Abstract
Nodulin genes are specifically expressed in the nitrogen-fixing root nodules. We have identified a novel type of DNA-binding protein (CPP1) interacting with the promoter of the soybean leghemoglobin gene Gmlbc3. The DNA-binding domain of CPP1 contains two similar Cys-rich domains with 9 and 10 Cys, respectively. Genes encoding similar domains have been identified in Arabidopsis thaliana, Caenorhabditis elegans, the mouse, and human. The domains also have some homology to a Cys-rich region present in some polycomb proteins. The cpp1 gene is induced late in nodule development and the expression is confined to the distal part of the central infected tissue of the nodule. A constitutively expressed cpp1 gene reduces the expression of a Gmlbc3 promoter–gusA reporter construct in Vicia hirsuta roots. These data therefore suggest that CPP1 might be involved in the regulation of the leghemoglobin genes in the symbiotic root nodule.
The symbiosis between legumes and the nitrogen-fixing Rhizobia bacteria results in the formation of a new organ (root nodule) on the roots of the legume. In this organ, the bacteria convert dinitrogen to ammonia. In soybean, it takes 12–14 days from the infection of the root until the nodule is fully functional and nitrogen fixation starts (1). During this time, a number of plant genes are specifically activated (2). Some of these genes are expressed shortly after the infection by the bacteria. The products encoded by these genes, early nodulins, are most likely involved in the formation of the nodule structure. After several days, other proteins (late nodulins) appear which primarily are involved in the metabolic activities in the nitrogen-fixing root nodule. The most predominant late nodulins are the leghemoglobins (Lbs) which constitute about 5% of the total protein content in the mature nodule. Lb facilitates oxygen transport to the respiring bacteria within the central infected zone of the nodule. In soybean, there are four sequentially expressed lb genes of which the Gmlbc3 gene is the first one to be activated (1). The Gmlbc3 gene transcript was detected in root nodules 8 days after infection and the expression remained low until a dramatic increase in the expression occurred about 12–14 days after infection. The high-level expression of the lb genes is confined to the cells in the central infected zone of the nodule.
Only a limited number of transcription factors serving a function in root nodule formation and function have been identified. Two MADS-box-containing genes, nmh5 and nmh7, from Medicago sativa are expressed in the root nodules and these two putative transcription factors might therefore be involved in the regulation of nodule-expressed genes (3, 4). Two AT-rich sequence motifs in the soybean Gmlbc3 promoter interact with a nuclear factor NAT2, present in soybean root nodules (5, 6). NAT2-binding activities are also present in Sesbania rostrata nodules, roots, and leaves (7). Binding sites for NAT2-like proteins are also present in the S. rostrata glbc3 promoter and the promoter of the nodule-enhanced gln-γ gene from the French bean (7, 8). Functional studies of the NAT2-binding sites in the soybean Gmlbc3 gene demonstrated that these sites are general cis-elements (9). Thus, NAT2 is most likely a general activator of transcription and is not directly involved in the specific activation of the lb genes.
To identify DNA-binding proteins regulating the lb genes, a soybean nodule λgt11 cDNA expression library was screened by using oligonucleotides covering the proximal promoter region from the soybean Gmlbc3 gene as probes. One of the isolated cDNA clones encoded a novel DNA-binding protein (CPP1), which binds to the Gmlbc3 promoter. In the nodule, cpp1 and lb transcripts appear in the same tissue, but in different locations. A plasmid expressing CPP1 was able to repress the expression of a Gmlbc3–gusA reporter construct in transgenic vetch roots. These data suggest that CPP1 is able to down-regulate the expression of a lb gene. CPP1 contains a region similar to a domain present in some polycomb group proteins which are known to suppress gene expression of developmentally important regulatory genes.
Materials and Methods
Fusion Protein Purification and Plasmid Constructs.
A cDNA clone, pcpp1418–896 encoding amino acids 418–896 was isolated from a soybean nodule cDNA expression library in the same way as Gmndx (10). A subfragment of pcpp1418–896, corresponding to amino acids 456–596 was subcloned into the pGEX-5X-3 expression vector (Amersham Pharmacia). The CPP1 peptide was expressed as a glutathione S-transferase fusion protein (11) and the glutathione S-transferase domain was removed by using factor Xa.
