Abstract
Arabidopsis and Medicago truncatula represent sister clades within the dicot subclass Rosidae. We used genetic map-based and bacterial artificial chromosome sequence-based approaches to estimate the level of synteny between the genomes of these model plant species. Mapping of 82 tentative orthologous gene pairs reveals a lack of extended macrosynteny between the two genomes, although marker collinearity is frequently observed over small genetic intervals. Divergence estimates based on non-synonymous nucleotide substitutions suggest that a majority of the genes under analysis have experienced duplication in Arabidopsis subsequent to divergence of the two genomes, potentially confounding synteny analysis. Moreover, in cases of localized synteny, genetically linked loci in M. truncatula often share multiple points of synteny with Arabidopsis; this latter observation is consistent with the large number of segmental duplications that compose the Arabidopsis genome. More detailed analysis, based on complete sequencing and annotation of three M. truncatula bacterial artificial chromosome contigs suggests that the two genomes are related by networks of microsynteny that are often highly degenerate. In some cases, the erosion of microsynteny could be ascribed to the selective gene loss from duplicated loci, whereas in other cases, it is due to the absence of close homologs of M. truncatula genes in Arabidopsis.
Comparative genetic mapping has revealed a high degree of conservation in genome structure among closely related plant species, in terms of gene content, order, and function (Paterson et al., 1995, 2000; Gale and Devos, 1998; Bennetzen, 2000). The best-documented cases of genome conservation are from the grass family (Poaceae), where rice (Oryza sativa) with its small genome has been selected as a nodal species to study the economically important cereal crops including corn (Zea mays), sorghum (Sorghum bicolor), wheat (Triticum aestivum), and barley (Hordeum vulgare; Devos and Gale, 2000). Extensive macrosynteny has also been observed within the Solanaceae (Tanksley et al., 1992), the Brassicaceae (Kowalski et al., 1994; Lagercrantz and Lydiate, 1996; Lagercrantz, 1998; O'Neill and Bancroft, 2000; Acarkan et al., 2000), and the Fabaceae (Weeden et al., 1992; Menacio-Hautea et al., 1993; Torres et al., 1993; H.-K. Choi and D.R. Cook, unpublished data).
In contrast to within-family comparisons, genome structure appears to be less conserved between distantly related species, where collinearity may only be apparent over small chromosomal intervals (Paterson et al., 1996, 2000). Paterson and colleagues have suggested that deciphering such relationships will require a very high density of genetic markers for comparison (Paterson et al., 2000). More recent analyses, however, reveal that plant genomes possess a dynamic microstructure (Ku et al., 2000; Vision et al., 2000) that may preclude establishing global relationships between distantly related plant species based on genetic map data alone. In lieu of whole-genome sequence data, an intermediate strategy involving complete sequencing and annotation of bacterial artificial chromosome (BAC)-size clones has been adopted by several groups to obtain a glimpse of genome similarities at the micro-level (Ku et al., 2000; Liu et al., 2001; Mayer et al., 2001; Rossberg et al., 2001). In the comparison of Arabidopsis and tomato (Lycopersicon esculentum), species that diverged early in the radiation of dicot plants, this strategy revealed an extensive network of microsynteny between a 105-kb segment of tomato chromosome 2 and Arabidopsis chromosomes 2 to 5 (Ku et al., 2000). It remains uncertain, however, whether such syntenic networks are a general feature of cross-family comparisons or only restricted to certain genome regions. Comparison of Arabidopsis and rice using a similar strategy revealed microsynteny at some loci but not others, leading to the conclusion that Arabidopsis may not be adequate as a model for the structure of grass genomes (Devos et al., 1999; van Dodeweerd et al., 1999; Liu et al., 2001; Mayer et al., 2001).
The Fabaceae, or legume species, make up the third largest family of flowering plants, including numerous important agricultural crops such as soybean (Glycine max), pea (Pisum sativum), beans (Phaseolus vulgaris), and alfalfa (Medicago sativa). Legumes are also distinguished by their unique property of symbiotic nitrogen fixation, providing one of the major sources of available nitrogen in the biosphere. The agronomic and ecologic importance of this group of plants has provided incentive to undertake genome analyses in species such as Medicago truncatula, soybean, and Lotus japonicus (Cook, 1999). An important near-term goal for these efforts is to determine the extent to which the complete sequence of the Arabidopsis genome will have predictive value for the structure of legume genomes. Here, we report the results from a comparison of genome structure between the two model species M. truncatula and Arabidopsis. Our results suggest that the two genomes share in common an eroded network of microsynteny, but that broad macrosyntenic relationships are not apparent.
RESULTS AND DISCUSSION
High Level of Macrosynteny Is Not Evident between M. truncatula and Arabidopsis
As a prelude to comparative genome analysis between M. truncatula and Arabidopsis, we analyzed approximately 300 mapped genes in M. truncatula to identify their highly conserved counterparts in Arabidopsis (see criteria in “Materials and Methods” and specific comparisons in online supplemental data, which can be viewed at www.plantphysiol.org). The rationale for focusing on highly conserved genes is as follows. We expect that conserved gene order will be most evident in cases where linkage is derived from the most recent common ancestor. By definition, such genes are orthologous between the species under comparison. A corollary to this logic is that poorly conserved genes will be of limited value to identify ancestral synteny, in particular because such genes are likely to represent ancient paralogs derived through a process of within-species duplication. Our approach contrasts with that of previous studies to compare a legume genome with that of Arabidopsis (Grant et al., 2000), where low similarity criteria were used to examine conserved gene order between soybean and Arabidopsis.
One hundred and ten of the mapped M. truncatula genes possess significant homology to Arabidopsis genes according to the criteria described in “Materials and Methods” and presented in the supplemental data. Thirty of these M. truncatula genes had only a single match in Arabidopsis, whereas the remaining 80 (approximately 73%) had two or more matches. This redundancy of homology is consistent with the duplicated nature of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000).
