Abstract
Comparative phenotypic analysis of pea (Pisum sativum) sym35 mutants and Lotus japonicus nin mutants suggested a similar function for the PsSym35 and LjNin genes in early stages of root nodule formation. Both the pea and L. japonicus mutants are non-nodulating but normal in their arbuscular mycorrhizal association. Both are characterized by excessive root hair curling in response to the bacterial microsymbiont, lack of infection thread initiation, and absence of cortical cell divisions. To investigate the molecular basis for the similarity, we cloned and sequenced the PsNin gene, taking advantage of sequence information from the previously cloned LjNin gene. An RFLP analysis on recombinant inbred lines mapped PsNin to the same chromosome arm as the PsSym35 locus and direct evidence demonstrating that PsNin is the PsSym35 gene was subsequently obtained by cosegregation analysis and sequencing of three independent Pssym35 mutant alleles. L. japonicus and pea root nodules develop through different organogenic pathways, so it was of interest to compare the expression of the two orthologous genes during nodule formation. Overall, a similar developmental regulation of the PsNin and LjNin genes was shown by the transcriptional activation in root nodules of L. japonicus and pea. In the indeterminate pea nodules, PsNin is highly expressed in the meristematic cells of zone I and in the cells of infection zone II, corroborating expression of LjNin in determinate nodule primordia. At the protein level, seven domains, including the putative DNA binding/dimerization RWP-RK motif and the PB1 heterodimerization domain, are conserved between the LjNIN and PsNIN proteins.
Legumes establish endosymbiosis with bacteria belonging to the genera Azorhizobium, Bradyrhizobium, Mesorhizobium, and Sinorhizobium a.o., collectively called rhizobia. The development of this symbiosis is a multistep process mediated by signal exchange between partners (Bladergroen and Spaink, 1998; Schultze and Kondorosi, 1998; Stougaard, 2000; Hirsch et al., 2001). Rhizobium spp. secretes lipochitin-oligosaccharide molecules triggering the compatible host to initiate development of specialized organs, root nodules, from already differentiated root cells (Downie and Walker, 1999). Afterward, the microsymbionts invade the nodule primordia, and intracellular compartments containing nitrogen-fixing endosymbionts, termed symbiosomes, are formed (Roth and Stacey, 1989). The infection process differs among legume species, and different legumes develop morphologically distinct nodule types. Two of these, the determinate and indeterminate nodules, have been described in detail. Determinate nodules are generally initiated by division of root cells in the outer cortex, but activity of the root nodule meristem will cease before the nodule becomes fully functional. Soybean (Glycine max), Lotus japonicus, and bean (Phaseolus vulgaris) follow this developmental pathway. Indeterminate nodules are founded by inner cortical cells, and the meristem remains active throughout the life time of the nodule giving rise to elongated root nodules, for example of pea (Pisum sativum), alfalfa (Medicago sativa), and clover. The meristem is located in the tip of indeterminate nodules, and the differentiation process is visible in nodule sections as a developmental zonation, with the youngest dividing cells in the nodule tip and the oldest senescent cells closest to the root.
Plant mutants incapable of forming nodules and mutants arrested during nodule development have been found in many legume species (Borisov et al., 2000; Harrison, 2000; Stougaard, 2001). Interestingly, some of the non-nodulating mutants were also unable to interact with mycorrhizal fungi, demonstrating that a set of “common genes” is required for initiating both bacterial symbiosis and endosymbiosis with vesicular arbuscular mycorrhizal fungi (Duc et al., 1989; Wegel et al., 1998; Bonfante et al., 2000; Stougaard, 2001). One of the legume plants traditionally used for gene mapping and investigations of the genetic basis of plant-microbe interactions is pea. As a result of this worldwide effort, more than 200 independent symbiotic pea mutant lines are known to date (for review, see Borisov et al., 2000). Complementation analysis involving around 100 symbiotic mutants has defined more than 40 symbiotic (Sym) loci (Borisov et al., 2000), but efficient methods for cloning and characterization of genetically defined loci still need to be developed in pea. Like other agriculturally important legumes (soybean and alfalfa) that for decades have been used in genetic analysis of symbiotic systems, pea has a large and complex genome and is difficult to transform. Such disadvantages make these species less suitable for molecular genetics and genomics (Udvardi, 2001). For this reason, L. japonicus has been adopted as model legume (Handberg and Stougaard, 1992). One of the ideas behind the model legume concept was the exploitation of synteny and microsynteny between genomes of traditional and model legume species to accelerate the isolation and comparative characterization of genes in traditional legumes. A variant of this approach has recently been used successfully to clone and characterize the PsSym19 gene (Endre et al., 2002; Stracke et al., 2002). An earlier illustration of the advantages of the model approach was the transposon tagging, cloning, and characterization of the L. japonicus Nin gene encoding a putative transcriptional regulator (Schauser et al., 1999). Using Nin as example, we have taken a direct approach to demonstrate how model legume knowledge can be effectively used for comparative studies on cultivated legumes. Focusing on a set of pea mutants with a phenotype comparable with L. japonicus nin mutants, we show that the pea Sym35 gene is the ortholog of LjNin. The role of the two genes encoding the same type of developmental regulator is compared between pea and L. japonicus, which develop indeterminate and determinate root nodules, respectively.
