Abstract
Numerous TCP genes (transcription factors with a TCP domain) occur in legumes. Genes of this class in Arabidopsis (TCP1) and snapdragon (Antirrhinum majus; CYCLOIDEA) have been shown to be asymmetrically expressed in developing floral primordia, and in snapdragon, they are required for floral zygomorphy (bilaterally symmetrical flowers). These genes are therefore particularly interesting in Leguminosae, a family that is thought to have evolved zygomorphy independently from other zygomorphic angiosperm lineages. Using a phylogenomic approach, we show that homologs of TCP1/CYCLOIDEA occur in legumes and may be divided into two main classes (LEGCYC group I and II), apparently the result of an early duplication, and each class is characterized by a typical amino acid signature in the TCP domain. Furthermore, group I genes in legumes may be divided into two subclasses (LEGCYC IA and IB), apparently the result of a duplication near the base of the papilionoid legumes or below. Most papilionoid legumes investigated have all three genes present (LEGCYC IA, IB, and II), inviting further work to investigate possible functional difference between the three types. However, within these three major gene groups, the precise relationships of the paralogs between species are difficult to determine probably because of a complex history of duplication and loss with lineage sorting or heterotachy (within-site rate variation) due to functional differentiation. The results illustrate both the potential and the difficulties of orthology determination in variable gene families, on which the phylogenomic approach to formulating hypotheses of function depends.
The considerable advances in plant developmental genetics from a few model species have provided a starting point for studying plant morphological diversity and evolution at the molecular level. Genes that control development have been implicated in the evolution of novel phenotypes (for review, see Baum, 1998; Doebley and Lukens, 1998; McSteen and Hake, 1998; Cronk, 2001; Shepard and Purugganan, 2002). There is now a growing interest in expanding this knowledge to other species less amenable to genetic studies but displaying patterns of morphological variation that could be accounted for by changes in the expression of developmental genes.
Comparative expression studies rely on a phylogenetic framework to help identify candidate genes (Eisen, 1998). This approach has been used to find putative orthologs of MADS-box genes in non-model species of basal eudicots (Kramer and Irish, 1999). We present here a study of the evolution of putative homologs of the floral symmetry gene CYCLOIDEA (CYC) in legumes, with particular emphasis on the subfamily Papilionoideae. Using relatively wide sampling within Leguminosae is potentially a useful way of identifying the different subgroups within a gene family, as represented in legumes.
In snapdragon (Antirrhinum majus L. [Lamiales, Veronicaceae]), floral dorsal identity is controlled by two closely related nuclear genes CYC and DICHOTOMA (DICH; Luo et al., 1996, 1999; Almeida et al., 1997). In floral meristems, CYC and DICH have overlapping expression patterns on the adaxial side, but they have diverged so that CYC is expressed slightly later in development than DICH but has a greater effect on phenotype. These two genes belong to a gene family of putative transcription factors characterized by a basic helix-loop-helix domain referred to as the TCP domain (Cubas et al., 1999a). In Arabidopsis, 24 members have been identified. A subclass of this gene family, to which CYC/DICH and the maize (Zea mays) architecture gene TEOSINTE BRANCHED 1 belong, also has a highly conserved Arg-rich R domain (Cubas, 2002). CYC-like genes have been implicated in the control of floral symmetry in other species in the Lamiales, such as Linaria vulgaris Miller (Veronicaceae; Cubas et al., 1999b). The homolog of CYC in Arabidopsis, TCP1, has recently been shown to be expressed transiently at the adaxial base of floral and axillary meristems (Cubas et al., 2001). This suggests that asymmetric expression of CYC-like genes may predate the divergence of the Asteridae (e.g. snapdragon) and the Rosidae (e.g. Arabidopsis and Leguminosae). Such asymmetrically expressed genes may have been recruited repeatedly for the evolution of zygomorphy in separate lineages.