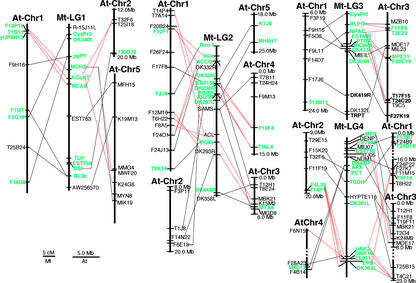
Alignment of the M. truncatula and Arabidopsis chromosomes reveals a complex relationship between the two genomes (Fig. 1). In particular, many mapped M. truncatula genes share high homology to several loci within the Arabidopsis genome. As a consequence, each of the eight M. truncatula linkage groups can be aligned to any of the five Arabidopsis chromosomes. In an effort to obtain a “one-to-one” comparison between the two genomes, we focused on tentative orthologous pairs of loci gained from the eukaryotic gene orthologs database of The Institute for Genomic Research (TIGR; http://www.tigr.org/tdb/tgi/ego/index.shtml). On the basis of this analysis, 82 pairs of loci were found to be tentative orthologs (see supplemental data). Comparing linkage relationships between tentative orthologs did not reveal a simplified picture of synteny between the M. truncatula and Arabidopsis genomes. In most cases, the Arabidopsis orthologs of linked M. truncatula genes are either not linked in Arabidopsis or linked but interrupted by orthologs that are not linked in M. truncatula (Fig. 1). This result is in contrast to that reported by Grant et al. (2000), where significant synteny was reported between soybean and Arabidopsis along the entire length of Arabidopsis chromosome 1 and soybean linkage group A2.
Figure 1.
Chromosomal alignments between the M. truncatula genetic linkage groups 1 to 4 and the Arabidopsis chromosomes. Homologs between the two genomes are connected with lines. The markers with green color are putative orthologous loci. Red lines indicate the possible syntenic relationships between the two genomes.
The convoluted relationship suggested by marker alignment can be explained by either the absence of extended synteny between the two genomes or synteny that is obscured by the duplication of individual genes or chromosomal segments followed by extensive reshuffling and selective gene loss in both genomes (Blanc et al., 2000; Ku et al., 2000). In cases of gene duplication, paralogs arising subsequent to separation of the M. truncatula and Arabidopsis lineages would be most complicating to synteny analysis because they increase the number of phylogenetically equidistant loci between the genomes. The TIGR ortholog prediction, used above, identifies the most conserved genes between genomes without regard to lineage relationship. As a counterpoint to the analysis of “orthologs” predicted by the eukaryotic gene ortholog analysis, we sought to predict the lineage relationships for all genes under comparison in Figure 1. Ratios of non-synonymous to synonymous nucleotide substitutions were similar within gene families and were substantially less than one for all gene pairs under analysis (data not shown). These results suggest both a high level of purifying selection and similar rates of evolution between paralogs. Thus, we used the rate of non-synonymous nucleotide substitution (Ka) as a measure of divergence for each conserved gene pair between M. truncatula and Arabidopsis, and between different homologs within the Arabidopsis genome (Table I). The average Ka for single-copy Arabidopsis genes compared with the corresponding marker gene in M. truncatula was 0.19 ± 0.09, with a range of 0.03 to 0.44. Similar aggregate values were obtained when Ka was calculated for M. truncatula genes with two or three homologs in Arabidopsis (i.e. for two homologs, Ka = 0.18 ± 0.12 with a range of 0.03–0.45; for three homologs, Ka = 0.17 ± 0.12 with a range of 0.05–0.50). The largely similar average Ka values reflect the fact that many of the duplicated Arabidopsis genes are approximately equally diverged from their M. truncatula counterpart. When between-species Ka values were compared with within-species Ka values, it was possible to infer lineage relationships for the different genes, as shown in Table I. Thus, 18 of the 28 marker genes with two homologs (lineage pattern A1) and seven of the 16 marker genes with three homologs (lineage patterns B1 and B3) are predicted to have arisen by duplication in Arabidopsis after separation of the M. truncatula and Arabidopsis lineages, whereas in other cases, the gene duplications in Arabidopsis are predicted to predate the speciation event. For the cases where duplication occurred after speciation, comparison of genome structure is complicated by the proliferation of multiple, phylogenetically equidistant loci. We suggest that in such cases a true ortholog cannot be defined between the genomes, and that the utility of the TIGR ortholog set is restricted to identifying the most highly conserved paralogs.
Table I.