RESULTS
Comparative Phenotypic Description of Pea Mutants and L. japonicus nin Mutants
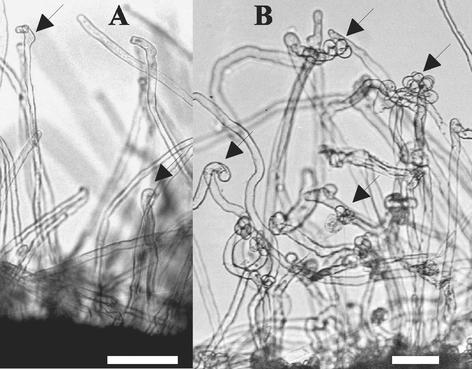
L. japonicus nin mutants are characterized by their excessive root hair deformation in response to Mesorhizobium loti, lack of infection thread formation, and lack of cortical cell divisions. In contrast, nin mutants have normal mycorrhizal interaction, suggesting a function of the Nin gene downstream of the “common genes” required for both rhizobial and mycorrhizal invasion (Schauser et al., 1999; Stougaard, 2001). In the collection of well-characterized pea symbiotic mutants, similar nodulation and mycorrhization phenotypes were observed in Pssym7, Pssym14, and Pssym35 mutants (Tsyganov et al., 1999; 2002). However, the excessive root hair curling response, as observed on Ljnin mutants, was only observed on Pssym35 mutants but not on Pssym7 or Pssym14 mutants, and further characterization suggested that the phenotype of Pssym35 mutants was identical to the Ljnin phenotype (Fig. 1). Three independent sym35 mutants (lines SGENod−-1, SGENod−-3 (Tsyganov et al., 1994, 1999), and RisNod8 (Engvild, 1987) matched the phenotype of Ljnin mutants because they were all (a) blocked in infection thread initiation, (b) characterized by absence of cortical cell divisions, (c) displaying “excessive” root hair curling, and (d) colonized by arbuscular mycorrhiza (Tsyganov et al., 1999, 2002).
Figure 1.
Root hair curling phenotype of wild-type SGE plant (A) and an SGENod−-1 (sym35) mutant (B). Both were inoculated with Rhizobium leguminosarum bv viciae, and root hairs were photographed 23 d after inoculation. Arrows point at curled root hairs; bar = 0.1 mm.
Cosegregation of PsNin and Pssym35
The phenotypic comparison suggested that the PsSym35 locus could be identical to the PsNin gene, and this hypothesis was first tested by genetic mapping and cosegregation analysis. A 2.5-kb fragment of the PsNin gene was isolated using degenerate primers designed from alignment and identification of conserved nucleotide sequences between LjNin and an Arabidopsis Nin-like gene. RFLP analysis in the parent lines, JI281 and JI388, of a pea recombinant inbred mapping population identified an EcoRV RFLP polymorphism. Subsequent mapping in the population placed the PsNin RFLP on the top of the pea linkage group I about 1 cM from the marker C2/2++− marker (Hall et al., 1997). In parallel, the linkage of sym35 and several classical morphology markers of the pea genetic map was tested in a sym35 (SGENod−-3) × NGB1238 mapping population. This analysis showed a weak linkage between sym35 and the d marker from linkage group I locating sym35 on the same chromosome arm as PsNin (sym35-d, linkage 37.5% ± 4.67%, P(0.5) < 0.05). The other possible candidates sym7 and sym14 map to pea linkage groups III and II, respectively (Weeden et al., 1998). Hence, mapping confirmed PsSym35 as the most likely candidate for PsNin and placed PsNin at the end of chromosome I reminiscent of the position of LjNin on the L. japonicus map (Hayashi et al., 2001; Pedrosa et al., 2002; Sandal et al., 2002).
To perform a cosegregation analysis between PsNin and Pssym35, a PCR strategy based on nucleotide polymorphisms within the PsNin gene of the parental lines NGB1238 and sym35 (SGENod−-3) was chosen. On the basis of the DNA sequence of the 2.5-kb PsNin fragment isolated from sym35 and NGB1238, respectively, mapping primers specific for either one or the other parent were designed. DNA of individual wild-type and mutant F3 plants of the sym35 × NGB1238 mapping population was subjected to PCR amplification using the allele-specific primers described above. Analysis of 121 plants homozygous for the Sym35 wild-type allele and 149 plants homozygous for the mutant sym35 allele in F3 showed 100% cosegregation of Sym35 and PsNin. Absence of recombination in 540 meiotic events maps sym35 less than 0.19 cM from PsNin. This result made it likely that PsSym35 is PsNin and encouraged us to isolate and sequence the complete PsNin gene and to characterize the alleles of PsNin in the three sym35 mutants.
Primary Structure of the PsNin Gene
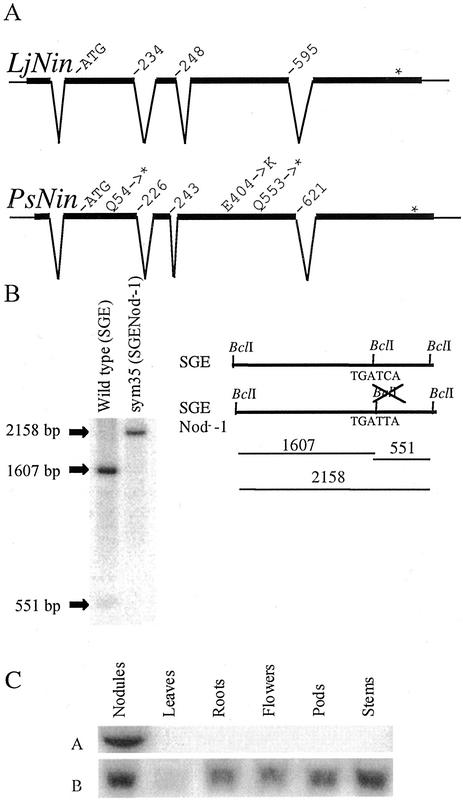
The PsNin gene was isolated from a genomic λ-library of the pea cv Alaska and an 8-kb region including approximately 3 kb of the promoter was sequenced. As a first step in the analysis of sym35 mutant alleles present in pea cv SGE or pea cv Finale genetic backgrounds, the pea cv Alaska PsNin sequence information was used to identify PCR amplification primers, and the PsNin gene from the pea line SGE and Finale variety was sequenced from the obtained PCR products. As a result, the wild-type PsNin sequence was determined in three pea varieties. The corresponding cDNA was isolated from a root hair enriched library of pea cv Finale. Comparison with the L. japonicus Nin cDNA and the pea genomic sequence showed that the longest 2,033-bp cDNA was incomplete. The 5′ end of PsNin was therefore isolated by 5′-RACE, and a full-length cDNA of 3 kb was assembled. Comparison of the PsNin gene sequence and the full-length cDNA showed that the exon/intron structure is conserved between L. japonicus and pea Nin genes (Fig. 2A). Even the length of the introns is very similar. At the nucleotide level, there is an overall 60% identity between the coding regions of the L. japonicus and pea Nin genes, whereas large blocks of several hundred nucleotides are more than 80% identical. The LjNIN and PsNIN proteins have 55% identical amino acids.
