Abstract
Bacterially derived Nod factor is critical in the establishment of the legume/rhizobia symbiosis. Understanding the mechanisms of Nod factor perception and signal transduction in the plant will greatly advance our understanding of this complex interaction. Here, we describe the identification of a new locus, nodulation-signaling pathway 2 (NSP2), of Medicago truncatula that is involved in Nod factor signaling. Mutants at this locus are blocked for Nod factor-induced gene expression and show a reduced root hair deformation response. nsp2 plants also show a complete absence of infection and cortical cell division following Sinorhizobium meliloti inoculation. Nod factor-induced calcium spiking, one of the earliest responses tested, is still functional in these mutant plants. We conclude that the gene NSP2 is a component of the Nod factor signal transduction pathway that lies downstream of the calcium-spiking response.
The symbiotic interaction between legumes and rhizobial bacteria accounts for a significant portion of biological nitrogen fixation worldwide. The site of fixation is the nodule, a unique plant organ located on the root, which functions to generate the aerobic environment essential for bacterial survival and nitrogenase activity. Nodule formation involves plant/bacterial signaling, with the bacterially generated signaling molecule Nod factor playing a critical role (Long, 1996; Downie and Walker, 1999; Oldroyd, 2001).
Purified Nod factor, when applied to the appropriate plant host, can induce many of the plant responses associated with exposure to the bacterial symbiont (Downie and Walker, 1999). Nod factors act predominantly on two cell types in the root: epidermal cells and inner cortical cells. In epidermal cells, Nod factor induces depolarization of the plasma membrane, oscillations in cytosolic Ca2+ referred to as calcium spiking, the induction of specific gene expression, and distortion of polar growth in root hairs (Ehrhardt et al., 1992, 1996; Pichon et al., 1992; Cardenas et al., 2000; Journet et al., 2001). Nod factor also induces mitotic activation of inner cortical cells that ultimately leads to the development of the nodule primordia. The formation of infection threads, that allow the invasion of bacteria into the root cortex, involves Nod factor, but also requires the presence of the rhizobial bacteria, suggesting the possible role of additional bacterial-signaling molecules (Dénarié et al., 1996; Oldroyd, 2001).
Genetic dissection of the Nod factor-signaling pathway has been limited by the availability of a genetically tractable legume system. Medicago truncatula and its symbiotic bacterial partner Sinorhizobium meliloti have been adopted as model organisms for the study of this symbiotic interaction (Cook, 1999). M. truncatula was selected as a model legume for its diploid genetics, relatively small genome, rapid life cycle, and ease of transformation. A number of studies in this species have identified genes critical for the establishment and regulation of the rhizobial symbiosis (Sagan et al., 1995; Penmetsa and Cook, 1997; Catoira et al., 2000, 2001).
Genetic studies in M. truncatula have identified a number of mutants defective in Nod factor signaling (Sagan et al., 1995; Catoira et al., 2000). These mutants fall into four complementation groups. Doesn't make infections (dmi1), dmi2, and dmi3 no longer show root hair deformation, gene expression, or mitotic induction of cortical cells but do show swelling at the tip of root hairs in response to Nod factor. nodulation-signaling pathway (nsp) mutants show root hair deformation and limited gene expression, but are blocked for cortical cell induction in response to Nod factor. Analysis of the calcium-spiking responses to Nod factor in these mutants revealed that dmi1 and dmi2 mutants are blocked for the induction of calcium spiking, whereas dmi3 and nsp mutants are able to induce calcium spiking after Nod factor application (Wais et al., 2000). These results suggest a simple model for Nod factor signaling in which DMI1 and DMI2 act upstream of calcium spiking and DMI3 functions downstream of calcium spiking but upstream of all other Nod factor responses. NSP would then be placed downstream of calcium spiking and root hair deformation but upstream of gene expression and cortical cell division.
Here, we describe the identification of a new complementation group, NSP2, involved in Nod factor signaling. Mutants at this locus are blocked for infection by the bacteria, show reduced nodulation gene expression, and show altered root hair deformation, but are unaffected for the induction of calcium spiking in response to Nod factor. These phenotypes suggest a role for the gene in the transduction of Nod factor signaling and a position in the Nod factor-signaling pathway downstream of calcium spiking.