The full-length Gmlbc3 promotor used for the trans-activation experiment was a 2-kb fragment (12). The gusA reporter gene was interrupted by the second intron (IV2) of the potato ST-LSl gene (13). The CaMV 35S promoter originated from the −829 promoter (14). The coding region of cpp1 constituted a fragment from 59 bp upstream to the initiator AUG to 243 bp downstream from the stop codon.
The gfp–cpp1 gene fusion was made in pZP211 (15) by fusing the CaMV 35S promoter to the full-length cpp1 cDNA with an in-frame green fluorescent protein (GFP)-encoding fragment modified from mRS-GFP (16). The control plasmid was identical to the latter plasmid except that the cpp1 sequence was deleted.
Electrophoretic Mobility Shift Assays.
A bacterial-expressed CPP1 peptide (amino acids 456–596) was incubated with a 32P-labeled Gmlbc3 promoter fragment (−284 to +44) in 20 mM Tris⋅HCl (pH 7.5)/0.5 mM DTT/0.1% Nonidet P-40/6% glycerol/630 mM KCl, in the presence of 0.5 μg of BSA and 0.5 μg of salmon sperm DNA at 25°C for 60 min. Control DNA fragments: a 368-bp fragment from the pPZP211 vector (GenBank accession no. V10490, nucleotides 8650–0004), and a 188-bp fragment of the CaMV 35S promoter (GenBank accession no. V00141, nucleotides 6,622–6,810). The complexes were separated in a native 4% polyacrylamide gel in 0.1 M Tris-borate buffer (pH 8.2).
RNase Protection.
The probe used for the detection of cpp1 transcripts was obtained from pcpp1418–896. The ribonuclease protection assay was performed as described (17).
In Situ Hybridization.
The plasmid pCPP1418–896 was used as a template for runoff transcription. Antisense of cpp1 was obtained by using a SP6 polymerase after digestion with EcoRI. Sense cpp1 was made by using a T3 polymerase after digestion with HindIII. The sections were made from 21-day-old soybean nodules. The antisense and sense probes were radioactively labeled with [35S]UTP and degraded to a length of about 150 nt before hybridization (18, 19). Sections were stained with 0.1% toluidine blue and mounted with DPX.
Nuclear Localization of CPP1.
A control plasmid expressing only GFP and a plasmid expressing CPP1-GFP was introduced and analyzed in onion epidermal cells (20).
Transformation of Vicia hirsuta.
The constructs were inserted into the binary vector pPZP211 (15). The vectors were electrotransformed into the Agrobacterium rhizogenes strain ARqual, which mediates the generation of transgenic hairy roots (21). To inoculate these roots with Rhizobium leguminosarum bv. viciae, the wild-type seed and roots were removed from the seedlings and composite plants were subcultured and infected with the bacteria (21). Plant material was cultured at 18°C 16 h light/8 h dark. Inoculated roots harvested 16 days after infection were used for the qualitative histochemical β-glucuronidase assay by using X-gluc and the quantitative fluorometric assay by using MUG as the enzyme substrates (14).
Results
CPP1 Contains Two Cys-Rich Repeats.
A cpp1 cDNA (gmN14) was isolated from a soybean nodule expression library (22). The full-length sequence of cpp1 was determined from the sequence of the isolated cDNA clone combined with the sequence of a 5′ rapid amplification of cDNA ends product (GenBank accession no. AJ010165). An ORF-encoding 896 aa was identified in cpp1 (Fig. 1A). It is likely that this ORF corresponds to the full-length product encoded by the cpp1 gene, because an in-frame stop codon is located 27 bases upstream of the potential initiator AUG in the rapid amplification of cDNA ends product.
Figure 1.
(A) Complete aa sequence of CPP1. The two Cys-rich regions are highlighted in black. A conserved aa sequence is highlighted in gray. The numbers refer to the aa numbers starting from the initiator Met. (B) Alignment of the two Cys-rich regions (C1 and C2). The positions of the first and last aa are indicated. Identical aa are highlighted in black and similar aa are indicated by a gray background.