Figure 2.
The intron-exon structure of LjNin and PsNin genes is conserved. A, The sequences of Sym35 from pea cv Finale and L. japonicus were compared with their respective cDNAs and aligned. Apart from short stretches in the promoter regions (Fig. 5), the genomic sequences show little or no similarity outside of the exons. Amino acid positions at the exon-intron boundaries and the changes in sym35 mutant alleles are indicated. B, Southern hybridization visualizing the RFLP generated by mutation of a BclI restriction site in the sym35 SGENod−-1 allele. Positions of the BclI sites in the mutant and wild-type alleles and the fragments generated by BclI digestion of genomic DNA are shown in the schematic drawing. The hybridization probe used covers 2 kb of the coding sequence. C, Northern analysis of PsNin expression in various pea organs. A visualizes the hybridization with the Sym35-specific probe. B shows the control hybridization with ubiquitin.
Identification of Mutations in Three Independent Psnin/sym35 Mutants
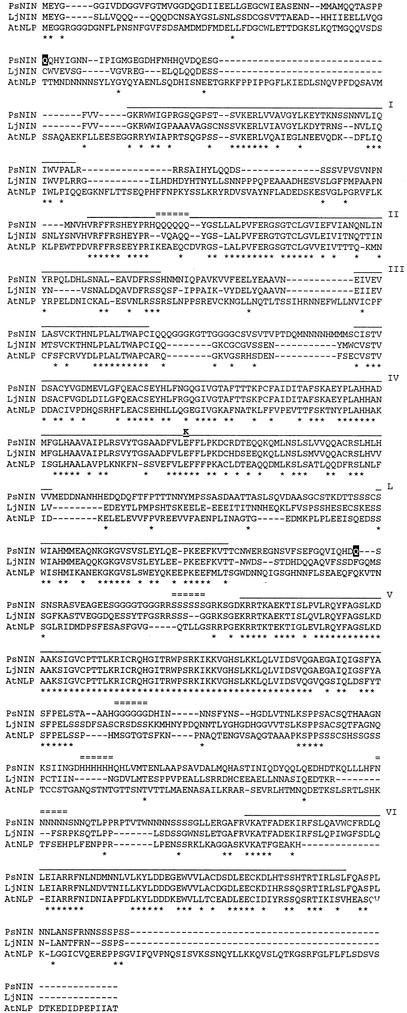
To verify that pea Sym35 is PsNin, a region of approximately 4 kb covering all exons and introns of PsNin was amplified by PCR and sequenced from the three allelic sym35 mutants of pea SGENod−-1, SGENod−-3 (Tsyganov et al., 1999), and RisNod8 (Engvild, 1987). In all three mutants, single-nucleotide substitutions were identified when compared with the PsNin gene sequence from the respective pea line SGE or cv Finale wild-type parents: SGENod−-1 (sym35) has a C to T transition in position 1,657 of the predicted coding sequence. This creates a stop codon (CAG to TAG) after D552. In addition, this point mutation destroys a BclI restriction site creating an RFLP shown in Figure 2B. The mutant allele SGENod−-3 (sym35) has a C to T transition in position 160 of the predicted coding sequence. This creates a stop codon (CAA to TAA) after P53, and finally RisNod8 (sym35) has a G to A change (GAG to AAG) in position 1,210 of the predicted coding sequence. This causes an amino acid substitution from E to K in position 404 of the protein. E404 is embedded in domain IV (Fig. 3) and is strictly conserved among all NIN-like proteins (NLPs; data not shown).
Figure 3.
Identification of conserved domains in LjNIN and PsNIN. The translation products of LjNin and PsNin cDNAs are aligned with the most homologous NLP from Arabidopsis using ClustalX. For assignment of protein domains, an alignment including all nine NLPs from Arabidopsis was carried out, but only the sequence of the most homologous NLP from Arabidopsis is shown in the figure. Six regions of high conservation between all 11 proteins are shown (domains I–VI) together with one region (L) conserved between LjNIN, PsNIN, and the most homologous NLP from Arabidopsis. Region V is the most conserved region and surrounds the putative DNA binding and dimerization, RWP-RK, motif. Region VI has similarity to the PB1 heterodimerization domain conserved in animals, fungi, and plants. Domains I to VI and L are overlined and identical amino acids marked by asterisks. Positions of stop codons (aa in black shadow) or amino acid changes caused by the three sym35 mutations are indicated in the PsNIN sequence. The small tracts of repeated amino acids are marked by double lines.
Domains of the PsNIN Protein
The open reading frame of the pea cDNA encodes a conceptual protein of 922 amino acids (101 kD) compared with the LjNIN of 878 amino acids (Fig. 3). Conserved domains shared by all known NLPs were identified by aligning LjNIN and PsNIN to the nine NLPs identified in Arabidopsis (L. Schauser, W. Wieloch, and J. Stougaard, unpublished data). Six regions of high conservation were identified (domains I–VI). At present, no function can be suggested for domains I through III. Domain IV contains the hydrophobic stretches suggested to be either membrane-spanning regions or hydrophobic pockets (Schauser et al., 1999). Domain V is the most conserved region and makes up the previously identified RWP-RK region, suggested to serve in dimerization and DNA binding in this family of putative transcriptional regulators (Schauser et al., 1999). Domain VI has similarity to the PB1 domain, a motif conserved in animals, fungi, and plants (Ponting et al., 2002). The tertiary structure of the PB1 domain has been determined and was shown to belong to the ubiquitin-like β-grasp fold-containing proteins. This domain is present in many eukaryotic cytoplasmic signaling proteins for example RasGTP-binding proteins. The function of this domain is the selective formation of PB1 domain heterodimers (Ito et al., 2001) In addition to these widely conserved domains, another domain (L) is conserved among LjNIN, PsNIN, and the Arabidopsis NLP translated from At4g35270. Altogether, this uneven distribution of conserved amino acids suggests a modular domain structure of the NIN proteins.
