1 transcripts were abundant at the root tip region of the primary and secondary roots (Figs. 5 and 6), confirming the results of RNA gel analyses. The cells expressing the GmEXP1 gene were generally located in the epidermis and underlying cell layers of the region of elongation of the primary root (Fig. 5). Furthermore, expression of the GmEXP1 gene was strongly up-regulated at the secondary root initials emerging from the primary root (Fig. 6). The expression patterns of the GmEXP1 gene imply that the GmEXP1 gene is root-specific and its root-specific expression is temporally and spatially regulated during root development in soybean.
Roles of the GmEXP1 Gene during Root Development in Soybean
As the cells in the root apical meristem divide, some of them differentiate and specify their own fates later on, whereas others remain undifferentiated. The cells to be differentiated are located in the region of cell elongation, which is only a few millimeters in length but cannot be sharply delimited from the region of cell division. Most of the increase in root length results from the cell elongation in the region.
After extensive studies, it has been proposed that expansins have rather diverse effects throughout the whole developmental process. For example, Cho and Cosgrove (2000) showed that expansins have versatile developmental roles that include control of organ size, morphology, and abscission. It has been also shown that local expression of expansin could induce the entire leaf development process (Pien et al., 2001). However, the role of expansins during root growth is still poorly understood despite much research effort. Nonetheless, a clue to accessing their function during root growth is the involvement of the proteins during reformations of the cell wall in the epidermal cells that play an important role in regulating elongation and maturation events of plants (Hasenstein and Evans, 1988). It is because as long as the epidermis of the root is extensible, the root is capable of growing; however, once cell elongation is complete, the primary cell wall is locked into shape (Carpita and Gibeaut, 1993).
We have previously shown that the SbHRGP3 gene of soybean is involved in the maturation event that leads not only to the cessation of cell elongation but also to the prevention of further elongation of soybean roots (Ahn et al., 1996, 1998). SbHRGP3 expression was specific to the epidermal cells of the region from which the secondary root was to be initiated in soybean and transgenic tobacco plants. This indicated that the SbHRGP3 gene has a special structural function not only in maturation but also in hardening of the epidermal cells to reduce the severe damage caused by the emerging secondary roots.
On the basis of the results obtained from this study, we suggest that GmEXP1 plays an important role in the soybean root elongation. This suggestion is based on the results of (a) the high levels of GmEXP1 expression in the epidermal cells and underlying cell layers in the region of elongation (Fig. 5), (b) its expression in emerging secondary root initials (Fig. 6), and (c) the accelerated growth caused by its ectopic expression in the transgenic plants (Figs. 7 and 8). The rapid root growth observed in the transgenic plants especially appears to provide a critical clue about the function of GmEXP1 in root development, because the plants showed elongated epidermal cells along the root axis (Table I) and enlarged cortex cells. We could not detect considerable difference in vascular tissues in the cross sections of the roots between the transgenic and wild-type plants; however, it is probable that the vascular tissues were also elongated to keep pace with the accelerated growth. We hence propose that overaccumulation of GmEXP1 proteins lead to cell elongation, which may result in accelerated growth of the roots of the transgenic plants. Nonetheless, although the roots overexpressing the GmEXP1 gene were much elongated, we still cannot rule out a possibility that the additional expansin in the roots is overriding any regulatable changes in expansin levels that might be involved in normal responses, because we used a heterologous system to study the soybean expansin gene.
Interestingly, the GmEXP1 overexpression differentially affected development of the transgenic plants. For instance, leaf palisade and spongy parenchyma cells of the overexpressing plants were affected and irregularly distributed, probably due to the overexpression (Fig. 7, A and B). However, xylem tissues in both petioles and roots of the transgenic plants were not significantly different from those of wild-type control plants (Fig. 7, G and H; data not shown), although xylem tissues were considerably thickened in the stems of the transgenic plants (Fig. 7, E and F). In contrast, numbers of pith cells and cortex cells are not significantly changed in the transgenic stem, implying that the GmEXP1 overexpression selectively affects plant development. These results support the contention that the different expansins may play different roles in various cell types during plant development, as inferred by the complexity of the expansin family (Li et al., 2002).