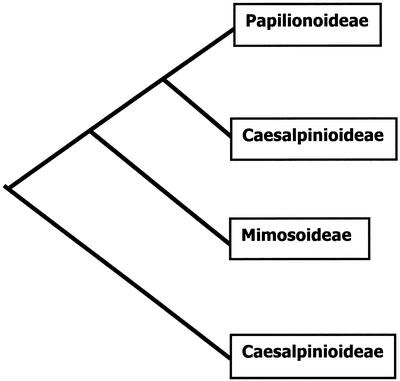
The Leguminosae is one such plant family where zygomorphy is believed to have evolved separately from the Lamiales (Stebbins, 1974; Donoghue et al., 1998). With approximately 18,000 species, it is one of the most species-rich angiosperm families, with the greatest number of species (approximately 12,000) found in the subfamily Papilionoideae. Papilionoids are characterized by highly zygomorphic flowers, with an enlarged dorsal (standard) petal, and lateral (wings) and ventral (keel) petals surrounding the reproductive organs. This highly specialized floral form, an adaptation to bee pollination, contrasts with that of the other two subfamilies Caesalpinioideae and Mimosoideae. Mimosoid flowers are typically actinomorphic, with reduced outer whorls, whereas Caesalpinioideae display more variation in floral morphology ranging from near radial symmetry to zygomorphy. Current molecular evidence suggests that mimosoids and papilionoids have evolved from different lineages of a paraphyletic caesalpinioid group (Doyle et al., 1997; Bruneau et al., 2001; Kajita et al., 2001; Fig. 1).
Figure 1.
Phylogenetic relationship of the three legume subfamilies based on current molecular evidence, with Mimosoideae and Papilionoideae derived from a paraphyletic Caesalpinoideae (Doyle et al., 1997; Bruneau et al., 2001; Kajita et al., 2001).
Within the Papilionoideae, a few taxa with atypical near radial symmetry have traditionally been considered basal members of this subfamily, even transitional between caesalpinioids and papilionoids (Polhill, 1981). However, recent molecular evidence suggests that these unusual taxa are derived from typical papilionoids (Pennington et al., 2000). These putative reversals from zygomorphy to actinomorphy provide a framework for studying the control of floral symmetry in legumes.
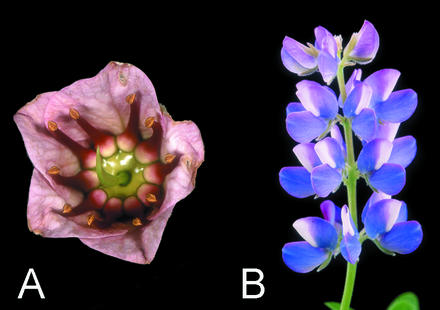
In the model legumes Lotus japonicus, soybean (Glycine max), and pea (Pisum sativum), CYC-like genes have been isolated, and in the case of L. japonicus, two genes have been found to be asymmetrically expressed in the developing flower (D. Luo, unpublished data). This study aims to expand these findings to other taxa from other major papilionoid groups such as the dalbergioid and genistoid clades as well as basal lineages (Pennington et al., 2001) where most of the morphological variation lies. This study comprises species with unusual flower morphology, such as Acosmium subelegans (Mohl.) Yakovlev and Cadia purpurea (Picc.) Aiton from the Genistoid clade, and Swartzia jorori Harms from the basal papilionoid group (Polhill, 1981; Pennington et al., 2001). C. purpurea, in particular, has open, near radial flowers, with equal free stamens arranged in a ring (Fig. 2a). This contrasts with typical papilionoids from the Genistoid group such as Lupinus (Lupinus nanus; Fig. 2b). Inclusion of legumes with unusual floral morphology is likely to be useful in studies of the origin of derived modifications in floral symmetry.
Figure 2.
a, Flower of Cadia purpurea, a near actinomorphic papilionoid legume. b, Inflorescence of Lupinus nanus bearing highly zygomorphic flowers typical of the Papilionoideae.
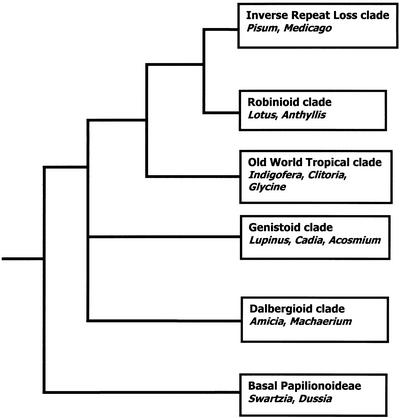
As functional gene studies expand from model organisms to related species, it becomes necessary to identify the functional counterparts of genes well-characterized in model species. The phylogenomic method proposes that orthology (i.e. common descent) is a likely predictor of functional equivalence (Eisen, 1998). Modern phylogenetic techniques now often permit robust determination of orthology relations of genes. We have thus taken a phylogenetic approach to investigate orthologs of CYC in legumes, with sampling that ensures coverage of all the main clades of papilionoid legumes (Fig. 3).
Figure 3.
Schematic representation of the relationship of some of the major groups in the Papilionoideae as defined by current molecular evidence (Doyle et al., 1997; Hu et al., 2000; Kajita et al., 2001; Pennington et al., 2001), with representative taxa used in the LEGCYC analyses.