Both the LjNIN and PsNIN proteins contain a hexa-Gln stretch, and although the position is not conserved, their occurrence in both is puzzling and may have functional significance. Additional accumulation of short tracts of particular amino acids is a peculiar feature observed in the pea NIN protein. Among the small tracts of Gln, His, Gly, Asn, and Ser (Fig. 3), only the hexa-His and hexa-Asn tracts are encoded by a triplet repeat reminiscent of the genetic expansion of poly-Gln repeats associated with several neurodegenerative disorders (Richards et al., 1992). The functional significance, if any, of these repeats is unclear but none of them are conserved in LjNIN or the other NLPs.
Analysis of Expression of PsNin
The similarity of the phenotype between Pssym35 and Ljnin mutants suggests that the orthologous genes are active with a similar temporal and spatial expression pattern. To determine the expression of the PsNin gene in various organs, a northern analysis was performed on RNA extracted from roots, leaves, nodules, flowers, pods, and stems. As seen in Figure 2C, expression was detected only in nodules. This expression pattern is comparable with the previously determined expression pattern of the LjNin gene where steady-state mRNA was only detected in nodules in northern hybridizations (Schauser et al., 1999).
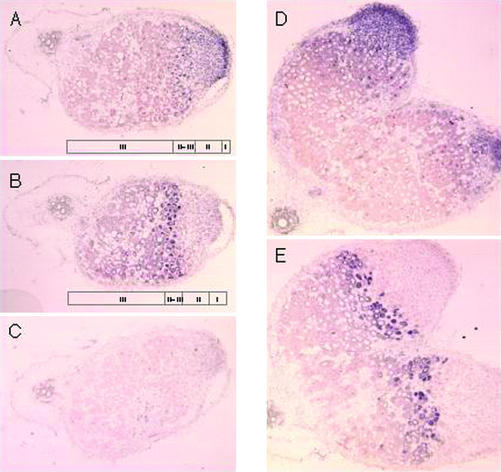
L. japonicus and pea develop determinate and indeterminate root nodules, respectively. It was therefore of interest to investigate if there were major differences in the cellular accumulation of PsNin transcripts in the pea nodules. In L. japonicus, Nin is expressed early in the nodule primordium (Schauser et al., 1999), and to determine the cellular expression pattern in pea, in situ hybridization was performed on sections of nodules harvested 4 weeks after inoculation with R. leguminosarum bv viciae. Figure 4, A and D, shows that PsNin is expressed in the meristematic cells (zone I) and cells of the infection zone (zone II). Cells of interzone (II–III) mark the boundary where transcripts were detectable, and in the nitrogen fixation zone (III), transcripts were not detected. These cells are either not expressing PsNin, or expression is below the level of detection for the in situ hybridization technique. Expression of the bacterial subunit of nitrogenase nifH (Fig. 4, B and E) is a diagnostic feature of cells in interzone (II–III; Yang et al., 1991; Kawashima et al., 2001). The PsNin expression in zone I and II was comparable with the expression of LjNin detected in the dividing cells and the later stages of nodule primordia in the determinate L. japonicus nodules (Schauser et al., 1999). The apparent down-regulation of expression in interzone II to III and zone III differed from the continued presence of LjNin transcripts in cells of the central zone of fully developed determinate nodules.
Figure 4.
In situ localization of PsNin transcripts in longitudinal sections of pea nodules. Nodules were harvested 4 weeks after inoculation with R. leguminosarum bv viciae, sectioned, and hybridized with digoxigenin-labeled RNA probes. Hybridizing transcripts are visualized as purple color. A, PsNin antisense probe. B, Bacterial nifH antisense probe. C, PsNin sense probe. D, PsNin antisense probe on bifurcated nodule. E, Bacterial nifH antisense on bifurcated nodule. nifH was used to define interzone II to III and nitrogen fixation zone (III) in the nodules.
The expression pattern for the two genes predicts that distinct DNA regulatory elements could be present in both the PsNin and LjNin promoter regions. A ClustalX alignment identifies several blocks of conserved sequence (Fig. 5). Assignment of functional significance to these sequences would require detailed deletion analysis and site-specific mutagenesis. However, the sequence content of two adjacent 14- and 12-nucleotide blocks located between position −384 and −352 of the LjNin promoter and between −385 and −352 of the PsNin promoter attracts attention. The proximal 14-nucleotide-long conserved sequence is a perfect match to the 3′ half (TTGTCTCTT) of the extended organ-specific element (AAAGATNNTTGTCTCTT) first identified in the soybean and Sesbania rostrata leghemoglobin promoters and subsequently found in other nodule expressed genes (Stougaard et al., 1987; Ramlov et al., 1993; Szczyglowski et al., 1994). Interestingly, the same sequence encompasses a TGTCTC sequence shown in Arabidopsis to be an auxin-responsive element binding the ARF1 transcription factor (Ulmasov et al., 1997). The 12-nucleotide conserved sequence is AT rich, and although the sequence itself is different, the position is reminiscent of the AT-rich binding sites for the NAT2 trans-factor located immediately 5′ to the organ-specific element of the soybean and S. rostrata leghemoglobin promoters (Laursen et al., 1994).
Figure 5.