Predicted lineage patterns of comparative genes between M. truncatula and Arabidopsis
|
M. truncatula Genes with Two Homologs in
Arabidopsis
|
M. truncatula Genes with Three
Homologs in Arabidopsis
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker | Ka1 Mt-At1 | Ka2 Mt-At2 | Ka At1-At2 | Inferred Lineage | Marker | Ka1 Mt-At1 | Ka2 Mt-At2 | Ka3 Mt-At3 | Ka At1-At2 | Ka At1-At3 | Ka At2-At3 | Inferred Lineage | |||||
| DK326L | 0.034 | 0.038 | 0.015 | A1 | MDH2 | 0.037 | 0.053 | 0.21 | 0.047 | 0.216 | 0.198 | B3 | |||||
| RBBP | 0.051 | 0.176 | 0.114 | A2 | KCOAT | 0.097 | 0.109 | 0.17 | 0.065 | 0.189 | 0.189 | B3 | |||||
| TRPT | 0.069 | ND | 0.078 | ND | ACCO | 0.163 | 0.218 | 0.223 | 0.226 | 0.247 | 0.08 | B2 | |||||
| TGDH | 0.079 | 0.084 | 0.023 | A1 | ACL | 0.077 | 0.077 | 0.149 | 0.026 | 0.142 | 0.134 | B3 | |||||
| RL3b | 0.087 | 0.108 | 0.098 | A2 | PGKI | 0.119 | 0.114 | 0.067 | 0.057 | 0.111 | 0.112 | B2 | |||||
| DK455 | 0.087 | 0.088 | 0.083 | A1 | 43A3R | 0.189 | ND | ND | 0.012 | 0.21 | 0.213 | B1 | |||||
| DK363 | 0.093 | 0.141 | 0.047 | A1 | RLPO | 0.103 | 0.107 | 0.098 | 0.013 | 0.107 | 0.112 | B2 | |||||
| NCAS | 0.093 | 0.108 | 0.054 | A1 | DNABP | 0.155 | 0.133 | 0.22 | 0.121 | 0.263 | 0.235 | B3 | |||||
| TUP | 0.093 | 0.153 | 0.172 | A2 | RNAH | 0.104 | 0.115 | 0.082 | 0.054 | 0.114 | 0.123 | B2 | |||||
| AAT | 0.097 | 0.122 | 0.044 | A1 | PDL | 0.111 | 0.291 | 0.233 | 0.299 | 0.211 | 0.341 | B4 | |||||
| PTRS | 0.106 | 0.115 | 0.036 | A1 | ENOL | 0.083 | 0.274 | 0.352 | 0.254 | 0.346 | 0.37 | B4 | |||||
| ppPF | 0.109 | 0.117 | 0.052 | A1 | EIF | 0.083 | 0.121 | 0.062 | 0.073 | 0.092 | 0.121 | B2 | |||||
| EPS | 0.113 | 0.117 | 0.01 | A1 | SQEX | 0.146 | 0.149 | 0.12 | 0.098 | 0.212 | 0.178 | B2 | |||||
| PPGM | 0.116 | 0.116 | 0.067 | A1 | UNK16 | 0.432 | ND | ND | 0.016 | ND | ND | B3 | |||||
| PCT | 0.121 | 0.144 | 0.224 | A2 | TE011 | 0.516 | 0.496 | 0.492 | 0.272 | 0.447 | 0.452 | B3 | |||||
| COA | 0.137 | 0.154 | 0.082 | A1 | |||||||||||||
| 12NIL | 0.143 | 0.161 | 0.161 | A2 | 
|
||||||||||||
| PFK | 0.151 | 0.179 | 0.203 | A1 | |||||||||||||
| DK351 | 0.182 | 0.271 | 0.197 | A2 | |||||||||||||
| EST718 | 0.201 | 0.291 | 0.232 | A2 | |||||||||||||
| EST158 | 0.204 | 0.211 | 0.199 | A1 | |||||||||||||
| PPDK | 0.212 | 0.44 | 0.458 | A2 | |||||||||||||
| UNK7 | 0.215 | 0.28 | 0.182 | A1 | |||||||||||||
| DK419 | 0.224 | 0.3 | 0.235 | A2 | |||||||||||||
| MPP | 0.26 | 0.269 | 0.096 | A1 | |||||||||||||
| LB1 | 0.366 | 0.548 | 0.455 | A2 | |||||||||||||
| CRS | 0.411 | 0.487 | 0.18 | A1 | |||||||||||||
| GH1 | 0.442 | 0.447 | 0.302 | A1 | |||||||||||||
Conserved Gene Order over Small Genetic Intervals
In contrast to the apparent absence of extended synteny between M. truncatula and Arabidopsis, we observed frequent conservation of gene order over shorter genetic intervals. For example, three M. truncatula genes (Mt-MDH2, Mt-KcoAT, and Mt-NCAS) are within a 5-centiMorgan (cM) interval on the M. truncatula linkage group 1 (Mt-LG1), whereas their tentative orthologs (F19P19/At1g04410, T1G11/At1g04710, and yUP8H12/At1g05150, respectively) are in the same order and resident within a 400-kb region on the Arabidopsis chromosome 1 (At-Chr1; Fig. 1). More distant homologs of Mt-KcoAT and Mt-NCAS, namely F25I18/At2g33150 and T32F6/At2g32450, respectively, are also tightly linked within a 400-kb block on the At-Chr2. These two Arabidopsis regions have been previously described to be homeologous (Blanc et al., 2000), and the absence of a homolog of Mt-MDH2 from the At-Chr2 region is consistent with the finding that homeologous regions of Arabidopsis can be highly degenerate, with less than one-half of the gene homologs retained (Arabidopsis Genome Initiative, 2000).
In some cases, homologs of tightly linked genes on the M. truncatula genetic map are resident in the same BAC or BAC contig in Arabidopsis, providing further evidence of conserved local gene content. A typical example is from Mt-LG4, where a tight cluster of five genes (Mt-UNK3, Mt-UNK16, Mt-TE011, Mt-ESP, and Mt-DK363L) within a 2-cM interval are collinear with a BAC contig (At-T14P1 and At-F4L23) on At-Chr2 that contains all five of the predicted orthologs in Arabidopsis. Similar to the situation described above (Fig. 1), BAC contigs with less completely conserved gene content are distributed over two additional Arabidopsis chromosomes. Thus, BAC At-T4C21 on At-Chr3 and At-F6N15 on At-Chr4 contain four and two homologs of the five genes, respectively. Consistent with the apparent absence of extended macrosynteny, the observed local synteny does not appear to extend over longer genome intervals. For example, of four closely linked genes within a 4-cM interval on Mt-LG3 (i.e. Mt-EST400, Mt-PTRS, Mt-DK360L, and Mt-DK473L), the syntenic pair of Mt-EST400 and Mt-PTRS (MPE11/BAB01073 and F20C19/At3g26340, respectively) is only loosely linked to that of Mt-DK360L and Mt-DK473L (T24C20/At3g48190 and T17F15/At3g48000, respectively) on At-Chr3. A similar conservation of gene content and order has been observed between a small region of the soybean and Arabidopsis genomes (Foster-Hartnett et al., 2002).