The encoded aa sequence contains two Cys-rich motifs of 34 and 35 aa located at positions 473–506 (C1) and between 559 and 593 (C2), respectively (Fig. 1B). C1 and C2 are 57% identical with the highest degree of similarity in the basic N-terminal region. All nine Cys present in C2 are conserved in C1. C1 has an additional Cys and by introducing a gap of 1 aa, this residue matches a His in C2 (Fig. 1B). Six of the Cys form CXC motifs. A homology search in the European Molecular Biology Laboratory database identified six genomic A. thaliana sequences on chromosomes 2, 3, and 4, respectively (accession nos. CAB43914, AB012247, AB022223, AAD24386, and Z97337), a genomic sequence from Caenorhabditis elegans (accession no. Z82274), tesmin genes from mouse and human (accession nos. U86074 and U67176), and a KLP3A-encoding gene from Drosophila melanogaster (L19117). In addition, a Cys-rich region present in some polycomb proteins, e.g., CURLY LEAF from A. thaliana (accession no. Y10580) and E(z) from D. melanogaster (accession no. U00180), showed some similarity to CPP1. An alignment of the CPP1 aa sequence to the indicated sequences showed that the Cys-rich domains are highly conserved in most of the sequences (Fig. 2). The N-terminal part of each Cys-rich motif which consists mainly of basic aa appears to be more conserved than the C-terminal part of the domains. The CPP1 aa sequence, RNPLAFAPK, located between the two Cys-rich domains and a basic region N-terminal to C1, is also conserved between the sequences, except for the polycomb proteins (Fig. 2). In the latter, the region between the Cys motifs appears to be absent. Apart from the conserved regions, no other sequence similarities were detected.
Figure 2.
Alignment of the CPP1 aa sequence to the following sequences: Translated genomic DNA sequences from A. thaliana and C. elegans. The names include the GenBank accession nos. Introns have been excised from the genomic sequences at appropriate positions. Tesmins from human (Hs) and mouse (Mm) and polycomb group proteins from Drosophila (Dm) and Arabidopsis (At). Identical aa are highlighted in black and similar aa are highlighted in gray.
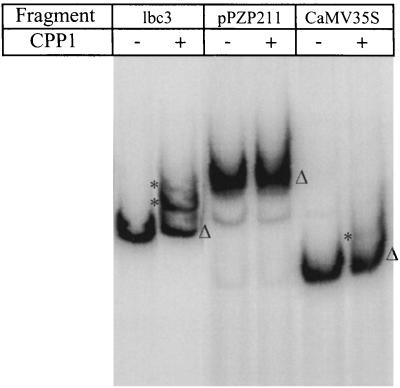
To investigate the DNA-binding properties of CPP1, a peptide containing the two Cys-rich motifs was expressed in Escherichia coli. After purifying the protein, an electrophoretic mobility shift assay was performed, by using a probe spanning from −284 to +44 of the Gmlbc3 promoter. This region contains the important regulatory elements WPE, OSE, and NE (12). Two distinct retarded complexes were observed in the presence of the CPP1 peptide (Fig. 3, lbc3), whereas control DNA fragments showed no or very little interaction with CPP1 (Fig. 3, pPZP211 and CaMV35S). Thus, the region containing the two Cys-rich motifs, constitutes a DNA-binding domain in CPP1 and the binding appears to require specific DNA motifs.
Figure 3.
Binding of a CPP1 subdomain to the Gmlbc3 promoter. A bacterial-expressed CPP1 peptide (amino acids 456–596) was incubated with a 32P-labeled Gmlbc3 promoter fragment (lbc3), a DNA fragment from pPZP211 (pPZP211), and a promoter fragment from CaMV35S (CaMV35S), respectively. The complexes were separated in a native 4% polyacrylamide gel. (−) No CPP1 protein. (▵) Free fragments and (*) DNA–protein complexes.
CPP1 Is Induced Late in Nodule Development.