RESULTS
The Identification of a New Nod− Complementation Group
A genetic screen for nodulation mutants in fast neutron-mutagenized M. truncatula seed identified 10 mutants that were unable to form nodules in the presence of S. meliloti (Nod−). These mutants were identified from five independent pools of M1 plants (C. Starker, L. Smith, G. Oldroyd, J. Doll, and S. Long, unpublished data). Two mutants, 0-2 and 0-4, isolated from separate pools, complemented mutants from all five previously identified Nod− complementation groups, indicating that 0-2 and 0-4 represent new complementation groups. Allelism tests between 0-2 and 0-4 indicated that these two mutants were allelic (C. Starker, L. Smith, G. Oldroyd, J. Doll, and S. Long, unpublished data). To ensure that these tests represented true crosses, a line of 0-4 carrying a β-glucuronidase (GUS) marker construct was used as the pollen donor in a cross to 0-2. The F1 of this cross were Nod− and GUS positive, verifying the previous allelism tests. A segregation ratio for the mutant of 53:18 (2.9:1) in the F2 of a mutant to wild-type cross indicates that the mutation is the result of a single recessive gene/locus. For reasons described below this new gene was called NSP2, with 0-2 defined as nsp2-1 and 0-4 as nsp2-2.
Root Hair and Infection Phenotypes of nsp2-1 and nsp2-2
To assess the degree to which nsp2 mutants were infected, we analyzed mutant plants inoculated with S. meliloti 1021 (pXLGD4), which constitutively expresses LacZ. We examined plants at 3 and 14 d postinoculation and stained for β-galactosidase activity to identify infection events. Wild-type plants showed infection threads containing bacteria at 3 d (Fig. 1E) and infected nodules at 18 d postinoculation (Fig. 2C). nsp2 mutant plants showed no indication of infection at either time point (Figs. 1F and 2D).
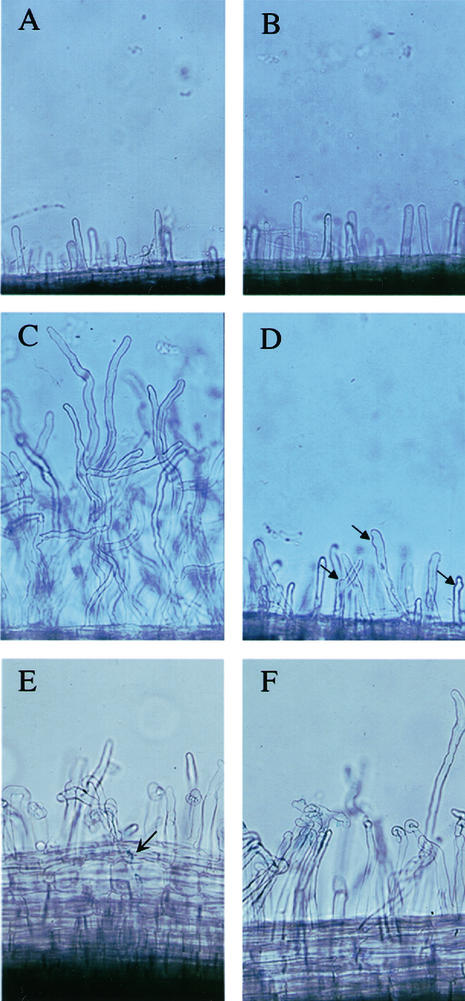
Figure 1.
Nod factor-induced root hair deformation is reduced in nsp2 mutants. Root hairs on wild-type (A) and nsp2-2 (B) plants grown in the absence of Nod factor show no differences. C, Wild-type plants grown in the presence of 10 pM Nod factor show extensive distorted root hair growth and root hair branching. D, This response is reduced in nsp2-2 plants. However root hairs of nsp2-2 still show some deformation, indicated by arrows, that is not present on untreated plants (B). Both wild type (E) and nsp2-2 (F) show root hair deformation when treated with S. meliloti, shown 3 d postinoculation with 1021 (pXLGD4). The arrow indicates an infection event visualized using the β-galactosidase activity of 1021 (pXLGD4). No infections were observed in nsp2 mutants.