It has been also proposed that expansin activity is regulated during root elongation by various environmental stresses. For example, Wu et al. (1996) previously reported that maintenance of cell elongation in the apical region of the maize primary root at low water potentials was associated with an increase in expansin activity. They further characterized spatial patterns of expansin gene expression and found that two α-expansins (Exp1 and Exp5) and two β-expansins (ExpB2 and ExpB8) are expressed specifically in the growing region, whereas expression of β-expansin ExpB6 is shifted basipetally (Wu et al., 2001). In this study, we have shown that the transgenic plants overexpressing the GmEXP1 gene exhibited rapid root growth under both the neutral and acidic conditions (Fig. 8A), supporting the acid growth hypothesis involving the pH-dependent activation of expansins for root elongation (Cosgrove, 1998).
When the plants are grown in soil, their roots face the obstacle-touching stress ad infinitum and have to modify their development to bypass and to find appropriate space for further growth. Practically, under the obstacle-touching stimulus such as incubating in an inclined position, the Arabidopsis roots begin to bend to realign themselves with gravity and encounter the agar surface. They consequently perceive the obstacle-touching stimulus, because the roots are unable to penetrate agar of high concentration (Okada and Shimura, 1990, 1992). Our results have shown that the GmEXP1 overexpression caused rapid growth and insensitivity to the obstacle-touching stimulus in the transgenic plants (Fig. 8B). Therefore, it appears that GmEXP1 is confined to the region of elongation and may play an important role in overcoming the obstacle-touching stress when soybean plants grow under the field conditions.
In most cases, isolation of a loss-of-function allele of a specific gene from its host plant is the best way to study the developmental function of the gene; however, this approach has not yet been established in soybean. Therefore, in this study, the in vivo function of GmEXP1 was implemented by a gain-of-function study in transgenic plants. We also tried to overexpress antisense GmEXP1 in transgenic tobacco plants, but no apparent phenotype was observed (data not shown). Further investigation showed that expression levels of the most homologous expansin gene in tobacco, NtEXP3, were only slightly changed in the transgenic plants overexpressing antisense GmEXP1, indicating that tobacco plants are not an appropriate system for the study of antisense expression of soybean expansin genes. This suggests that the RNA-mediated interference approach to produce inheritable gene silencing in soybean would be an alternative way to examine the function of GmEXP1 gene in the future (Chuang and Meyerowitz, 2000; Kennerdell and Carthew, 2000).
In summary, we have isolated a root-specific expansin gene, GmEXP1, from soybean and compared its expression patterns with those of an extensin gene, SbHRGP3. It has been suggested that both proteins play distinct roles in root development. The primary root is elongated in the region of GmEXP1 expression, whereas it ceases its elongation and undergoes maturation in the region of SbHRGP3 expression. In the region of maturation in the primary root, SbHRGP3 reforms the cell walls in the secondary root initiation region to prevent severe damage from emerging secondary roots that burrow their way with increased extensibility due to the GmEXP1 expression. Further investigations including loss-of-function studies and the interaction of GmEXP1 and SbHRGP3 will shed light on the role of expansins and extensins during root development in soybean.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Soybean (Glycine max cv Paldal) and tobacco (Nicotiana tabacum cv Xanthi) were grown at 26°C under a 16-h-light/8-h-dark cycle. For DNA and RNA gel-blot analyses, soybean seeds were germinated on two sheets of moist paper (Whatman, Clifton, NJ) in petri dishes in darkness at 26°C for 2 d. After germination, soybean seedlings were transferred in a modified 0.5× Hoagland solution (Hoagland and Arnon, 1950) under a 16-h-light/8-h-dark cycle at 26°C. Roots of 20-d-old seedlings were used to isolate total RNA to analyze organ-specific expression patterns of expansin genes. For the temporal expression patterns of the expansin genes, total RNA was extracted from the roots of 1-, 2-, 3-, 4-, 5-, 7-, 9-, and 13-d-old seedlings. For the spatial expression patterns of the expansin genes, the primary roots were serially dissected along the root axis of 5-d-old seedlings, and the 5-cm-long secondary roots were serially dissected from of 9-d-old seedlings. For in situ hybridization, the roots from the 5-d-old seedlings were used.