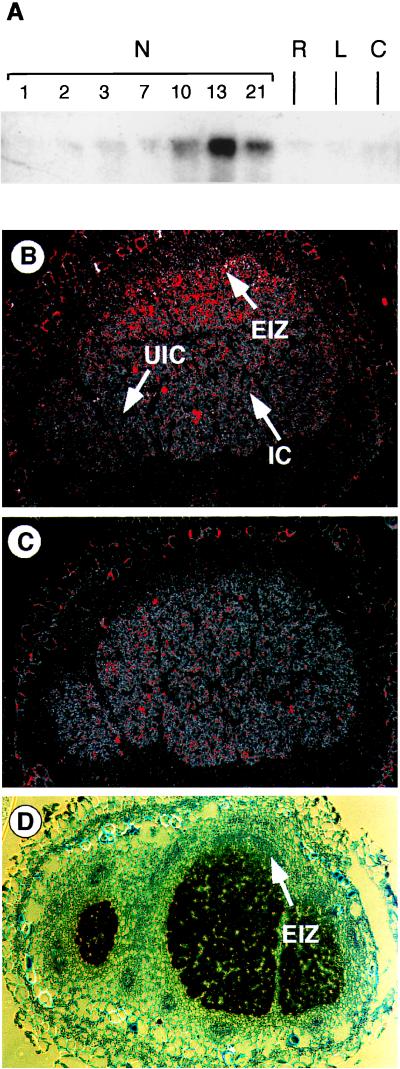
The expression of cpp1 was studied by RNase protection analysis. Identical amounts of RNA were used for each reaction as measured by an ethidium bromide-stained gel. In root nodules, the cpp1 transcripts appeared in significant amounts 10 days after the infection, reaching a maximum 3 days later (Fig. 4A). Subsequently, the level of the transcripts declines. In root or callus, cpp1 was expressed at low levels. Thus, the cpp1 gene is induced late in root nodule development.
Figure 4.
Expression analysis of cpp1. (A) RNase protection analysis by using a transcript of cpp1 corresponding to amino acid numbers 757–811 as a probe. The RNA used in this analysis was isolated from uninfected soybean roots (R), leaves (L), callus (C), and nodules harvested 1–21 days after infection by Bradyrhizobium japonicus (N1–N21). (B) In situ hybridization with cpp1 antisense, (C) cpp1 sense, and (D) Gmlbc3 antisense transcripts as probes to cross sections of a 13-day-old soybean nodule. Infected cells (IC), uninfected cells (UIC), and early infection zone (EIZ) are indicated. The silver grains were visualized by epipolarization microscopy through a red filter (B and C) or directly by bright-field microscopy (D). The images were adjusted in Adobe photoshop to enhance the weak red color.
The expression of cpp1 is confined to the infected region of the soybean root nodules as shown by the sensitive epipolarization image of an in situ hybridization experiment (Fig. 4B). The level of expression is higher in the distal part of the nodule, whereas almost no expression is observed at the base of the nodule. The expression of Gmlbc3 is similarly restricted to the infected region, but in contrast to cpp1, the expression level is low at the tip and high from the center toward the base of the nodule (Fig. 4D). The level of Gmlbc3 expression was very high in comparison to cpp1 expression and could therefore easily be visualized by bright-field microscopy. In conclusion, the cpp1 and Gmlbc3 genes are expressed in the central infected region, but their expression appears to be mutually exclusive in this region.
CPP1 Is Localized in the Nucleus.
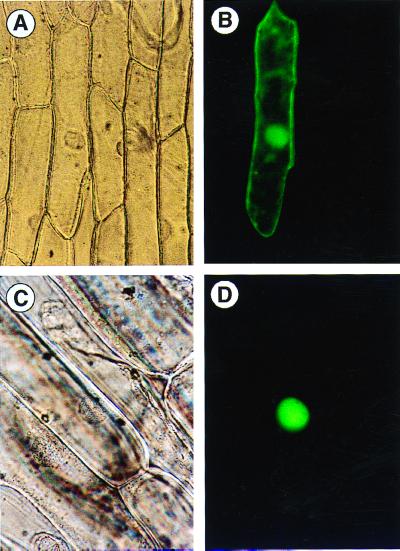
To investigate the cellular localization of CPP1, the coding region of cpp1 was fused in-frame to the GFP-coding sequence. The expression of the gene construct was driven by a constitutively expressed CaMV 35S promoter. A construct without the cpp1-coding region served as a control. The two constructs were delivered into an onion epidermal cell layer by a particle inflow gun, and after an overnight incubation, the cells were inspected by fluorescence microscopy. The control plasmid gave rise to a uniform distribution of the GFP within the cell (Fig. 5 A and B), whereas the in-frame fusion to cpp1 resulted in a strictly nuclear localization of the GFP fusion protein (Fig. 5 C and D). This implies that CPP1 is located in the nucleus.
Figure 5.