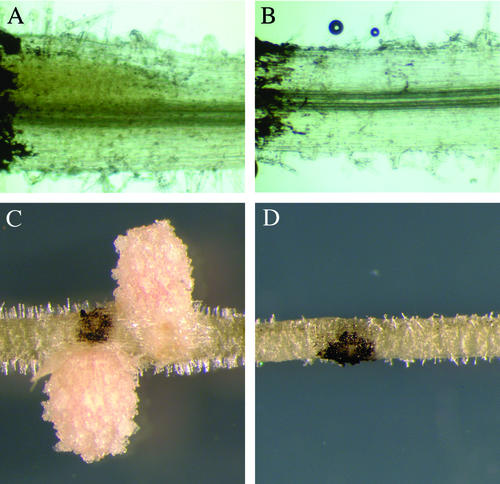
Figure 2.
The mitotic induction of cortical cells by S. meliloti is absent in nsp2. Plants were spot inoculated with S. meliloti and assessed at 3 d postinoculation (A and B) and 18 d postinoculation (C and D). At 3 d postinoculation, roots were stained with 0.1 m potassium iodide that causes a diffuse yellow staining within dividing cortical cells in wild-type plants (A) that is completely absent in nsp2-2 plants (B). C, Nodules form at later time points within the restricted area of the inoculation in wild type; D, no response is apparent at these later time points in nsp2. The dark coloration in all the images is lamp black that was applied to mark the point of inoculation.
Both mutant and wild-type plants showed similar levels of root hair deformation in the previous infection assay (Fig. 1, E and F). However, further analysis revealed differences in root hair deformation in nsp2 plants after Nod factor application. nsp2 mutants show a reduction in the level of root hair growth and deformation in response to Nod factor relative to wild type (Fig. 1, C and D). Untreated root hairs are identical in wild-type and nsp2 mutant plants (Fig. 1, A and B).
Nodulin Gene Expression Is Greatly Reduced in nsp2 Mutants
A number of genes are induced in response to rhizobia and Nod factor. We chose to examine the expression of two such genes RIP1 and ENOD11, as representatives of early nodulin genes (Cook et al., 1995; Journet et al., 2001). We assessed RIP1 expression using northern analysis. In wild-type plants, RIP1 is induced within a few hours of rhizobial inoculation and reaches a maximum expression at 24 h (Fig. 3A). In both alleles of nsp2, RIP1 induction is greatly compromised (Fig. 3A). RIP1 is not induced in nsp2 plants until 48 h postinoculation, and even then, expression levels are low. To assess ENOD11 expression we used transgenic plants in which the ENOD11 promoter drives the expression of GUS (Journet et al., 2001). This transgene was crossed into nsp2-2 plants. F2 plants that were homozygous for both the transgene and the nsp2-2 mutation were assayed for ENOD11 expression in response to both S. meliloti and Nod factor. In wild-type plants carrying the ENOD11-GUS fusion, GUS activity could be seen in the root epidermis behind the root tip at 6, 24, and 48 h post-treatment with either bacteria or Nod factor (Fig. 3B; data not shown). In nsp2-2 plants, no ENOD11-inducible expression could be seen under these conditions, although non-symbiotic root tip expression was observed (Fig. 3B; data not shown).
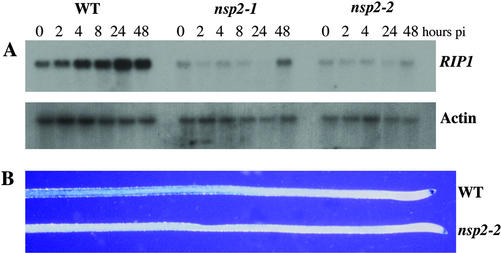
Figure 3.
Early nodulin gene expression is blocked in nsp2 mutants. A, Expression levels of RIP1 were assessed at time points postinoculation with S. meliloti using northern analysis. Wild-type plants show a gradual induction of RIP1 expression that peaks at 24 h postinoculation. nsp2-1 and nsp2-2 show only a slight induction of RIP1 at 48 h postinoculation. pi, Postinoculation. B, Expression of ENOD11 was assessed using a construct that places uidA under the regulation of the ENOD11 promoter. Roots were treated with 1 nm Nod factor for 6 h and then assessed for GUS activity. Both wild type and nsp2-2 show non-symbiotic root cap expression, but only wild type shows the Nod factor-inducible GUS activity in a region behind the root tip.