RESULTS
Legume CYC Sequence Characterization
Thirty-eight sequences with a TCP and R domain were amplified using primers LEGCYC/F1 and R1 in 16 different taxa. Sequence number per taxon ranged from one to four, with only one sequence isolated from non-papilionoid taxa. However, basal papilionoid taxa, such as S. jorori and Dussia macroprophyllata Harms, had multiple copies comparable in number with more derived papilionoid species (see Table I for summary and GenBank accession no.). No evident sequence modifications (e.g. premature stop codons) were observed in papilionoids with unusual floral morphology.
Table I.
List of sequences obtained with primers LEGCYC-F1 and R1, and corresponding GenBank accession number
| Sequence | GenBank Accession No. | Sequence | GenBank Accession No. |
|---|---|---|---|
| Ceratonia 1 | AY225810 | Lupinus sp. 1 | AY225832 |
| Dialium 1 | AY225811 | Lupinus sp. 2 | AY225834 |
| Zapoteca 1 | AY225812 | Lupinus sp. 3 | AY225833 |
| Pisum 1 | AY225813 | Lupinus sp. 4 | AY225835 |
| Anthyllis 1 | AY225814 | Lupinus nanus 1 | AY225836 |
| Anthyllis 2 | AY225815 | Lupinus nanus 2 | AY225837 |
| Anthyllis 3 | AY225816 | Lupinus nanus 3 | AY225838 |
| Lotus berthelotii 1 | AY225817 | Lupinus angustifolius 1 | AY225839 |
| Lotus berthelotii 2 | AY225818 | Lupinus angustifolius 2 | AY225840 |
| Indigofera 1 | AY225819 | Machaerium 1 | AY225841 |
| Indigofera 2 | AY225820 | Machaerium 2 | AY225842 |
| Indigofera 3 | AY225821 | Amicia 1 | AY225843 |
| Clitoria 1 | AY225822 | Amicia 2 | AY225844 |
| Clitoria 2 | AY225823 | Dussia 1 | AY225845 |
| Clitoria 3 | AY225824 | Dussia 2 | AY225846 |
| Cadia 1 | AY225825 | Dussia 3 | AY225847 |
| Cadia 2 | AY225826 | Swartzia 1 | AY225848 |
| Cadia 3 | AY225827 | Swartzia 2 | AY225849 |
| Cadia 4 | AY225828 | Swartzia 3 | AY225850 |
| Acosmium 1 | AY225829 | ||
| Acosmium 2 | AY225830 | ||
| Acosmium 3 | AY225831 |
Fragment length ranged from 274 bp (Pisum 1) to 427 bp (Clitoria 1), with a mean length of 333.81 (± 40.2) bp. These fragments were also highly variable in sequence (at the amino acid and nucleotide level), with numerous substitutions and indel events in the region between the TCP and R domain. As a result, unambiguous sequence alignment for all legume CYC-like sequences was only possible in the TCP and R domains.
Position of Legume CYC-Like Sequences in the TCP Gene Family
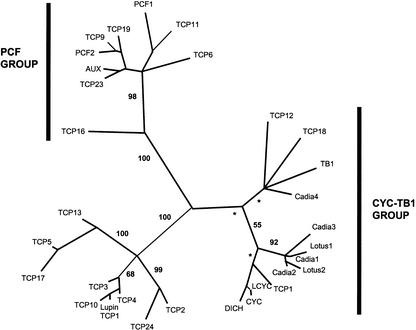
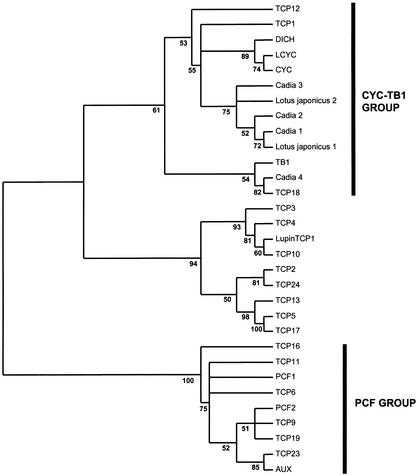
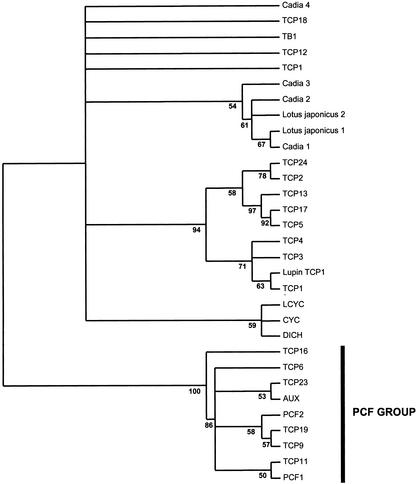
TCP domains of seven legume CYC-like protein sequences from two species, C. purpurea and L. japonicus, were analyzed in the context of the TCP gene family. Analysis of the TCP domain peptide matrix using protein distance, parsimony, maximum likelihood (ML), and Bayesian methods resulted in congruent trees with strong support values for the major groups. Figure 4 shows the protein ML unrooted phylogram, with support values obtained by Bayesian analysis of the data. The 50% majority rule (MR) protein distance and maximum parsimony trees are also shown for comparison (Figs. 5 and 6, respectively). All analyses strongly suggest that the TCP gene family can be divided into two main groups: the PCF group (recovered in every analyses with 100% support values) and a second group containing CYC/TB1 and, among others, the five Arabidopsis genes (TCP1, TCP12, TCP18, TCP2, and TCP24) with an R domain. These results confirm the conclusions of Cubas (2002), but with greater sampling and more comprehensive phylogenetic analysis. Within the latter group, CYC/TB1 genes form a separate group from another well-supported clade (in all analyses) of yet uncharacterized proteins. Although unrooted trees are difficult to interpret evolutionarily, because the point of origin is uncertain, these trees strongly suggest that the legume sequences here are the best candidates for CYC/TCP1 orthologs.