Nuclear localization of CPP1. (A) Onion epidermal cell expressing GFP viewed by bright-field microscopy. (B) Same as A but viewed by epifluorescence microscopy. (C) Onion epidermal cell expressing CPP1–GFP fusion protein viewed by bright-field microscopy. (D) Same as C but viewed by epifluorescence microscopy.
CPP1 Represses the Expression of a Leghemoglobin Gene in Vetch Roots.
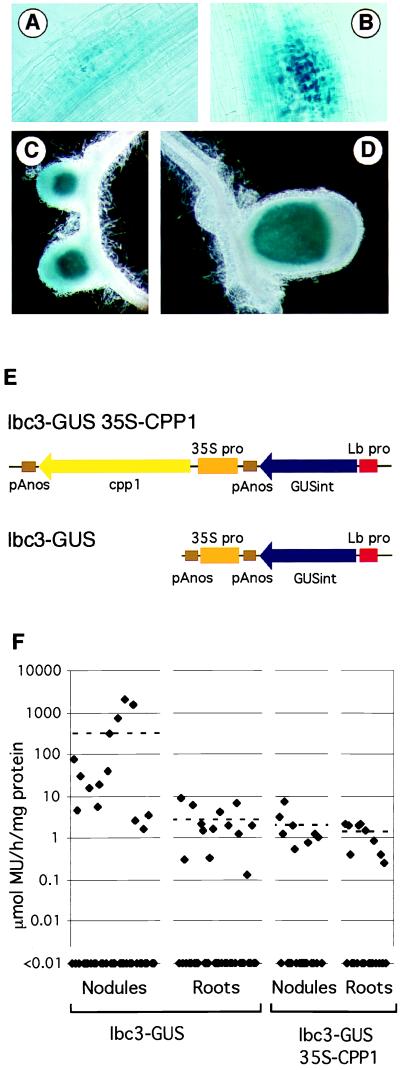
CPP1 interacts in vitro with a promoter fragment of the Gmlbc3. To investigate the function of CPP1, the expression of a Gmlbc3–gusA fusion was studied in transgenic V. hirsuta roots in the presence of a constitutively expressed cpp1 gene. The Gmlbc3 promoter is specifically expressed in Lotus corniculatus root nodules (12). A 2-kb Gmlbc3 promoter was fused to a gusA reporter gene. A CaMV 35S promoter was placed upstream to the Gmlbc3–gusA reporter gene (Fig. 6E, lbc3-GUS). The construct was transformed into V. hirsuta by using A. rhizogenes, and the emerging transgenic hairy roots were inoculated with Rhizobium leguminosarum bv. viciae. As observed (12), the Gmlbc3 promoter was active in young and mature nodules (Fig. 6 C and D). However, β-glucuronidase (GUS) activity was also detected in nodule primordia (Fig. 6 A and B). In contrast to the expression pattern observed in L. corniculatus, a few V. hirsuta roots also showed a low level of GUS activity in the vascular tissue. To regulate the expression of the Gmlbc3 promoter, the coding region of cpp1 was fused to the CaMV 35S promoter upstream to the Gmlbc3–gusA reporter gene. The orientation of the two chimeric genes are shown in Fig. 6E. After transformation into V. hirsuta, 40 independent roots with the control construct were obtained in addition to 23 roots with the cpp1-coding region containing construct. To quantify GUS expression from the constructs, visible nodules were dissected from the roots and GUS activity was determined by a fluorometric assay for each individual plant of the dissected nodules and the remaining roots which contained also small nodules and nodule primordia. The fluorometric values related to the amount of protein in the extracts as determined by Coomassie blue staining (23) are shown in Fig. 6F. A number of plants had very low GUS activities (below 0.01 μmol/h per mg protein) probably because the plants only contained the T-DNA from the Ri-plasmid. These plants were excluded when calculating the mean values. The mean values of the control plants were 118 μmol MU/h per mg protein in nodules and 1.2 units in the root fraction. However, when the CPP1 was present, the expression in nodules was reduced more than 100-fold to a mean value of 0.7 units. The expression in the roots was only slightly reduced to a mean value of 0.5 units. A lbc3-GUS 35S-CPP1 construct in which the lbc-GUS was inverted gave essentially the same results (data not shown). These data show that the presence of the constitutively expressed cpp1 gene leads to a more than a 100-fold reduction in expression of the Gmlbc3 promoter in nodules. This indicates that CPP1 functions as a repressor of the soybean Gmlbc3 gene.