Segmental duplication has been well documented in the Arabidopsis genome (Vision et al., 2000). Here we observe (a) that many close homologs between the M. truncatula and Arabidopsis genomes have undergone duplication subsequent to separation of the two lineages, and (b) that retained synteny is confined to small genome segments that are often repeated in the Arabidopsis genome. Thus, frequent segmental duplication and rearrangement after speciation are primary factors contributing to divergence between the M. truncatula and Arabidopsis genomes.
Microsynteny between M. truncatula and Arabidopsis at the BAC Clone Level
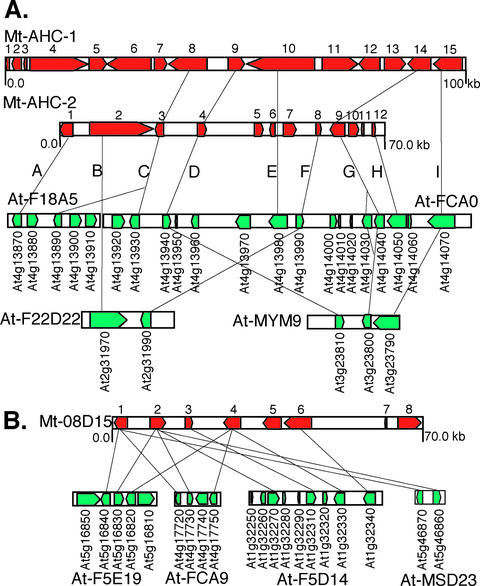
Two M. truncatula BAC contigs, Mt-AHC1 (AY224188) and Mt-AHC2 (AY224189), together with the BAC clone Mt-08D15 (AC087771) were subjected to complete sequencing. Mt-AHC1 and Mt-AHC2 were selected because they represent a segmental duplication in M. truncatula, initially identified based on the presence of highly conserved adenosylhomocysteinase (AHC) gene paralogs. The resulting sequences for Mt-AHC1, Mt-AHC2, and Mt-08D15 are approximately 100, 68, and 70 kb and contain 15, 12, and eight predicted genes, respectively. The average gene density of the three sequenced BACs is about one gene per 6.8 kb. Detailed analysis of these sequences and comparison with Arabidopsis are summarized in Table II and Figure 2.
Table II.
Matches of predicted M. truncatula genes with Arabidopsisa
| Predicted Open Reading Frames | Expressed Sequence Tag (EST) Matchesb | Matches of Arabidopsis
Genes
|
Putative Function | ||||
|---|---|---|---|---|---|---|---|
| At-chr1 | At-chr2 | At-chr3 | At-chr4 | At-chr5 | |||
| AHC1.8 | TC25520 | At1g36370 | At4g13890, At4g13930 | At5g26780 | |||
| At1g22020 | At4g32520, At4g37930 | Hydroxymethyltransferase | |||||
| AHC1.9 | TC21970 | AT3g23810 | At4g13940 | Adenosylhomocysteinase | |||
| AHC1.10 | TC16577 | At4g13980 | Heat shock transcription factor | ||||
| AHC1.11 | NA | At4g21110 | G10-like protein | ||||
| AHC1.12 | NA | A large gene family | Hypothetical protein | ||||
| AHC1.13 | TC18388 | At4g08580 | At5g17900 | Microfibril-associated protein | |||
| AHC1.14 | AL383790 | At3g23800 | At4g14030, At4g14040 | Selenium-bindingprotein | |||
| AHC1.15 | NA | At3g23790 | At4g14070 | AMP-binding protein | |||
| AHC2.1 | BE999775 | At4g13870 | Unknown protein | ||||
| AHC2.2 | AW736328 | At2g31970 | RAD50 DNA repairprotein | ||||
| AHC2.3 | TC22017 | Same as those of AHC1.8 | Hydroxymethyltransferase | ||||
| AHC2.4 | TC21843 | At3g23810 | At4g13940 | Adenosylhomocysteinase | |||
| AHC2.5 | NA | A large gene family | Polygalacturonase | ||||
| AHC2.6 | NA | No match | |||||
| AHC2.7 | NA | A large gene family | Polygalacturonase | ||||
| AHC2.8 | NA | At2g31990 | At4g13990 | Hypothetical protein | |||
| AHC2.9 | TC19298 | At3g23800 | At4g14030, At4g14040 | Selenium-bindingprotein | |||
| AHC2.10 | NA | At2g25010 | Unknown protein | ||||
| AHC2.11 | NA | No significant match | |||||
| AHC2.12 | NA | At4g14050 | Hypothetical protein | ||||
| 08D15.1 | TC22974 | At1g67950 | At3g01210 | At4g17720 | At5g16840 | ||
| At5g46870 | Hypothetical protein | ||||||
| 08D15.2 | TC17359 | At1g32270 | At4g17730 | At5g16830 | |||
| At5g46860 | Syntaxin | ||||||
| 08D15.3 | AW585392 | At1g32310 | Unknown protein | ||||
| 08D15.4 | TC17232 | At1g32330 | At3g02990 | At4g17750 | At5g16820 | Heat shock transcription factor HSF1 | |
| 08D15.5 | NA | No significant match | |||||
| 08D15.6 | BE247890 | At1g32340 | RING finger protein | ||||
| 08D15.7 | NA | No significant match | |||||
| 08D15.8 | NA | At1g10680 | At4g25960 | P-Glycoprotein-2 | |||
For genes with multiple hits, only representatives are listed; for AHC1 locus, only genes that are within the syntenic range are listed.
NA, EST match not available.
Figure 2.