Identification of putative conserved regulatory sequences in promoter regions of LjNin and PsNin. A region of 472 nucleotides upstream of the transcript start of LjNin was compared with the 486 nucleotides upstream of the transcript start of PsNin. A conserved block, indicated with a line, contains the 3′ half of the two-motif nodulin consensus AAAGAT-TTGTCTCTT (Stougaard et al., 1987; Ramlov et al., 1993; Szczyglowski et al., 1994) overlapping a sequence identical to auxin-responsive element TGTCTC (Ulmasov et al., 1997). ●, Transcription start site for LjNin and PsNin; ○, a minor transcription start in LjNin.
DISCUSSION
We have identified and characterized the PsNin gene and shown that the PsSym35 locus is PsNin. A simple sequence alignment between the L. japonicus Nin gene and an Arabidopsis Nin-like gene present in the databases allowed primer design and PCR amplification of the pea Nin ortholog. The PsNin gene structure and the position of mutations in the three available sym35 alleles were subsequently determined. This exemplifies how comparative phenotypic analysis and mapping makes it possible to transfer the achievements of molecular genetics from model legumes into agriculturally important crop species such as pea. Ongoing experiments are taking this one step further using the L. japonicus transformation system. Detailed characterization of the PsSym35 gene and functional analysis of conserved protein domains is approached by complementation of Ljnin mutants. With the rapidly accumulating expressed sequence tag and genome sequences from members of the legume family, this approach will be even simpler in the future. Candidate genes can be isolated, and exploiting the recently released L. japonicus genome sequences mapping approximately 1,300 genes along the chromosomes (Nakamuro et al., 2002), new gene-specific legume markers can be developed to start saturating the crop legume maps. Such markers can also be used to determine the level of synteny in the legume family. Marker-assisted breeding in legumes is bound to benefit from this development of markers, genome, and map information achieved from model legumes.
In the course of characterizing the PsNin gene and identifying the sequence differences in the three Pssym35 mutant alleles, the wild-type PsNin gene was sequenced from three varieties of pea. Alignment of the sequences demonstrates a low level of sequence polymorphism between the varieties. Pea cvs Alaska and Finale are very similar with only 18 single-nucleotide polymorphisms and four triplet indels in 3.9 kb, whereas the pea SGE line appears to be more distant with 33 single-nucleotide polymorphisms and three triplet indels of three, six, and 15 nucleotides, respectively, toward pea cv Alaska. This presents a small glimpse of the genetic variation in the European breeding stocks.
This comparative study between L. japonicus and pea led to the cloning of a symbiotic gene first identified after chemical mutagenesis and description of three mutant alleles. All three mutants of Pssym35 have the same phenotype, although the mutations are quite different. The allele in SGENod−-3 creates a stop codon and predicts that only a short protein of 53 amino acids without the RWP-RK domain and the PB1 dimerization domain is synthesized. This would most likely be a null mutation. SGENod−-1 has a stop codon after D552 resulting in a protein without the RWP-RK domain. The amino acid change in RisNod8 exchanges an acidic E for a basic K residue in an 8-amino acid motif conserved between all NLPs presently in the databases. This motif lies embedded in domain IV (Fig. 3). Apart from characterizing the allele, such missense mutations will eventually contribute to the understanding of NIN protein function. Five domains were found to be conserved between PsNIN and LjNIN. Four of the domains (I, II, III and IV) appear to be family “specific,” present only in NLPs, whereas the RWP-RK (V) and PB1 (VI) domains are conserved in other proteins (L. Schauser, W. Wieloch, and J. Stougaard, unpublished data). Interestingly, one additional domain (L) was found to be shared between LjNIN, PsNIN, and the most closely related Arabidopsis protein.
In L. japonicus, Nin expression was detectable only in nodules by northern hybridization, whereas more sensitive RNase protection assays were necessary to detect expression in other organs. Analyzed by northern hybridization, the expression pattern of PsNin appears to be comparable. Interestingly, the PsNin cDNA was found in a root hair-enriched library, indicating that PsNin is already expressed in the root hairs of pea. In 4-week-old pea nodules, the expression of PsNin was detected in cells of the meristem (zone I) and the infection zone (II). Across the interzone (II–III) and in the nitrogen fixation zone (III), transcripts were no longer detected. This expression pattern is in accordance with the mutant phenotype that suggests a function for PsNin in nodule inception and infection thread formation downstream of lipochitin-oligosaccharide signal perception. Continued expression in the nodule meristem and the infection zone of indeterminate nodules indicate that PsNIN is necessary for these developmental stages also after the onset of the organogenic process. In determinate L. japonicus nodules, expression of Nin was detected in the nodule primordia, central tissues, parenchyma, and vascular tissues in the mature nodules (Schauser et al., 1999). In the indeterminate pea nodule, expression was not detected in the more differentiated cell types, and although there might be differences in the sensitivity of the in situ hybridization techniques used, this appears to be a difference between determinate and indeterminate nodules. The overlapping expression pattern of the LjNin and PsNin genes is to some extent reflected in the sequences of their promoter regions. A promoter sequence identical to the 3′ half of the organ-specific element of the late nodulin leghemoglobin is conserved in both PsNin and LjNin. This may constitute a DNA regulatory element mediating Nin gene expression during nodulation. In addition, an auxin regulatory element appears to be embedded in this sequence. The involvement of auxin in initiating root nodule organogenesis (Mathesius et al., 1998) has been a long-standing hypothesis, and interestingly, the LjNin gene-promoter appears to respond to auxin addition in certain cell types (Y. Umehara and J. Stougaard, unpublished data). Together, these results demonstrate a comparable function for the PsNIN and LjNIN regulatory proteins and constitute another example of conservation of components in the development of determinate and indeterminate root nodules. Following this line of investigation in future comparative genomics, the numerous classical pea mutants and the well-established pea physiology will complement the functional analysis of model legume genes.