Calcium Spiking Is Induced in nsp2-2
M. truncatula root hair cells respond rapidly to Nod factor by undergoing cytosolic calcium oscillations, termed calcium spiking (Ehrhardt et al., 1996). Previous characterization of Nod− mutants for calcium spiking has indicated that dmi1 and dmi2 appear to be blocked upstream of calcium spiking, whereas dmi3 and nsp retain their ability to induce calcium spiking (Wais et al., 2000). We assessed the calcium-spiking response of nsp2-2 mutants. We found that calcium spiking was induced in 30 of 34 cells on seven nsp2-2 plants (Fig. 4), with no detectable differences in the calcium-spiking response relative to wild type (data not shown).
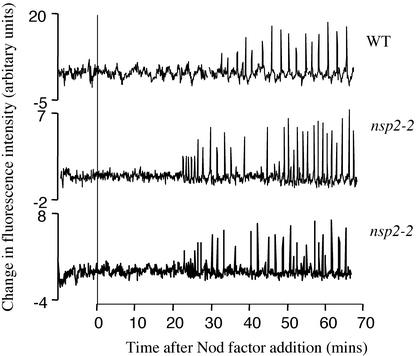
Figure 4.
Calcium spiking is functional in nsp2-2 mutant plants. Individual root hairs of wild-type and nsp2-2 plants were assessed for Nod factor-induced calcium spiking. Each trace represents changes in calcium levels of individual root hair cells as evidenced by fluctuations in the fluorescence of the calcium responsive dye Oregon green. Subtle differences in the lag to induction of spiking following Nod factor application and the period between spikes are not significant when a larger number of cells are compared.
Mitotic Induction of Inner Cortical Cells Is Blocked in nsp2 Mutants
Spot inoculation of S. meliloti onto wild-type M. truncatula resulted in the formation of nodule primordia and ultimately nodules within an isolated region (Fig. 2, A and C). Similar treatments on nsp2 mutants had no apparent effect on the inner cortical cells, as indicated by the absence of any swelling (Fig. 2, B and D). To analyze the early induction of inner cortical cell division, we assessed the roots 3 d postinoculation after potassium iodide staining. In wild-type plants, the dividing region of the cortex at the point of inoculation is apparent from swelling of the root and an inner densely cellular region with diffuse yellow staining (Fig. 2A) that is completely absent in nsp2-2 mutants (Fig. 2B). Small starch granules are visible at higher magnification, however, the diffuse yellow coloration in wild-type plants is most likely a result of iodine staining of densely cytoplasmic cells rather than starch granules (Alison Smith, personal communication). We conclude that cortical cells are not induced for cell division after bacterial application in nsp2 mutants.
NSP2 Is Positioned on Linkage Group 3
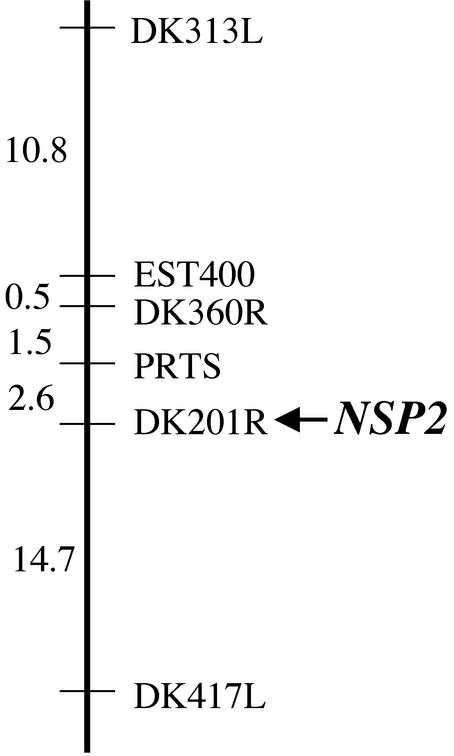
To establish the framework for the positional cloning of NSP2, we identified the map position of the gene. Mapping populations between nsp2-1 and nsp2-2 and the M. truncatula ecotype A20 were established. Twenty-two nsp2 mutant plants from the F2 of this mapping population were analyzed with representative markers from each linkage group. Initial linkage was found with DK313L located on linkage group 3. Further analysis of 94 nsp2 mutant plants from the mapping population indicated that NSP2 was 15.4 cM from DK313L (Fig. 5). These plants were also assessed with other genetic markers surrounding DK313L. Tight linkage was found with marker DK201R (no recombinants identified in the 94 mutant plants).