Screening of a Soybean Root cDNA Library
Two degenerate primers were synthesized based on the amino acid sequences conserved in plant expansins: the forward primer named EXP1 [5′-NNGGATCCGA(T/C) GCNTCNGGNACNATGGG(T/C) GG(T/C) GCTG(T/C) G(G/T) TANGG-3′] and the reverse primer named EXP2 [5′-NNGGATCCTT(T/G)(G/C)(A/T)(C/T) TGCCA(G/A) TTNN(C/G) NCCCCA(A/G) TTNC(G/T)-3′]. The PCR was carried out using soybean genomic DNA as a template in a volume of 50 μL containing 1× PCR buffer, 200 μm of each dNTP, 1 μm of each primer, 1.5 mm MgCl2, and 2.5 units of Taq DNA polymerase (Bioneer, Daejon, Korea). After PCR, the amplified product was labeled by using the Prime-a-Gene system (Promega, Madison, WI) and used for screening a cDNA library constructed in the Uni-ZAP XR vector (Stratagene, La Jolla, CA) as described previously (Sambrook et al., 1989).
DNA and RNA Gel-Blot Analyses
Genomic DNA was isolated from soybean leaves and 10 μg of the genomic DNA was digested with EcoRV, HindIII, or NcoI. Blotting and hybridization procedures were performed under standard conditions (Sambrook et al., 1989). The GmEXP1 gene-specific probe was a 389-bp fragment containing the C-terminal coding region of 213 bp and the 3′-untranslated region of 176 bp that was generated by digestion of the GmEXP1 cDNA with HindIII and BamHI. The GmEXP2 gene-specific probe was a 350-bp fragment containing the 3′-untranslated region of the GmEXP2 cDNA obtained after digestion with DraI and HindIII.
Total RNAs were isolated from soybean leaves, roots, stems, and hypocotyls, and RNA gel-blot analysis was performed as described by Ahn et al. (1998). An equivalent amount (10 μg) of total RNA was electrophoresed in a 1% (w/v) agarose gel containing formaldehyde and transferred onto Hybond N+ membrane (Amersham Biosciences AB, Uppsala). For quantification, the membrane was analyzed with a bioimaging analyzer (BAS-1500, Fuji, Tokyo).
In Situ Hybridization
In situ hybridization experiments were performed as previously described (Glick and Thompson, 1993; Cho and Kende, 1998). Roots of 5-d-old seedlings were immediately immersed in the formaldehyde-acetic acid-fixation solution containing 50% (v/v) ethanol, 5% (v/v) acetic acid, and 3.7% (v/v) formaldehyde. The tissues were subsequently fixed and embedded in paraplast. The embedded tissues were sliced into 8-μm sections using a microtome (Leica Instruments GmbH, Wetzlar, Germany). The probe for in situ hybridization was the GmEXP1 gene-specific probe, as confirmed by DNA and RNA gel-blot analyses. DIG-labeling RNA probes (antisense and sense) were generated in a reaction mixture containing digoxigenin-UTP using SP6 or T7 polymerase depending on the orientation of the insert in the pSPT18 vector (Roche Diagnostics, Mannheim, Germany). For slide mounting, the color-reacted slides were dehydrated in the ethanol solution, dipped in xylene, and mounted with Permount (Fisher Scientific Co., Fair Lawn, NJ). Photographs were taken under bright field illumination using a microscope (Optiphot-2, Nikon, Tokyo).