MATERIALS AND METHODS
Plant Material
The following pea (Pisum sativum) lines were used in the study: three allelic but independently obtained non-nodulating pea mutants SGENod−-1 (sym35) and SGENod−-3 (sym35; Tsyganov et al., 1994, 1999) and RisNod8 (sym35; Engvild, 1987; gene symbol was assigned according to Drs. G. Duc and M. Sagan [personal communication]) as well as the initial wild-type lines pea SGE line (Kosterin and Rozov, 1993) and pea cv Finale (Engvild, 1987). Also, the multiply marked genetic line NGB1238 (catalog no. of Nordic GenBank) or JI73 (catalog no. of Pisum Genetic Stocks Collection, John Innes Centre) was used in the experiments. Recombinant inbred lines from a cross between the lines JI281 and JI399 were used for mapping (Hall et al., 1997).
Bacterial Strains
For nodulation tests, the commercial, symbiotically effective strain CIAM 1026 (Collection of All-Russia Research Institute for Agricultural Microbiology) of Rhizobium leguminosarum bv viciae was used as the inoculant. For expression analysis, R. leguminosarum bv viciae R418 strain (kindly provided by Herman Spaink) was used as the inoculant.
Isolation of PsNin
A number of Arabidopsis homologs of LjNin can be found in the databases. Alignment between LjNin and an Arabidopsis homolog (F23E12.170; accession no. T06130) was used to design degenerate primers based on conserved regions. Combinations of the forward primers 5′-GCCCTTCCTGTYTTCGAAAGAGG-3′ and the two reverse primers 5′-GGCAAKCTTTTGGAATGAAAAACTCC-3′ and 5′-AGGCTTCCTGCAAAGTAYTGTC-3′ gave PCR products in the expected size ranges from the pea lines JI399 and JI281. The PCR products were cloned with the Topo TA cloning kit (Invitrogen, Carlsbad, CA) and sequenced. The overlapping sequences representing a pea ortholog showed high similarity to LjNin.
Mapping Experiments and Cosegregation Analysis
To carry out the initial mapping of the Sym35 gene, the mutant SGENod−-3 (sym35) was crossed to the line NGB1238 (wb, b, k, s, r, tl, gp, d, le, Fs, and Ust) and F1 plants were grown under greenhouse conditions with full mineral nutrition to obtain the seeds for F2 progeny. The F2 generation was analyzed for the segregating morphological markers, and the seeds for the F3 generation were collected from individual F2 plants. The symbiotic phenotype (presence/absence of nodules and signs of nitrogen starvation) of the plants in the F3 generation (grown in sand without combined nitrogen [Borisov et al., 1997]) was analyzed to reveal homozygous (Sym35/Sym35 or sym35/sym35), and heterozygous (Sym35/sym35) plants in the F2. Data on segregation were processed to position PsNin on the pea genetic map with the use of computer programs “Plant” and “Cross” developed by Serge M. Rozov (Institute of Cytology and Genetics, Siberian Branch of Russian Academy of Sciences, Novosibirsk, Russia).
For cosegregation analysis, the pea mutant SGENod−-3 (sym35) was crossed with the line NGB1238, and F2 plants were obtained. Homozygous Sym35/Sym35 and sym35/sym35 F2 plants were identified by phenotypic analysis of the F3 offspring. In the F3, only wild-type plants descending from homozygous F2 plants were used for cosegregation analysis together with sym35/sym35 mutant plants. Plants of the F3 population were grown under greenhouse conditions with full mineral nutrition. Two leaves from each plant were collected for DNA isolation, and the genotype of individual plants was determined using the parent-specific primers: NGB1238fw, 5′-GAAAGAGGAAGCGGACTTGT-3′; NGB1238rev, 5′-TGGTCTTCTCCGCCTTGG-3′; SN-3fw, 5′-GTGTCATTGATGTTGTTATCGCA-3′; and SN-3rev, 5′-GATGATGACCTGCGTCCACCA-3′.
Gene Libraries
Three pea gene libraries were used in the experiments: a nodule cDNA library (λ gt11 vector, pea cv Finale), a cDNA library enriched for root hairs (λ Zap II vector, pea cv Finale), and a genomic library (λ Dash II vector, pea cv Alaska).
Using a probe covering an approximately 2-kb fragment of the pea Nin gene, one million clones of each library were screened and processed according to the Stratagene (La Jolla, CA) protocols for λ-phage custom libraries.
DNA and RNA Techniques and Conditions of PCR
Pea genomic DNA was prepared essentially according to Saghai-Maroof et al. (1984). Total RNAs from different pea tissues were prepared using Trizol (Invitrogen) followed by LiCl precipitation or CsCl centrifugation and used for northern analysis and 5′-RACE. Isolation of phage λ-DNA and subcloning of the insert into plasmid vector (pBluescript SK±) were carried out using standard procedures. DNA sequences were produced using the Thermo Sequenase Dye Terminator Cycle Sequencing kit (Amersham Biosciences AB, Uppsala) and analyzed on an ABI prism 310 Genetic analyzer (Applied Biosystems, Foster City, CA). To get full-length cDNA, a combination of RT-PCR and 5′-RACE was performed with nodule RNA (cv Finale) using the SMART RACE kit according to supplier's instructions (BD Biosciences Clontech, Palo Alto, CA). Primers for RT-PCR: reverse primer, 5′-GAATGCTGTAATGTCGATTGCG-3′; forward primer, 5′-GCGGTGTATTTGGGACCATGG-3′. Primer for DNA synthesis for RT-PCR: 5′-CAGCTGCAGAGCCAGTGTAG-3′. Primers for 5′-RACE: 5′-ACCACTTTCTTGATCAACTTG-3′ and 5′-GTTGGATTGAAGCTAGTGAGAA-3′. Southern blot, northern blot, and hybridization was performed as described (Sambrook et al., 1989) using a 2-kb probe produced by PCR amplification using the primers 5′-CTAAGGAGGAGATCGGCAATTCA-3′ and 5′-GAACAGAATCTATCACCAGTTG-3′ and final washing at 0.3 × SSC and 0.1% (w/v) SDS at 65°C. The probe covers the coding sequence between positions 503 and 2,084.