Figure 5.
NSP2 maps to marker DK201R on linkage group 3. A representation of the significant region of linkage group 3 where linkage to NSP2 was found. Genetic markers are shown with the genetic distances (centiMorgans) between the markers indicated. The genetic distance was assessed by the relative distance between the marker and NSP2.
DISCUSSION
Genetic dissection of nodulation in M. truncatula has identified four genes that appear to have a role in Nod factor signal transduction (Catoira et al., 2000; Wais et al., 2000). Here, we describe the identification of a fifth gene NSP2. Mutations in this gene are blocked in multiple responses to Nod factor, implying a role in Nod factor signaling. The phenotypes of these mutants are very similar to previously described mutants in the complementation group NSP (Catoira et al., 2000). In concordance with this nomenclature, we have chosen to call this new gene NSP2, for nodulation-signaling pathway 2, and hence the gene previously called NSP, should be renamed NSP1.
Plants carrying mutations in NSP2 show defects in the Nod factor induction of root hair deformation and gene expression. Furthermore, these mutants are completely blocked for the formation of infection threads and cortical cell division. However, the induction of calcium spiking, the earliest response analyzed here, appears to be functional in these mutants. This suggests that the gene lies downstream of calcium spiking in Nod factor signal transduction, but upstream of gene expression. The fact that the mutants appear to induce root hair deformation, albeit much reduced after Nod factor application, implies a more complex role in the induction of this response. It is possible that this gene lies upstream of root hair deformation but that there is some level of genetic redundancy for this response.
A comparison of mutant phenotypes between nsp1 and nsp2, although very similar, does reveal some differences. nsp1 mutants were reported to be completely functional for the induction of root hair deformation (Catoira et al., 2000). However, when nsp1-1 was tested in the assay used here for Nod factor-induced root hair deformation, a response similar to nsp2 mutants was observed: The nsp1-1 mutant showed much reduced root hair deformation compared with wild-type plants (data not shown). It appears that the mode of Nod factor presentation is important, and the assay used here reveals a defect in the ability of nsp1 and nsp2 mutants to induce root hair deformation. The induction of gene expression after Nod factor application is slightly different in nsp1 and nsp2 mutants. The nsp1 mutants show reduced RIP1 and ENOD11 expression (Catoira et al., 2000), whereas nsp2 mutants show a complete block in ENOD11 expression and greatly reduced RIP1 expression. The gene expression profiles of nsp2 are more similar to those of the dmi mutants than the nsp1 mutant (Catoira et al., 2000). However, these gene expression differences between nsp1 and nsp2 could be explained by the severity of the mutant alleles.
It is possible that the NSP1 and NSP2 genes function at similar or parallel positions in the Nod factor signal transduction pathway. It is also possible they are genetically redundant to each other for the induction of root hair deformation. We are currently generating plants that carry mutations in both to assess the redundancy of these two genes for the root hair deformation response.
The fast neutron mutagenesis screen performed here identified a number of Nod− mutants. Two of these were in the previously characterized complementation group dmi1 (C. Starker, L. Smith, G. Oldroyd, J. Doll, and S. Long, unpublished data). However, two mutants represented a new complementation group not previously identified NSP2. Furthermore, a number of complementation groups in Nod factor-signaling mutants are represented by single or few alleles (Catoira et al., 2000). This suggests that the nodulation screens have not yet saturated the Nod factor signal transduction pathway. More thorough genetic screens, or alternative modes of screening should identify new genes involved in this signaling pathway.
The development of model legume systems has allowed work to progress toward understanding the Nod factor-signaling pathway at the molecular and biochemical levels. Here, we describe the identification of a new gene involved in Nod factor signal transduction. Further studies with NSP2, particularly in understanding its molecular identity, will greatly advance our understanding of the Nod factor signal transduction pathway and its role in the development of the complex interaction between legumes and their symbiotic bacterial partners.