Figure 4.
Unrooted phylogram of protein ML analysis using TREEPUZZLE v5.0 (Schmidt et al., 2000) of the TCP domain data set including representative legume sequences. Support values were obtained using MrBayes (Huelsenbeck and Ronquist, 2001); asterisks indicate that a clade was recovered in <50% of Bayesian trees. Results support a LEGCYC clade (excluding Cadia 4) as sister to the CYC/TCP1 clade. All TCP genes unless otherwise indicated, Arabidopsis; PCF, rice; TB1, maize; LCYC, L. vulgaris; CYC and DICH, snapdragon; AUX, cotton.
Figure 5.
Fifty percent MR consensus tree of the protein distance analysis using the PAM-Dayhoff model of protein substitution (PROTDIST; Felsenstein, 1993) of the TCP domain. Values >50% of the 100 jackknife replicates are given at branch nodes. Taxa as in legend to Figure 4.
Figure 6.
Fifty percent MR consensus tree of protein maximum parsimony analysis (PROTPARS; Felsenstein, 1993) of the TCP domain. Support values above 50% from the 100 jackknife replicates are shown. Maximum parsimony fails to resolve groups recovered in protein, ML, Bayesian, and protein distance analyses. Although it does not contradict any of the results from other methods, it offers no support for a CYC/TB1 clade.
All analyses suggest that the legume CYC (LEGCYC) sequences from C. purpurea and L. japonicus (with the exception of Cadia 4) form a strongly supported group (found in 92% of Bayesian trees). This monophyletic group (here called LEGCYC) is sister to the CYC-TCP1 clade in the ML, Bayesian (Fig. 4) and distance (Fig. 5) trees. LEGCYC genes are therefore putative orthologs of CYC and TCP1. Cadia 4 is recovered in ML (Fig. 4) and distance (Fig. 5) analyses in the clade containing TB1, TCP12, and TCP18. The parsimony analysis is not informative because the relationship between the LEGCYC clade, Cadia 4, the CYC/LCYC/DICH clade, TCP1, TCP12, TCP18, and TB1 collapses in a 50% MR consensus tree (Fig. 6).
Evolution of LEGCYC Genes: Partial TCP and R Nucleotide Analyses
To recover major groups within the LEGCYC genes, we analyzed a matrix of 29 legume nucleotide sequences, rooted using snapdragon CYC and DICH, chosen to represent the full range of papilionoid legume taxa and sequence variation. The legume sequences could only be aligned with the snapdragon sequences using the highly conserved TCP and R domains. Parsimony analysis of the 67 informative sites out of 145 in the partial TCP and R nucleotide sequences produced 168 trees with a minimal length of 278 steps (additional branch swapping did not recover any more maximum parsimony trees), a consistency index (CI) of 0.424 and a retention index (RI) of 0.636, indicating fairly high homoplasy (parallel evolution) in the data. A strict consensus tree (Fig. 7), rooted on snapdragon genes CYC and DICH, resolves only one large supported clade within the ingroup (corresponding to group II, see below). Otherwise, only the relationship between sequences from different species of the same genus (e.g. Lupinus spp.) or related genera (e.g. Anthyllis and Lotus spp.) were supported in this analysis.