Figure 6.
Regulation of a Gmlbc3 promoter in V. hirsuta roots by CPP1. (A–D) Histochemical GUS staining of V. hirsuta nodule primordia, roots, and nodules. Different developmental stages of nodules from plants transformed with a Gmlbc3 promoter fused to gusA: (A and B) nodule primordia; (C) young nodules; and (D) a mature nodule. (E) Schematic presentation of the lbc3–GUS and lbc3–GUS 35S-CPP1 constructs. (F) Fluorometric measurement of GUS activity expressed from a Gmlbc3 promoter in the absence of CPP1 (lbc3–GUS) or in the presence of CPP1 (lbc3–GUS 35S-CPP1). The GUS activities determined in nodules and roots for each individual plant are indicated by diamonds on a logarithmic scale. Dashed lines indicate the mean values.
Discussion
CPP1 Contains a Cys-Rich DNA-Binding Domain.
Here, we report the identification and characterization of a DNA-binding protein CPP1 which consists of 896 aa with two similar Cys-rich domains, C1 and C2. The amino acid sequences of the two motifs are very similar in the N-terminal basic region, whereas only the Cys residues are conserved in the C-terminal region. Six A. thaliana genomic sequences and a sequence from C. elegans showed a high similarity to the two Cys-rich domains of CPP1. All but one Cys residue are conserved between all these sequences and also noncysteine residues are conserved in the N-terminal part of both C1 and C2. The sequence alignment also revealed a basic stretch of aa located N-terminal to C1 in several of the peptides and a highly conserved short aa sequence (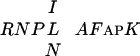 ) between C1 and C2. The sequences outside these conserved regions do not show any significant similarities. All of the identified A. thaliana and C. elegans sequences contain both C1 and C2, as well as the
) between C1 and C2. The sequences outside these conserved regions do not show any significant similarities. All of the identified A. thaliana and C. elegans sequences contain both C1 and C2, as well as the 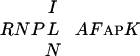 sequence. This region is able to bind DNA and we name this novel DNA-binding domain CRC (
sequence. This region is able to bind DNA and we name this novel DNA-binding domain CRC (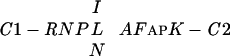 ).
).
A family of human and murine genes (tesmins) also encodes a region showing high similarity to the Cys-rich region of CPP1, including the conserved 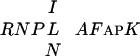 sequence (24). However, the C1 region in the tesmins is truncated in the conserved N-terminal part. The tesmins are believed to play a role in the early events of male germ cell differentiation, because in mice, the transcripts are restricted to spermatocytes undergoing meiosis.
sequence (24). However, the C1 region in the tesmins is truncated in the conserved N-terminal part. The tesmins are believed to play a role in the early events of male germ cell differentiation, because in mice, the transcripts are restricted to spermatocytes undergoing meiosis.
A region in the kinesin-like protein KLP3a contains nine Cys in a pattern similar to one found in the C2 motif of CPP1 (25). Similarity is also observed to a few noncysteine residues in C2, as well as to three conserved residues in the 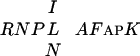 sequence. No region similar to C1 is present in KLP3a. Mutations in the KLP3A gene cause male and female sterility in Drosophila melanogaster. The female sterility is probably caused by an arrest of the male and female pronuclei preventing them from coming together. An additional effect of a KLP3A mutation is aberrations in meiosis and mitosis indicating a role of KLP3A in spindle function (25). Thus, both KLP3A and tesmins appear to control the development of the reproductive organs. However, no molecular information is available about the functions of tesmin and KLP3A.
sequence. No region similar to C1 is present in KLP3a. Mutations in the KLP3A gene cause male and female sterility in Drosophila melanogaster. The female sterility is probably caused by an arrest of the male and female pronuclei preventing them from coming together. An additional effect of a KLP3A mutation is aberrations in meiosis and mitosis indicating a role of KLP3A in spindle function (25). Thus, both KLP3A and tesmins appear to control the development of the reproductive organs. However, no molecular information is available about the functions of tesmin and KLP3A.