MATERIALS AND METHODS
Plant Growth Conditions and Bacterial Strains
Sterilized seedlings germinated overnight were plated onto buffered nodulation media (BNM; Ehrhardt et al., 1992) with 12 g/L agar and 0.1 μm l-α-(2-aminoethoxyvinyl)-Gly and were allowed to grow for a minimum of 3 d before inoculation. A17 cv Jemalong was used as the wild type. Sinorhizobium meliloti strain Rm1021 was grown in liquid Luria Broth overnight in the presence of the appropriate antibiotic. Bacteria were pelleted at 14,000 rpm for 10 min and resuspended in 10 mm MgSO4. This bacterial suspension was diluted 1/50 in 10 mm MgSO4 and inoculated onto plants by flooding the roots with the inoculum. Nod factor preparations were isolated as described by Ehrhardt et al., 1996.
Assessment of Nodulation and Infections
Analysis of infection was performed using the S. meliloti strain Rm1021 (pXLGD4) that constitutively expresses lacZ (Penmetsa and Cook 1997). At 3 and 12 d postinoculation, roots were fixed for 1 h in 50 μL mL− gluteraldehyde and 200 mm sodium cocadylate, washed twice in 200 mm sodium cocadylate, and stained overnight in 200 mm sodium cocadylate, 5 mm potassium ferrocyanide, 5 mm potassium ferricyanide, and 0.8 g L−1 X-gal. Roots were destained in a 1:5 dilution of commercial bleach for 5 min and imaged using an Optiphot microscope (Nikon, Tokyo). Spot inoculations were performed by placing a small drop, approximately 0.2 μL, of bacterial suspension onto the surface of the root using pulled pipette tips. The point of inoculation was marked using lamp black. Analysis of cortical cells at 3 d postinoculation was assessed on a Nikon Optiphot microscope following staining with 0.1 m potassium iodide. Roots were first cleared in 1:5 dilution of commercial bleach for 5 min and then stained in 0.1 m potassium iodide for 2 min.
Root Hair Deformation Assay
To assess the deformation of root hairs, plants were sandwiched between two thin slabs of BNM media containing 12 g L−1 agar and 0.1 μm l-α-(2-aminoethoxyvinyl)-Gly and in some cases 10 pM Nod factor. Plants were allowed to grow through the media slabs for up to 4 d. Root hairs at equivalent positions on the root were imaged using a Nikon Diaphot inverted microscope (Technical Instruments, San Francisco) set for Nomarski differential interference contrast optics and a Nikon 10× fluor objective.
RNA Gel-Blot Analysis
Plants were grown on BNM and inoculated with S. meliloti at an OD600 of 0.1. At set time points postinoculation, roots from 15 plants were isolated, frozen in liquid nitrogen, and stored at −80°C. RNA was isolated using TRIZOL Reagent (Invitrogen, Carlsbad, CA), according to the manufacturer's protocol. RNA samples were separated on a 12 g L−1 agarose gel containing 62 mL L−1 formaldehyde. The RNA was transferred to a nylon membrane (Hybond N, Amersham Biosciences UK, Ltd, Little Chalfont, Buckinghamshire, UK) and hybridized with a 280-bp fragment from exon 2 of RIP1 (Cook, Dreyer et al., 1995) and secondarily with a 720-bp fragment that represents a full-length cDNA clone of an actin homolog identified from the M. truncatula expressed sequence tag database (Covitz et al., 1998).
The GUS Assay
The ENOD11-GUS construct (Journet et al., 2001) was introduced into nsp2-2 plants by crossing with wild-type plants carrying the construct. Nod− plants were identified in the F2 of this cross and analyzed for the non-symbiotic ENOD11-GUS expression at the root tip. Sixteen progeny from the Nod−/GUS+ plants were assessed for GUS staining. If all 16 plants were GUS+, then the parent plant was considered homozygous for the ENOD11-GUS construct. nsp2-2, ENOD11-GUS plants were grown on media for 4 d. The plants were then transferred to liquid media containing 1 nm Nod factor for 6 h. The plants were fixed in 3 mL L−1 formaldehyde and 0.1 m potassium phosphate, pH 7.0, for 1 h on ice. GUS staining was performed overnight at 37°C with 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 5 mm EDTA, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, and 0.1 m potassium phosphate, pH 7.0.