Plant Transformation
The coding region (765 bp) of the GmEXP1 cDNA was amplified by PCR using the forward primer, EXPP1 (5′-ACCAAGCTTCAACCTCTCATCATTAGGC-3′) and the reverse primer, EXPP2 (5′-ACCAAGCTTGGAGTTGATGGGAATAATCA-3′). The PCR-amplified product was digested with HindIII and then inserted into the HindIII site of the pGA643 vector containing the cauliflower mosaic virus 35S promoter and NOS terminator (An et al., 1988). The resulting transgene was introduced into Agrobacterium tumefaciens cells according to the method of Holsters et al. (1978). A. tumefaciens-mediated tobacco plant transformation was performed as described (Horsch et al., 1985). Transformed shoots were selected on Murashige and Skoog basal medium supplemented with 200 mg L−1 kanamycin and 500 mg L−1 carbenicillin.
Transgenic Plant Assay under the Acidic and Obstacle-Touching Conditions
For the functional analysis of GmEXP1 in transgenic tobacco plants, transgenic tobacco seeds (T1and T2) were obtained by self-pollination. Seeds of transgenic tobacco and wild-type plants were sterilized with 10% (v/v) bleach solution for 10 min, washed several times with sterile water, and sown on 1.5% (w/v) agar plates containing 0.5× Murashige and Skoog basal medium (Invitrogen, Carlsbad, CA).
For investigation of the role of GmEXP1 in the acid growth of roots, after vernalization, transgenic seedlings in the T2 generation and wild-type seedlings on 1.5% (w/v) agar plates at pH 7.0 and pH 4.5 were incubated in a vertical position under a 16-h-light/8-h-dark cycle at 26°C. Their primary root lengths were measured at 17 d after germination.
To study the role of GmEXP1 in root growth under the obstacle-touching stress, transgenic seedlings in the T2 generation and wild-type seedlings were incubated on 1.5% (w/v) agar plates (pH 4.5) in a vertical position (90°), after cold treatment for 2 d in the dark. After 3 d, one-half of the plates containing both samples were inclined at 45°, and the rest were continually incubated in a vertical position under a 16-h-light/8-h-dark cycle at 26°C. Their primary root lengths were measured at 17 d after germination.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subjected to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
Supplementary Material
ACKNOWLEDGMENTS
We thank C.W. Jeong for his great assistance with the tissue sections and an anonymous reviewer for perceptive comments on the implications of the results.
Footnotes
This work was supported by the Korea Science and Engineering Foundation through the Plant Metabolism Research Center of Kyung Hee University and by the Crop Functional Genomics Center of the 21C Frontier Program of the Ministry of Science and Technology of Korea (grant to J.S.L.).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.009902.
LITERATURE CITED
- Aeschbacher RA, Schiefelbein JW, Benfey PN. The genetic and molecular basis of root development. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:25–45. [Google Scholar]
- Ahn JH, Choi Y, Kim S-G, Kwon YM, Choi YD, Lee JS. Expression of a soybean hydroxyproline-rich glycoprotein gene is correlated with maturation of roots. Plant Physiol. 1998;116:671–679. doi: 10.1104/pp.116.2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JH, Choi Y, Kwon YM, Kim S-G, Choi YD, Lee JS. A novel extensin gene encoding a hydroxyproline-rich glycoprotein requires sucrose for its would-inducible expression in transgenic plants. Plant Cell. 1996;8:1477–1490. doi: 10.1105/tpc.8.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Ebert PR, Mitra A, Ha SB. Binary vectors. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. pp. A31–A319. [Google Scholar]
- Baluska F, Volkmann D, Barlow PW. Specialized zones of development in roots: view from the cellular level. Plant Physiol. 1996;112:3–4. doi: 10.1104/pp.112.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P. Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell. 1999a;11:2203–2216. doi: 10.1105/tpc.11.11.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Dunsmuir P. Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Mol Biol. 1999b;39:161–169. doi: 10.1023/a:1006130018931. [DOI] [PubMed] [Google Scholar]