Some polycomb group proteins like Enhancer of Zeste from Drosophila and CURLY LEAF from A. thaliana also contain a region with similarity to C1 and C2 (26, 27). In CURLY LEAF, the positions of 8 of the 10 Cys are conserved in C1 and similarly 7 of 9 Cys are conserved in C2. However, in these polycomb proteins, the C1 and C2 regions are located adjacent to each other in contrast to CPP1 in which approximately 60 aa separate C1 and C2. Polycomb group gene products are responsible for the maintenance of a silent stage of repressed developmentally important regulatory genes including the homeotic genes (28). This is normally achieved through an interaction with DNA-binding proteins which bind to chromatin in the regions of the target genes. However, one example of a DNA-binding polycomb group protein has been reported, namely Pleiohomeotic (PHO) from Drosophila (29). PHO contains a DNA-binding zinc-finger domain similar to the one present in the mammalian YY1 transcription factor (30).
CPP1 Is a Potential Repressor of Leghemoglobin Gene Expression.
Repression of a minimal Gmlbc3 promoter in vetch root nodules by a constitutively expressed cpp1 suggests that CPP1 is involved in the expression of lb genes in the nodule. Nevertheless, it still cannot entirely be ruled out that a high level of CPP1 is toxic and we therefore select for integration events in relatively silent regions when cpp1 is present on the construct, resulting in a lower expression level of the gusA gene. However, the reduced expression of gusA in nodules is not observed in the corresponding roots. Furthermore, overexpressing CPP1 in transgenic Lotus japonicus plants does not impair proper root or nodule development (data not shown). For these reasons, we consider it highly unlikely that the observed results are caused by a toxic effect of CPP1.
In all legumes analyzed so far, the expression of the lb genes is restricted to the central infected part of the nodule and the amount of lb transcripts increases until the onset of nitrogen fixation (1, 2, 31). Gmlbc3 transcripts were not detected at the periphery of the infected zone whereas cpp1 transcripts showed the highest level of expression in this region. This observation supports the hypothesis that CPP1 is a repressor of lb gene expression or alternatively maintains a repressed state of the lb genes in these cells, similar to the function of the polycomb group proteins. Until recently, it was believed that cells in the central infected zone of a mature determinate nodule were all at the same developmental stage. However, an analysis of the expression of symbiotic bacterial genes in Phaseolus vulgaris nodules indicated that the cells at the periphery of the infected zone contain bacteria which are not differentiated into the nitrogen-fixing form (32). The authors therefore suggest that there are developmental zones in the determinate nodule with the less differentiated cells located at the periphery. Recent expression studies of a homeobox gene, Gmndx, also suggest that the soybean nodule has a zone with less differentiated cells (10). These cells are smaller and are not highly packed with bacteria as compared with large cells in the center. It is therefore possible that, in contrast to the lb genes, cpp1 is expressed in cells which are not fully differentiated.
Acknowledgments
The GFP derivative was a gift from Dr. Gerard van der Krogt, Wageningen Agricultural University, The Netherlands. This work was supported by the Danish Biotechnology Program, the Human Frontier Science Program, and European Union Biotech Contract ERBSC1CT920827. K.P.-M. was supported by The Joint Committee of the Nordic Natural Science Councils (NOS-N).
Abbreviations
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- lb
leghemoglobin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ010165 for cpp1).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090468497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090468497
References
- 1.Marcker A, Lund M, Jensen E O, Marcker K A. EMBO J. 1984;3:1691–1695. doi: 10.1002/j.1460-2075.1984.tb02033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nap J-P, Bisseling T. Science. 1990;250:948–954. doi: 10.1126/science.250.4983.948. [DOI] [PubMed] [Google Scholar]