Calcium-Spiking Experiments
Analysis of calcium spiking was performed as described by Ehrhardt et al. (1996), with slight modifications as described by Wais et al. (2000). Seedlings were grown overnight on BNM. The tip of the root was removed to avoid disturbances caused by growth of the root during the imaging, and then the plant was transferred to a liquid media bath on a large coverslip. Three to eight root hairs were injected with the calcium-sensitive dye Oregon green-dextran (Molecular Probes, Eugene, OR). Root hairs on two or three plants were injected for each experiment. Cells were allowed to recover from the injections for approximately 30 min, before imaging. Root hair cells were imaged for 10 min in the absence of Nod factor, and then Nod factor was added to the bath to a concentration of 1 nm. The cells on the first plant were imaged for 60 min after the addition of Nod factor. After this 60-min period, the cells on the second plant in the experiment were imaged. To aid in the identification of spiking, the raw fluorescence data was transformed by the equation Y = X(n+1) − Xn, which accentuates rapid changes in the calcium levels. Root hair cells that spike within approximately 60 min after the addition of Nod factor were considered positive for calcium spiking.
ACKNOWLEDGMENTS
We thank Raka Mitra for critically reading the manuscript and Cindy Smith for all of her help. We also thank David Barker for providing ENOD11-GUS plants before publication.
Footnotes
This work was supported in part by the Howard Hughes Medical Institute and the Department of Energy (grant no. DE–FG03–90ER20010).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.010710.
LITERATURE CITED
- Cardenas L, Moldaway-Clarke TL, Sanchez F, Quinto C, Feijó JA, Kunkel JG, Mepler PK. Ion changes in legume root hairs responding to Nod factor. Plant Physiol. 2000;123:443–451. doi: 10.1104/pp.123.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Denarie J. Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell. 2000;12:1647–1665. doi: 10.1105/tpc.12.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Timmers AC, Maillet F, Galera C, Penmetsa RV, Cook D, Denarie J, Gough C. The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development. 2001;128:1507–1518. doi: 10.1242/dev.128.9.1507. [DOI] [PubMed] [Google Scholar]
- Cook D, Dreyer D, Bonnet D, Howell M, Nony E, VandenBosch K. Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell. 1995;7:43–55. doi: 10.1105/tpc.7.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR. Medicago truncatula: a model in the making! Curr Opin Plant Biol. 1999;2:301–304. doi: 10.1016/s1369-5266(99)80053-3. [DOI] [PubMed] [Google Scholar]
- Covitz PA, Smith LS, Long SR. Expressed sequence tags from a root-hair-enriched Medicago truncatula cDNA library. Plant Physiol. 1998;117:1325–1332. doi: 10.1104/pp.117.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dénarié J, Debelle F, Prome JC. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- Downie JA, Walker SA. Plant responses to nodulation factors. Curr Opin Plant Biol. 1999;2:483–489. doi: 10.1016/s1369-5266(99)00018-7. [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Atkinson EM, Long SR. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science. 1992;256:998–1000. doi: 10.1126/science.10744524. [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell. 1996;85:673–681. doi: 10.1016/s0092-8674(00)81234-9. [DOI] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V. Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant-Microbe Interact. 2001;14:737–748. doi: 10.1094/MPMI.2001.14.6.737. [DOI] [PubMed] [Google Scholar]
- Long SR. Rhizobium symbiosis: Nod factors in perspective. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED. Dissecting symbiosis: developments in Nod factor signal transduction. Ann Bot. 2001;87:709–718. [Google Scholar]
- Penmetsa RV, Cook DR. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science. 1997;275:527–530. doi: 10.1126/science.275.5299.527. [DOI] [PubMed] [Google Scholar]
- Pichon M, Journet E-P, Dedieu A, de Billy F, Truchet G, Barker DG. Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell. 1992;4:1199–1211. doi: 10.1105/tpc.4.10.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan M, Morandi D, Tarenghi E, Duc G. Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn.) after γ-ray mutagenesis. Plant Science. 1995;111:63–71. [Google Scholar]
- Wais RJ, Galera C, Oldroyd G, Catoira R, Penmetsa RV, Cook D, Gough C, Denarie J, Long SR. Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc Natl Acad Sci USA. 2000;97:13407–13412. doi: 10.1073/pnas.230439797. [DOI] [PMC free article] [PubMed] [Google Scholar]