Microsynteny between sequenced M. truncatula BACs and segments of Arabidopsis chromosomes. The orientations of predicted genes are indicated by arrows. Letters A through I in A represent the nine unique gene homologies found between Arabidopsis and M. truncatula and thus define the minimum structure of the ancestral chromosome segment from which all the syntenic blocks were derived. A detailed description of gene annotations and homologies is given in Table II. The maps are drawn to scale.
Mt-AHC1 and Mt-AHC2 contain three pairs of homologous genes with the same order and orientation (i.e. Mt-AHC1.8 versus Mt-AHC2.3, Mt-AHC1.9 versus Mt-AHC2.4, and Mt-AHC1.14 versus Mt-AHC2.9; Table II). The three genes span 57 kb in Mt-AHC1 but only 40 kb in Mt-AHC2 (Fig. 2A). The increased physical interval in Mt-AHC1 is primarily due to longer introns in Mt-AHC1 genes rather than to extended intergenic regions. Comparing M. truncatula genes on Mt-AHC1 and Mt-AHC2 with their counterparts in Arabidopsis reveals a network of synteny (Fig. 2A), similar to that observed between tomato and Arabidopsis (Ku et al., 2000). Gene order is conserved among syntenic blocks, with a reversal of orientation observed only in two cases (Mt-AHC2.12 versus FCA0/At4g14050 and FCA0/At4g13990 versus F22D22/At2g31990). The BAC contig made up of At-F18A5 and At-FCA0 on At-Chr4 contains two cases of gene duplication: One is a tandem duplication (FCA0/At4g14030 and FCA0/At4g14040), whereas the other (F18A5/At4g13890 and FCA0/At4g13930) is a direct duplication separated by three cf-like disease resistance gene homologs (see below). Interestingly, this At-Chr4 BAC contig appears to be most representative of the ancestral chromosomal segment from which the other syntenic blocks are predicted to have originated, because it contains all but one (Mt-AHC2.2) of the syntenic genes observed both in M. truncatula and Arabidopsis. The less complete syntenic blocks on At-MYM9 and At-F22D22 in Arabidopsis probably arose from segmental duplication followed by gene deletion events (Ku et al., 2000).
Similar results were obtained when comparing genes on M. truncatula BAC clone Mt-08D15 with their homologs in Arabidopsis (Fig. 2B). Five of the eight predicted genes from Mt-08D15 (Mt-08D15.1, Mt-08D15.2, Mt-08D15.3, Mt-08D15.4, and Mt-08D15.6) can be aligned with genes on four different Arabidopsis BAC clones (At-F5D14, At-F5E19, At-FCA9, and At-MSD23). All syntenic genes are in the same order and orientation, and the intergenic spaces and intron sequences are generally longer in M. truncatula than in Arabidopsis.
Microsynteny between BAC Clones Is Highly Degenerate
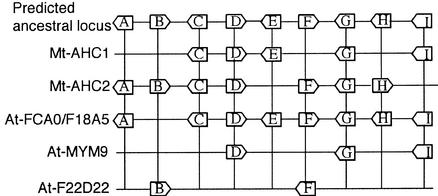
The 42 syntenic genes identified in the AHC and 08D15 contigs correspond to 14 unique gene homologies that are shared by at least one of the syntenic loci in both Arabidopsis and M. truncatula (Fig. 2). In the case of the AHC contigs, nine homologs are predicted to define the minimal structure of the ancestral locus (Fig. 2A, letters A–I). Interestingly, none of the conserved homologs were conserved in all of the duplicated regions, suggesting that the structure of the ancestral locus is degenerate in all of these modern day descendants. In cases where absent homologs are flanked by a conserved framework (e.g. homolog E in Mt-AHC2 and homolog F in Mt-AHC1), selective gene loss after duplication is a likely cause of degenerate microsynteny (Ku et al., 2000). At the extremity of syntenic regions, the absence of conserved homologs is indicative of either (a) small size of the segmental duplication, or (b) degenerate synteny that extends beyond the sequenced region in M. truncatula. Alignment of the structures of the AHC loci from Arabidopsis and M. truncatula (Fig. 3) indicates that each of the five loci (two in M. truncatula and three in Arabidopsis) has experienced selective gene loss. If the loss of adjacent genes is considered to be a single event, then the overall rate of gene loss for M. truncatula and Arabidopsis are similar (i.e. five events per 18 genes [28%] and seven events per 27 genes [26%], respectively).
Figure 3.
Reconstruction of the predicted ancestral AHC locus representing five segments from the M. truncatula and Arabidopsis genomes. The transcriptional orientations of the genes are indicated by arrows. Letters A through I are consistent with those in Figure 2.
In some cases, non-syntenic genes within a conserved framework are members of large gene families. For example, the syntenic segment on At-Chr4 contains a cluster of four cf-like disease resistance genes (F18A5/At4g13880, F18A5/At4g13900, F18A5/ At4g13910, and FCA0/At4g13920) within a 20-kb interval, three of which are flanked by two highly similar hydroxymethyltransferase (HMT) genes (F18A5/At4g13890 and FCA0/At4g13930; Fig. 2A). This nested duplication structure is reminiscent of the tomato cf-4/9 locus, where the cf resistance gene homologs are flanked by two conserved lipoxygenase genes (Parniske et al., 1997). Such genomic microstructure may provide substrates for unequal crossing-over events and thus contribute to expansion or contraction of local gene redundancy. In the case of resistance gene homologs, unequal crossing-over is implicated in the fast-evolving nature of these genes, which are often poorly conserved even between closely related genomes (Leister et al., 1998). FCA0/At4g13960, a member of F-box gene family, is also not represented in the M. truncatula AHC syntenic regions.
We also observed interspersion of low copy genes between syntenic genes both within and between M. truncatula and Arabidopsis (Fig. 2). This might be due to the loss of different subsets of genes from a common ancestor, independent gain of genes through transposition events, or reshuffling of small DNA segments (Blanc et al., 2000; Ku et al., 2000; Bancroft, 2001).