- Caderas D, Muster M, Vogler H, Mandel T, Rose JK, McQueen-Mason S, Kuhlemeier C. Limited correlation between expansin gene expression and elongation growth rate. Plant Physiol. 2000;123:1399–1414. doi: 10.1104/pp.123.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Cho H-T, Cosgrove DJ. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:9783–9788. doi: 10.1073/pnas.160276997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-T, Kende H. Expansins in deepwater rice internodes. Plant Physiol. 1997a;113:1137–1143. doi: 10.1104/pp.113.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-T, Kende H. Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell. 1997b;9:1661–1671. doi: 10.1105/tpc.9.9.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-T, Kende H. Tissue localization of expansins in deepwater rice. Plant J. 1998;15:805–812. doi: 10.1046/j.1365-313x.1998.00258.x. [DOI] [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civello PM, Powell ALT, Sabehat A, Bennett AB. An expansin gene expressed in ripening strawberry fruit. Plant Physiol. 1999;121:1273–1279. doi: 10.1104/pp.121.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Plant cell enlargement and the action of expansins. BioEssays. 1996;18:533–540. doi: 10.1002/bies.950180704. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Relaxation in a high-stress environment: the molecular bases of extensible cell walls and cell enlargement. Plant Cell. 1997;9:1031–1041. doi: 10.1105/tpc.9.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Cell wall loosening by expansins. Plant Physiol. 1998;118:333–339. doi: 10.1104/pp.118.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Enzymes and other agents that enhance cell wall extensibility. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:391–417. doi: 10.1146/annurev.arplant.50.1.391. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Expansive growth of plant cell walls. Plant Physiol Biochem. 2000a;38:109–124. doi: 10.1016/s0981-9428(00)00164-9. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000b;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM. Group I allergens of grass pollen as cell wall loosening agents. Proc Natl Acad Sci USA. 1997;94:6559–6564. doi: 10.1073/pnas.94.12.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ, Li Z-C. Role of expansin in cell enlargement of oat coleoptiles. Plant Physiol. 1993;103:1321–1328. doi: 10.1104/pp.103.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell DN. Cytokinin regulation of a soybean pollen allergen gene. Plant Mol Biol. 1994;25:829–835. doi: 10.1007/BF00028877. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organization of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Esau K. Anatomy of Seed Plants. Ed 2. New York: John Wiley and Sons; 1977. pp. 215–242. [Google Scholar]
- Fleming AJ, Caderas D, Wehrli E, McQueen-Mason S, Kuhlemeier C. Analysis of expansin-induced morphogenesis on the apical meristem of tomato. Planta. 1999;208:166–174. [Google Scholar]
- Fleming AJ, McQueen-Mason S, Mandel T, Kuhlemeier C. Induction of leaf primordia by the cell wall protein expansin. Science. 1997;276:1415–1418. [Google Scholar]
- Glick BR, Thompson JE. Methods in Plant Molecular Biology and Biotechnology. Boca Raton, FL: CRC Press; 1993. pp. 179–205. [Google Scholar]
- Hasenstein KH, Evans ML. Effects of cations on hormone transport in primary roots of Zea. Plant Physiol. 1988;86:890–894. doi: 10.1104/pp.86.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI. The water culture method for growing plants without soil. Calif Agric Exp Stn Circ. 1950;347:1–32. [Google Scholar]
- Holsters M, de Waele D, Depicker A, Messens E, van Montagu M, Schell J. Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet. 1978;163:181–187. doi: 10.1007/BF00267408. [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann N, Wallroth M, Eicholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Howell SH. Molecular Genetics of Plant Development. Cambridge, UK: Cambridge University Press; 1998. pp. 208–311. [Google Scholar]