In Situ Hybridization and Microscopic Analysis
In situ hybridization was performed as described by Kouchi and Hata (1993). RNA probes covering approximately 1 kb of PsNin (between positions 213 and 1,662 of the cDNA) were prepared with digoxigenin-11-UTP (Roche Diagnostics, Basel). Hybridization signals were detected by antidigoxigenin-alkaline phosphatase conjugate with nitroblue tetrazolium salt and 5-bromo-4-chloro-3-indolyl phosphate toluidinium salt (Roche Diagnostics). Longitudinal sections (8 μm) of the nodules were hybridized. nifH of R. leguminosarum bv vicea (kindly provided by Dr. H. Kouchi) was used for reference (Yang et al., 1991).
ACCESSION NUMBERS
The accession numbers from this study are as follows: PsNin cv Alaska, AJ493063; PsNin cv Finale, AJ493064; PsNin line SGE, AJ493065; PsNin mRNA cv Finale, AJ493066; and LjNin promoter region, AJ493067.
ACKNOWLEDGMENTS
We are very grateful to L.E. Dvoryaninova for her excellent technical assistance. We thank Henk Franssen for making the pea libraries available.
Footnotes
This study was supported by the Russian Foundation for Basic Research (grant nos. 01–04–49643 and 01–04–48580) and by the European INTAS program (grant no. 2322). A.Y.B.'s stay at Aarhus University was supported by the European Molecular Biology Organization (short-term fellowship no. ASTF 9556) and N.S.'s stay at the John Innes was supported by The Danish Agricultural and Veterinary Research Council.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.016071.
LITERATURE CITED
- Bladergroen MR, Spaink HP. Genes and signal molecules involved in the Rhizobia-Leguminosae symbiosis. Curr Opin Plant Biol. 1998;1:353–359. doi: 10.1016/1369-5266(88)80059-1. [DOI] [PubMed] [Google Scholar]
- Bonfante P, Genre A, Faccio A, Martini I, Schauser L, Stougaard J, Webb J, Parniske M. The Lotus japonicus LjSym4 gene is required for the successful symbiotic infection of root epidermal cells. Mol Plant-Microbe Interact. 2000;13:1109–1120. doi: 10.1094/MPMI.2000.13.10.1109. [DOI] [PubMed] [Google Scholar]
- Borisov AY, Barmicheva EM, Jacobi LM, Tsyganov VE, Voroshilova VA, Tikhonovich IA. Pea (Pisum sativum L.) mendelian genes controlling development of nitrogen-fixing nodules and arbuscular mycorrhiza. Czech J Genet Plant Breed. 2000;36:106–110. [Google Scholar]
- Borisov AY, Rozov SM, Tsyganov VE, Morzhina EV, Lebsky VK, Tikhonovich IA. Sequential functioning of Sym-13 and Sym-31, two genes affecting symbisome development in root nodules of pea (Pisum sativum L.) Mol Gen Genet. 1997;254:592–598. doi: 10.1007/s004380050456. [DOI] [PubMed] [Google Scholar]
- Downie JA, Walker SA. Plant responses to nodulation factors. Curr Opin Plant Biol. 1999;2:483–489. doi: 10.1016/s1369-5266(99)00018-7. [DOI] [PubMed] [Google Scholar]
- Duc G, Trouvelot A, Gianinazzi-Pearson V, Gianinazzi S. First report of non-mycorrhizal plant mutants (Myc-) obtained in pea (Pisum sativum L.) and Fababean (Vicia faba L.) Plant Sci. 1989;60:215–222. [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kaló P, Kiss GB. A receptor kinase gene regulating symbiotic nodule development. Nature. 2002;417:962–966. doi: 10.1038/nature00842. [DOI] [PubMed] [Google Scholar]
- Engvild Nodulation and nitrogen fixation mutants of pea (Pisum sativum) Theor Appl Genet. 1987;74:711–713. doi: 10.1007/BF00247546. [DOI] [PubMed] [Google Scholar]
- Hall KJ, Parker JS, Ellis THN, Turner L, Knox MR, Hofer JMI, Lu J, Ferrandiz C, Hunter PJ, Taylor JD et al. The relationship between genetic and cytogenetic maps of pea: II. Physical maps of linkage mapping populations. Genome. 1997;40:755–769. doi: 10.1139/g97-798. [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J. Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 1992;2:487–496. [Google Scholar]
- Harrison M. Molecular genetics of model legumes. Trends Plant Sci. 2000;5:414–415. doi: 10.1016/s1360-1385(00)01748-9. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Miyahara A, Sato S, Kato T, Yoshikawa M, Taketa M, Hayashi M, Pedrosa A, Onda R, Imaizumi-Anraku H et al. Construction of a genetic linkage map of the model legume Lotus japonicus using an intraspecific F2 population. DNA Res. 2001;8:301–310. doi: 10.1093/dnares/8.6.301. [DOI] [PubMed] [Google Scholar]
- Hirsch AM, Lum MR, Downie JA. What makes the rhizobia-legume symbiosis so special? Plant Physiol. 2001;127:1484–1492. [PMC free article] [PubMed] [Google Scholar]
- Ito M, Matusui Y, Ago T, Ota K, Sumimoto H. Novel modular domain PB1 recognizes PC motif to mediate functional protein-protein interactions. EMBO J. 2001;20:3947–3956. doi: 10.1093/emboj/20.15.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima K, Suganuma N, Tamaoki M, Kouchi H. Two types of pea leghemoglobin genes showing different O2-binding affinities and distinct patterns of spatial expression in nodules. Plant Physiol. 2001;125:641–651. doi: 10.1104/pp.125.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosterin OE, Rozov SM. Mapping of the new mutation blb and the problem of integrity of linkage I. Pisum Genet. 1993;25:27–31. [Google Scholar]