Divergence Estimates and Rearrangements to Gene Structure within Segmental Duplications
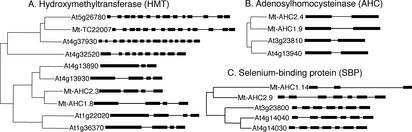
To understand the evolution of duplicated genes within and between species, we compared gene structures and phylogeny of the three genes present in both members of the M. truncatula AHC segmental duplication (i.e. AHC, HMT, and selenium-binding protein [SBP]; Table I). In the Arabidopsis genome, highly similar homologs of SBP and AHC are confined to the segmental duplications shown in Figure 2. The HMT gene family, on the other hand, is represented by several additional homologs elsewhere in the Arabidopsis genome. To determine the probable order of speciation versus segmental duplication, we conducted phylogenetic analysis by means of the neighbor-joining method. The resulting tree topologies, which are supported by 100% of 1,000 bootstrap replications in all cases of syntenic homologs, suggest that these segmental duplications derive from independent events that occurred subsequent to separation of the M. truncatula and Arabidopsis lineages (Fig. 4).
Figure 4.
Phylogeny versus gene structure of syntenic and non-syntenic homologous genes. Phylogenetic analyses of protein sequences were performed using the ClustalX program (Thompson et al., 1997), and trees were constructed using the neighbor-joining method. The intron-exon structures of genes are drawn to scale. Exons are shown as boxes and introns are shown as lines. Intron-exon structures were inferred based on comparison of EST and genomic sequence data and based on gene prediction tools, as described in “Materials and Methods.” The predicted structure of Mt-TC22007 is from TIGR database, and the intron sizes are unknown.
As a further measure of diversification, we compared intron-exon structures for the three genes described above (Fig. 4). As shown diagrammatically in Figure 4, the position and number of introns is largely conserved, whereas intron lengths are variable both within and between M. truncatula and Arabidopsis, with intron length generally longer in M. truncatula. Despite the overall conservation of gene structure, intron loss is evident for some of these duplicated genes. For example, the HMT gene At4g13890, which is a close paralog of At4g13930 on At-Chr4, is predicted to have lost the first intron by fusion of the first and second exons from the At4g13930 (Fig. 4A). The SBP gene At4g14040, a tandem duplication of At4g14030, is similarly missing the third intron (Fig. 4C), whereas SBP Mt-AHC1.14 exhibits a truncated N terminus and an enlarged second intron. Despite this fact, the Mt-AHC1.14 represents an expressed gene, as evidenced by a matching M. truncatula EST in the M. truncatula gene index (MtGI) database (AL383790).
In the case of the HMT gene family, the intron-exon structures of syntenic compared with non-syntenic homologs are highly diverged. Phylogenetic analysis of HMT genes (Fig. 4A) indicates that the syntenic and non-syntenic homologs belong to separate lineages, with similar intron-exon structures within a lineage but substantially different intron-exon structures between lineages. Because these HMT homologs have a generally conserved global alignment, we suggest that exon splicing has contributed to the differences in intron/exon number and has thus played a role in the divergence of this small gene family. In other instances, exon shuffling is implicated as a mechanism of divergence; for example, two HMT homologous genes, At1g22020 and At1g36370, are distinguished by a approximately 120-amino acid domain of unknown origin at their N terminus. The existence of M. truncatula HMT genes (e.g. TC22007) with a similar predicted intron-exon structure and amino acid sequence to those found in Arabidopsis (i.e. At4g37930 and At5g26780) demonstrates that this divergence is likely to predate separation of the M. truncatula and Arabidopsis lineages.
Frequency of Microsynteny between M. truncatula and Arabidopsis
An important question regarding microsynteny between distantly related species is whether the microsynteny observed in case studies of completely sequenced BAC clones is indicative of a wider conservation of genome structure. To address this issue, we analyzed 40 M. truncatula BAC clones/contigs that were subject to either survey sequencing or lower coverage sequence analysis based on a combination of BAC end and targeted internal sequence analysis. The short gene sequences obtained from BAC survey sequencing can be easily anchored to longer tentative consensus sequences (TCs) by means of querying MtGI using BLASTN, thus permitting a better informed analysis of conserved microsynteny between M. truncatula and Arabidopsis. The existence of microsynteny was inferred when close homologs of genes identified on a M. truncatula BAC (or contig) were also resident on a single Arabidopsis BAC or adjacent BACs. Through this analysis, 17 of the 40 loci show some level of microsynteny with one or more regions in Arabidopsis (see supplemental data), underscoring the conclusion that the genomes of M. truncatula and Arabidopsis are related by numerous regions of degenerate microsynteny.
The Potential Role of Species-Specific Genes in Genome Divergence
BAC survey sequencing indicates that the erosion of microsynteny is in part due to the absence of close homologs of some M. truncatula genes in Arabidopsis. For example, BAC end sequences from a M. truncatula BAC contig (BAC clones 21E21, 21L09, 33P03, and 79M20) identified three genes: nodulin25 (AJ277858), MtN22 (CAA75576; Gamas et al., 1996), and TC23093. A search of the Medicago truncatula GeneIndex (www.tigr.org/tdg/mtgi) reveals that all three genes are expressed to a higher level, exclusively in root nodules. Nodulin25 and MtN22 genes have no close homologs in Arabidopsis, while TC23093 is a member of a calmodulin multigene family. In addition to contributing to divergent genome structure, genes with taxonomically restricted homologies are prime candidates for important roles in determining lineage-specific phenotypes, such as symbiotic nitrogen fixation in legumes. A specific example of this situation is the case of the nodulation receptor kinase (NORK) locus (dmi2 in M. truncatula) that was recently cloned in M. truncatula, alfalfa, pea, and L. japonicus based on their conserved syntenic positions (Endre et al., 2002; Stracke et al., 2002). The NORK protein is a putative receptor kinase that is highly conserved among all legume species analyzed (Endre et al., 2002). Although the genome region that contains dmi2 is microsyntenic between M. truncatula and Arabidopsis, the NORK homolog is missing from the syntenic region of Arabidopsis, and only very distant homologs are evident elsewhere in the genome.