- Jacobs T. Why do plant cells divide? Plant Cell. 1997;9:1021–1028. doi: 10.1105/tpc.9.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E, Cosgrove DJ. Expansins in growing tomato leaves. Plant J. 1995;8:795–802. doi: 10.1046/j.1365-313x.1995.8060795.x. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol. 2002;128:854–864. doi: 10.1104/pp.010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z-C, Durachko DM, Cosgrove DJ. An oat coleoptile wall protein that induces wall extension in vitro and that is antigenically related to a similar protein from cucumber hypocotyls. Planta. 1993;191:349–356. [Google Scholar]
- Link BM, Cosgrove DJ. Acid-growth response and α-expansin in suspension cultures of bright yellow 2 tobacco. Plant Physiol. 1998;118:907–916. doi: 10.1104/pp.118.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyndon RF. Plant Development: The Cellular Basis. London: Unwin Hymann; 1990. [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induces wall extension. Proc Natl Acad Sci USA. 1994;91:6574–6578. doi: 10.1073/pnas.91.14.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz EM. Genetic control of cell division patterns in developing plants. Cell. 1997;88:299–308. doi: 10.1016/s0092-8674(00)81868-1. [DOI] [PubMed] [Google Scholar]
- Michael AJ. A cDNA from pea petals with sequence similarity to pollen allergen, cytokinin induced and genetic tumor specific genes: identification of a new family of related sequences. Plant Mol Biol. 1996;30:219–224. doi: 10.1007/BF00017818. [DOI] [PubMed] [Google Scholar]
- Okada K, Shimura Y. Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science. 1990;250:274–276. doi: 10.1126/science.250.4978.274. [DOI] [PubMed] [Google Scholar]
- Okada K, Shimura Y. Aspects of recent developments in mutational studies of plant signaling pathways. Cell. 1992;70:369–372. doi: 10.1016/0092-8674(92)90159-a. [DOI] [PubMed] [Google Scholar]
- Pezzotti M, Feron R, Mariani C. Pollination modulates expression of the PPAL gene, a pistil-specific β-expansin. Plant Mol Biol. 2002;49:187–197. doi: 10.1023/a:1014962923278. [DOI] [PubMed] [Google Scholar]
- Pien S, Wyrzykowska J, McQueen-Mason S, Smart C, Fleming A. Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc Natl Acad Sci USA. 2001;98:11812–11817. doi: 10.1073/pnas.191380498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven PH, Evert RF, Eichhorn SE. Biology of Plants. 6th Ed. New York: Worth Publishers; 1999. [Google Scholar]
- Reinhardt D, Wittwer F, Mandel T, Kuhlemeier C. Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell. 1998;10:1427–1437. doi: 10.1105/tpc.10.9.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Cosgrove DJ, Albersheim P, Darvill AG, Bennett AB. Detection of expansin proteins and activity during tomato fruit ontogeny. Plant Physiol. 2000;123:1583–1592. doi: 10.1104/pp.123.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Lee HH, Bennett AB. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA. 1997;94:5955–5960. doi: 10.1073/pnas.94.11.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scheres B, McKhann HI, van den Berg C. Roots redefined: anatomical and genetic analysis of root development. Plant Physiol. 1996;111:959–964. doi: 10.1104/pp.111.4.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW, Masucci JD, Wang H. Building a root: the control of patterning and morphogenesis during root development. Plant Cell. 1997;9:1089–1098. doi: 10.1105/tpc.9.7.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason SJ, Shieh M, Cosgrove DJ. Molecular cloning and sequence analysis of expansins: a highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc Natl Acad Sci USA. 1995;92:9245–9249. doi: 10.1073/pnas.92.20.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varner JE, Lin L-S. Plant cell wall architecture. Cell. 1989;56:231–239. doi: 10.1016/0092-8674(89)90896-9. [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Martinez-Vilchez IM, Verbelen J-P, Miller JG, Fry SC. In vivo co-localization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell. 2000;12:1229–1237. doi: 10.1105/tpc.12.7.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sharp RE, Durachko DM, Cosgrove DJ. Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiol. 1996;111:765–772. doi: 10.1104/pp.111.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Thorne ET, Sharp RE, Cosgrove DJ. Modification of expansin transcript levels in the maize primary root at low water potentials. Plant Physiol. 2001;126:1471–1479. doi: 10.1104/pp.126.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.