- 3.Heard J, Caspi M, Dunn K. Mol Plant–Microbe Interact. 1997;10:665–676. doi: 10.1094/MPMI.1997.10.5.665. [DOI] [PubMed] [Google Scholar]
- 4.Heard J, Dunn K. Proc Natl Acad Sci USA. 1995;92:5273–5277. doi: 10.1073/pnas.92.12.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen E Ø, Marcker K A, Schell J, Bruijn F D. EMBO J. 1988;7:1265–1271. doi: 10.1002/j.1460-2075.1988.tb02940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobsen K, Laursen N B, Jensen E O, Marcker A, Poulsen C, Marcker K A. Plant Cell. 1990;2:85–94. doi: 10.1105/tpc.2.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metz B A, Welters P, Hoffmann H J, Jensen E O, Schell J, de Bruijn F J. Mol Gen Genet. 1988;214:181–191. doi: 10.1007/BF00337709. [DOI] [PubMed] [Google Scholar]
- 8.Forde B G, Freeman J, Oliver J E, Pineda M. Plant Cell. 1990;2:925–939. doi: 10.1105/tpc.2.9.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laursen N B, Larsen K, Knudsen J Y, Hoffmann H J, Poulsen C, Marcker K A, Jensen E O. Plant Cell. 1994;6:659–668. doi: 10.1105/tpc.6.5.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jørgensen J-E, Grønlund M, Pallisgaard N, Larsen K, Marcker K A, Jensen E O. Plant Mol Biol. 1999;40:65–77. doi: 10.1023/a:1026463506376. [DOI] [PubMed] [Google Scholar]
- 11.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 12.Stougaard J, Sandal N N, Grøn A, Kühle A, Marcker K A. EMBO J. 1987;6:3565–3569. doi: 10.1002/j.1460-2075.1987.tb02686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vancanneyt G, Schmidt R, O'Connor-Sanchez A, Willmitzer L, Rocha-Sosa M. Mol Gen Genet. 1990;220:245–250. doi: 10.1007/BF00260489. [DOI] [PubMed] [Google Scholar]
- 14.Jefferson R A, Kavanagh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajdukiewicz P, Svab Z, Maliga P. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 16.Davis S J, Vierstra R D. Plant Mol Biol. 1998;36:521–528. doi: 10.1023/a:1005991617182. [DOI] [PubMed] [Google Scholar]
- 17.Christiansen H, Hansen A C, Vijn I, Pallisgaard N, Larsen K, Yang W-C, Bisseling T, Marcker K A, Jensen E O. Plant Mol Biol. 1996;32:809–821. doi: 10.1007/BF00020479. [DOI] [PubMed] [Google Scholar]
- 18.Scheres B, Van De Wiel C, Zalensky A, Horvath B, Spaink H, Van Eck H, Zwartkruis F, Wolters A M, Gloudemans T, Van Kammen A, et al. Cell. 1990;60:281–294. doi: 10.1016/0092-8674(90)90743-x. [DOI] [PubMed] [Google Scholar]
- 19.van de Wiel C, Scheres B, Franssen H, van Lierop M J, van Lammeren A, van Kammen A, Bisseling T. EMBO J. 1990;9:1–7. doi: 10.1002/j.1460-2075.1990.tb08073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Arnim A G, Deng X W, Stacey M G. Gene. 1999;221:35–43. doi: 10.1016/s0378-1119(98)00433-8. [DOI] [PubMed] [Google Scholar]
- 21.Quandt H-J, Pühler A, Broer I. Mol Plant–Microbe Interact. 1993;6:699–706. doi: 10.1094/mpmi-6-055. [DOI] [PubMed] [Google Scholar]
- 22.Jensen E Ø, Pallisgaard N, Christiansen H, Vijn I, Bisseling T, Grønbæk M, Nielsen K, Jørgensen J-E, Larsen K, Hansen A C, et al. In: Biological Fixation of Nitrogen for Ecology and Sustainable Agriculture. Legocki A, Bothe H, Pühler A, editors. G 39. Berlin: Springer; 1997. pp. 87–90. [Google Scholar]
- 23.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 24.Sugihara T, Wadhwa R, Kaul S C, Mitsui Y. Genomics. 1999;57:130–136. doi: 10.1006/geno.1999.5756. [DOI] [PubMed] [Google Scholar]
- 25.Williams B C, Dernburg A F, Puro J, Nokkala S, Goldberg M L. Development (Cambridge, UK) 1997;124:2365–2376. doi: 10.1242/dev.124.12.2365. [DOI] [PubMed] [Google Scholar]
- 26.Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz E M, Coupland G. Nature (London) 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- 27.Jones R S, Gelbart W M. Mol Cell Biol. 1993;13:6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennison J A. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 29.Brown J L, Mucci D, Whiteley M, Dirksen M L, Kassis J A. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y, Seto E, Chang L S, Shenk T. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 31.Szczyglowski K, Szabados L, Fujimoto S Y, Silver D, de-Bruijn F J. Plant Cell. 1994;6:317–332. doi: 10.1105/tpc.6.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patriarca J E, Taté R, Fedorova E, Riccio A, Defez R, Iaccarino M. Mol Plant–Microbe Interact. 1996;9:243–251. [Google Scholar]