Summary
Comparative genome analysis between the model legume M. truncatula and Arabidopsis reveals a lack of extensive macrosynteny between these two genomes, whereas genetic evidence as well as both complete and low coverage sequencing of M. truncatula BAC clones support a model of degenerate microsynteny. Our data suggest that diversification between these two genomes is driven by at least three factors occurring subsequent to separation of the M. truncatula and Arabidopsis lineages: (a) extensive segmental duplication, accompanied by (b) chromosomal rearrangement and (c) extensive erosion of the structure of duplicated regions. On the basis of a comparison of homologous segmental duplications within each genome, we infer that the nature and extent of within-genome degradation of microsynteny are largely similar between these two species. Divergence between these two genomes was also evident in the form of genes in M. truncatula that lack close homologs in Arabidopsis. Interestingly, some of these genes appear to be expressed specifically during symbiotic nitrogen fixation in M. truncatula, a process that is characteristic of legumes, and thus may contribute to legume-specific phenotypes. On the basis of these results, it is evident that Arabidopsis cannot serve as a specific model for the structure of legume genomes. Instead, we anticipate that sequencing efforts on legume genomes, including M. truncatula and L. japonicus for which genome sequencing projects are currently under way, will provide the basis for a detailed view of legume genome structure.
MATERIALS AND METHODS
Sequencing of BAC clones (Nam et al., 1999) was performed by a shotgun approach. Completion of individual BAC clones involved paired-end sequencing from subclones to obtain a 7- to 8-fold average depth of coverage. For BAC survey sequencing, 96 randomly selected subclones were sequenced. The sequences were assembled with PHRAP software package (Gordon et al., 1998; Ewing et al., 1998). The completed BAC sequences were analyzed for gene prediction using GENSCAN (Burge and Karlin, 1997) and GENEMARK.HMM (Lukashin and Borodovsky, 1998) with Arabidopsis settings.
A Medicago truncatula genetic map (H.-K. Choi and D.R. Cook, unpublished data; http://www.medicago.org) composed primarily of cleaved amplified polymorphic sequences markers was used to evaluate the level of global synteny between the M. truncatula and Arabidopsis genomes. All template sequences of mapped markers (ESTs or BAC end sequences) were first analyzed against the MtGI (http://www.tigr.org/tdb/mtgi) to obtain TCs for individual ESTs or to correlate BAC end sequences with expressed genes. Assignment of a genomic sequence to a TC or an EST was allowed when two sequences overlapped for at least 40 bp.
BLASTX searches (Altschul et al., 1997) were performed against the Arabidopsis protein database (http://www.Arabidopsis.org). Where possible, M. truncatula TCs were used as query sequences. For the annotated BAC sequences, the predicted genes were used as query sequences for BLAST searches.
We used several criteria to filter the BLAST results. Significant matches were claimed only when (a) the BLAST expected value was less than e−30, (b) the amino acid sequence alignment showed at least 50% identity, and (c) the BLAST output suggested low copy number in the Arabidopsis genome (typically less than three; see supplemental data). In cases of short query sequences (<500 bp), hits with an e value < e−30 and an identity of >60% were considered to be significant. The physical positions of Arabidopsis genes on chromosomes were obtained by querying Map Viewer (http://www.Arabidopsis.org) with the Arabidopsis BAC clone IDs.
Putative orthologs between M. truncatula and Arabidopsis were obtained from the TIGR MtGI database and review of the corresponding orthologous gene alignments (Lee et al., 2002). Sequence alignments and phylogenetic analysis were performed using the ClustalX (Thompson et al., 1997). Phylogenetic trees were constructed using the neighbor-joining method as implemented in ClustalX with 1,000 bootstrap sampling steps. The non-synonymous nucleotide substitution rates (Ka) were calculated using MEGA v2.1, using the method of Kumar et al. (2001).
Supplementary Material
ACKNOWLEDGMENTS
We thank Nevin Young, Randy Shoemaker, Gary Stacey, and Steve Tanksley for critical review of the manuscript.
Footnotes
This work was supported by the National Science Foundation Plant Genome Research Program (grant no. 9872664 to D.R.C.).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.016436.