- Kouchi H, Hata S. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- Laursen NB, Larsen K, Knudsen JY, Hoffmann HJ, Poulsen C, Marcker KA, Jensen EØ. A protein binding AT-rich sequence in the soybean leghemoglobin c3 promoter is a general cis element that requires proximal DNA elements to stimulate transcription. Plant Cell. 1994;6:659–668. doi: 10.1105/tpc.6.5.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U, Schlaman HRM, Spaink HP, Sautter C, Rolfe BG, Djordjevic MA. Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 1998;14:23–34. doi: 10.1046/j.1365-313X.1998.00090.x. [DOI] [PubMed] [Google Scholar]
- Nakamuro Y, Kaneko T, Asamizu E, Kato T, Sato S, Tabata S. Structural analysis of a Lotus japonicus genome: II. Sequence features and mapping of sixty-five TAC clones which cover the 6.5-Mb region of the genome. DNA Res. 2002;9:63–70. doi: 10.1093/dnares/9.2.63. [DOI] [PubMed] [Google Scholar]
- Pedrosa A, Sandal N, Stougaard J, Schweitzer D, Bachmair A. Chromosomal map of the model legume Lotus japonicus. Genetics. 2002;161:1661–1672. doi: 10.1093/genetics/161.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Ito T, Moscat J, Diaz-Meco MT, Inagaki F, Sumimoto H. OPR, PC and AID: all in the PB1 family. Trends Biochem Sci. 2002;27:10. doi: 10.1016/s0968-0004(01)02006-0. [DOI] [PubMed] [Google Scholar]
- Ramlov KB, Laursen NB, Stougaard J, Marcker KA. Site-directed mutagenesis of the organ-specific element in the soybean leghemoglobin lbc3 promoter. Plant J. 1993;4:577–580. doi: 10.1046/j.1365-313x.1993.04030577.x. [DOI] [PubMed] [Google Scholar]
- Richards RI, Holman K, Friend K, Kremer E, Hillen D, Staples A, Brown WT, Goonewardena P, Schwartz C. Evidence of founder chromosomes in fragile X syndrome. Nat Genet. 1992;1:257–260. doi: 10.1038/ng0792-257. [DOI] [PubMed] [Google Scholar]
- Roth LE, Stacey G. Bacterium release into host cells of nitrogen-fixing soybean nodules: The symbiosome membrane comes from three sources. Eur J Cell Biol. 1989;49:13–23. [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance chromosomal location and population dynamics. Proc Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis TG. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sandal N, Krusell L, Radutoiu S, Olbryt M, Pedrosa A, Stracke S, Parniske M, Bachmaier A, Sato S, Tabata S et al. A genetic linkage map of the model legume Lotus japonicus and strategies for fast mapping of new loci. Genetics. 2002;161:1673–1683. doi: 10.1093/genetics/161.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- Schultze M, Kondorosi A. Regulation of symbiotic root nodule development. Annu Rev Genet. 1998;32:33–57. doi: 10.1146/annurev.genet.32.1.33. [DOI] [PubMed] [Google Scholar]
- Stougaard J. Regulators and regulation of legume root nodule development. Plant Physiol. 2000;124:531–540. doi: 10.1104/pp.124.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard J. Genetics and genomics of root symbiosis. Curr Opin Plant Biol. 2001;4:328–335. doi: 10.1016/s1369-5266(00)00181-3. [DOI] [PubMed] [Google Scholar]
- Stougaard J, Sandal NN, Grøn A, Kühle A, Marcker KA. 5′ Analysis of the soybean leghemoglobin lbc3 gene: regulatory elements required for promoter activity and organ specificity. EMBO J. 1987;6:3565–3569. doi: 10.1002/j.1460-2075.1987.tb02686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- Szczyglowski K, Szabados L, Fujimoto SY, Silver D, de Bruijn F. Site-specific mutagenesis of the nodule-infected cell expression (NICE) element and the AT-rich element ATRE-BS2* of the Sesbania rostrata leghemoglobin glb3 promoter. Plant Cell. 1994;6:317–322. doi: 10.1105/tpc.6.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsyganov VE, Borisov AY, Rosov SM, Tikhonovich IA. New symbiotic mutants of pea obtained after mutagenesis of laboratory line SGE. Pisum Genet. 1994;V.26:36–37. [Google Scholar]
- Tsyganov VE, Voroshilova VA, Kukalev AS, Azarova TS, Yakobi LM, Borisov AY, Tikhonovich IA. Pisum sativum L. genes Sym14 and Sym35 control infection thread growth initiation during the development of symbiotic nodules. Russ J Genet. 1999;35:284–291. [Google Scholar]
- Tsyganov VE, Voroshilova VA, Priefer UB, Borisov AY, Tikhonovich IA. Genetic dissection of the initiation of the infection process and nodule tissue development in the Rhizobium-pea (Pisum sativum L.) symbiosis. Ann Bot (Lond) 2002;89:357–366. doi: 10.1093/aob/mcf051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK. Legume models strut their stuff. Mol Plant-Microbe Interact. 2001;14:6–9. doi: 10.1094/MPMI.2001.14.1.6. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- Weeden NF, Ellis THN, Timmerman-Vaughan G, Swiecicki WK, Rozov SM, Berdnikov VA. A consensus linkage map for Pisum sativum. Pisum Genet. 1998;30:1–4. [Google Scholar]
- Wegel E, Schauser L, Sandal N, Stougaard J, Parniske M. Mycorrhiza mutants of Lotus japonicus define genetically independent steps during symbiotic infection. Mol Plant-Microbe Interact. 1998;9:933–936. [Google Scholar]
- Yang W-C, Horvath B, Hontelez J, Van Kammen A, Bisseling T. In situ localization of Rhizobium mRNAs in pea root nodules: nifA and nifH localization. Mol Plant-Microbe Interact. 1991;4:464–468. [Google Scholar]