LITERATURE CITED
- Acarkan A, Rossberg M, Koch M, Schmidt R. Comparative genome analysis reveals extensive conservation of genome organisation for Arabidopsis thaliana and Capsella rubella. Plant J. 2000;23:55–62. doi: 10.1046/j.1365-313x.2000.00790.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bancroft I. Duplicate and diverge: the evolution of plant genome microstructure. Trends Genet. 2001;17:89–93. doi: 10.1016/s0168-9525(00)02179-x. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. Comparative sequence analysis of plant nuclear genomes: microcolinearity and its many exceptions. Plant Cell. 2000;12:1021–1029. doi: 10.1105/tpc.12.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. Extensive duplication and reshuffling in the Arabidopsisgenome. Plant Cell. 2000;12:1093–1101. doi: 10.1105/tpc.12.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Cook DR. Medicago truncatula: a model in the making! Curr Opin Plant Biol. 1999;2:301–304. doi: 10.1016/s1369-5266(99)80053-3. [DOI] [PubMed] [Google Scholar]
- Devos KM, Beales J, Nagamura Y, Sasaki T. Arabidopsis-rice: Will colinearity allow gene prediction across the eudicot-monocot divide? Genome Res. 1999;9:825–829. doi: 10.1101/gr.9.9.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Gale MD. Genome relationships: the grass model in current research. Plant Cell. 2000;12:636–646. doi: 10.1105/tpc.12.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB. areceptor kinase gene regulating symbiotic nodule development. Nature. 2002;417:962–966. doi: 10.1038/nature00842. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred: I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Foster-Hartnett D, Mudge J, Larsen D, Yan H, Denny R, Penuela S, Young ND. Comparative genomic analysis of sequences sampled from a small region on soybean (Glycine max) molecular linkage group G. Genome. 2002;45:634–645. doi: 10.1139/g02-027. [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM. Comparative genetics in the grasses. Proc Natl Acad Sci USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P, Nieble FC, Lescure N, Cullimore J. Use of a subtractive hybridization approach to identify new Medicago truncatulagenes induced during root nodule development. Mol Plant-Microbe Interact. 1996;9:233–242. doi: 10.1094/mpmi-9-0233. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Grant D, Cregan P, Shoemaker RC. Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc Natl Acad Sci USA. 2000;97:4168–4173. doi: 10.1073/pnas.070430597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski SP, Lan T-H, Feldmann KA, Paterson AH. Comparative mapping of Arabidopsis thaliana and Brassica oleraceachromosomes reveals islands of conserved organization. Genetics. 1994;138:499–510. doi: 10.1093/genetics/138.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku H-M, Vision T, Liu J, Tanksley SD. Comparing sequenced segments of the tomato and Arabidopsisgenomes: Large-scale duplication followed by selective gene loss creates a network of synteny. Proc Natl Acad Sci USA. 2000;97:9121–9126. doi: 10.1073/pnas.160271297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetic analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Lagercrantz U. Comparative mapping between Arabidopsis thaliana and Brassica napus indicates that Brassicagenomes have evolved through extensive genome replication accompanied by chromosome fusion and frequent rearrangements. Genetics. 1998;150:1217–1228. doi: 10.1093/genetics/150.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U, Lydiate DJ. Comparative genome mapping in Brassica. Genetics. 1996;144:1903–1910. doi: 10.1093/genetics/144.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Sultana R, Pertea G, Cho J, Karamycheva S, Tsai J, Parvizi B, Cheung F, Antonescu V, White J et al. Cross-referencing eukaryotic genomes: TIGR orthologous gene alignments (TOGA) Genome Res. 2002;12:493–502. doi: 10.1101/gr.212002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D, Kurth J, Laurie DA, Yano M, Sasaki T, Devos K, Graner A, Schulze-Lefert P. Rapid reorganization of resistance gene homologues in cereal genomes. Proc Natl Acad Sci USA. 1998;95:370–375. doi: 10.1073/pnas.95.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sachidanandam R, Stein L. Comparative genomics between rice and Arabidopsisshows scant collinearity in gene order. Genome Res. 2001;11:2020–2026. doi: 10.1101/gr.194501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark. hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K, Murphy G, Tarchini R, Wambutt R, Volckaert G, Pohl T, Dusterhoft A, Stiekema W, Entian KD, Terryn N et al. Conservation of microstructure between a sequenced region of the genome of rice and multiple segments of the genome of Arabidopsis thaliana. Genome Res. 2001;11:1167–1174. doi: 10.1101/gr.161701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menacio-Hautea D, Fatokum CA, Kumar L, Danesh D, Young ND. Comparative genome analysis of mungbean (Vigna radiata L. Wilczek) and cowpea (V. unguiculata) using RFLP analysis. Theor Appl Genet. 1993;86:797–810. doi: 10.1007/BF00212605. [DOI] [PubMed] [Google Scholar]
- Nam Y, Penmetsa RV, Endre G, Uribe P, Kim D, Cook DR. Construction of a bacterial artificial chromosome library of Medicago truncatulaand identification of clones containing ethylene-response genes. Theor Appl Genet. 1999;98:638–646. [Google Scholar]
- O'Neill CM, Bancroft I. Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J. 2000;23:233–243. doi: 10.1046/j.1365-313x.2000.00781.x. [DOI] [PubMed] [Google Scholar]
- Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell. 1997;91:821–832. doi: 10.1016/s0092-8674(00)80470-5. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Burow MD, Draye X, Elsik CG, Jiang CX, Katsar CS, Lan TH, Lin YR, Ming R et al. Comparative genomics of plant chromosomes. Plant Cell. 2000;12:1523–1540. doi: 10.1105/tpc.12.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Lan TH, Reischmann KP, Chang C, Lin YR, Liu SC, Burow MD, Kowalski SP, Katsar CS, DelMonte TA et al. Toward a unified genetic map of higher plants, transcending the monocot-dicot divergence. Nat Genet. 1996;14:380–382. doi: 10.1038/ng1296-380. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Lin Y-R, Li Z, Scherta KF, Doebley JF, Pinson SRM, Liu S-C, Stensel JW, Irvine JE. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science. 1995;269:1714–1717. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- Rossberg M, Theres K, Acarkan A, Herrero R, Schmitt T, Schumacher K, Schmitz G, Schmidt R. Comparative sequence analysis reveals extensive microcolinearity in the lateral suppressor regions of the tomato, Arabidopsis, and Capsella genomes. Plant Cell. 2001;13:979–988. doi: 10.1105/tpc.13.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB. High density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AM, Weeden NF, Martin A. Linkage among isozyme, RFLP, and RAPD markers in Vicia faba. Theor Appl Genet. 1993;85:937–945. doi: 10.1007/BF00215032. [DOI] [PubMed] [Google Scholar]
- van Dodeweerd AM, Hall CR, Bent EG, Johnson SJ, Bevan MW, Bancroft I. Identification and analysis of homoeologous segments of the genomes of rice and Arabidopsis thaliana. Genome. 1999;42:887–892. [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Weeden NL, Muehlbauer FJ, Ladizinsky G. Extensive conservation of linkage relationships between pea and lentil genetic maps. J Hered. 1992;83:123–129. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.