Figure 7.
Maximum parsimony analysis of the legume partial TCP and R domain nucleotide sequences. Strict consensus of 168 most parsimonious trees (CI = 0.424, RI = 0.636), with bootstrap values shown, rooted on snapdragon CYC and DICH.
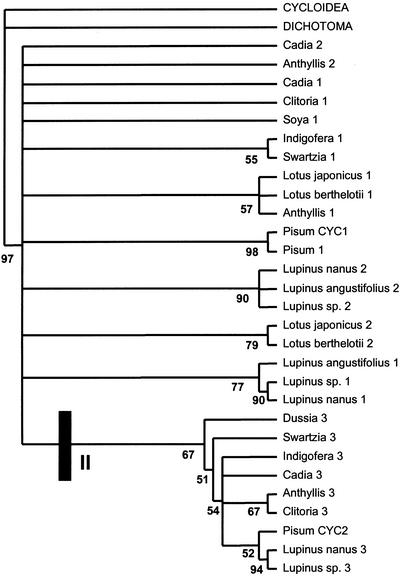
Model-based methods, such as Bayesian inference, are less sensitive to long-branch attraction and may therefore be better alternatives for analyzing homoplastic data. Bayesian analysis (Fig. 8) recovered two groups of legume sequences with support values (called here group I and group II). Group II had very high (97%) Bayesian support, whereas group I had weak support of 52%. Both groups include species from basal as well as more derived papilionoids and would appear to represent an early duplication event. However, relationships between sequences other than from closely related species or genera (e.g. Lupinus spp.) were difficult to interpret.
Figure 8.
Bayesian analysis MR tree of the legume TCP and R nucleotide sequences allowing for codon-specific nucleotide substitution, rooted on snapdragon CYC and DICH. Major clades I and II within LEGCYC are indicated with high Bayesian support.
Therefore although parsimony analysis of this small data set did not resolve relationships between LEGCYC genes well, Bayesian analysis gave a more fully resolved tree. The poor performance of parsimony analysis was probably due to high homoplasy in the data set coupled with the low number of informative characters with consequent low phylogenetic signal.
Evolution of LEGCYC Genes: Inclusion of Sequence Data between the TCP and R Domains
The region between the TCP and R domains was then added to the initial data set, together with additional legume sequences. Due to the high length and sequence variability of this region, it could not be aligned with nonlegume sequences, and so all analyses are unrooted. Furthermore, because of length variability, alignment was difficult even within legumes. For this reason some of the positions in which the alignment was ambiguous were excluded from the analysis (300 aligned positions). Eight LEGCYC sequences were excluded altogether from this analysis for the same reason. The remaining 38 sequences covered 292 unambiguously aligned characters, which required the insertion of 34 gaps of 1- to 6-bp triplets for alignment.
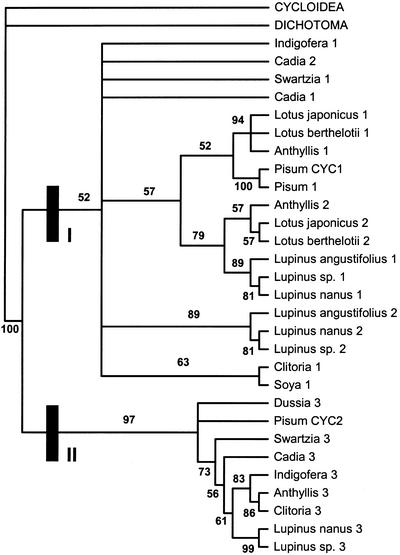
Parsimony analysis of the resulting 153 parsimony informative characters from the extended data set resulted in a single most parsimonious tree of 748 steps, with CI = 0.452 and RI = 0.601. The tree recovered two clades (groups I and II from the previous analyses) with a bootstrap value of 65%, although sequence relationship within these groups had little bootstrap support with the exception of sequences from closely related taxa (Fig. 9). The topology of the ML tree and the 50% MR consensus tree from the Bayesian analysis was identical, with only three nodes collapsing in the Bayesian consensus tree. The topology of those trees was also similar to the tree from the parsimony analysis, but the level of support for the nodes (estimated by Bayesian inference) was much higher in the model-based analysis. For instance, group I and II were recovered in the Bayesian analysis with high support (Fig. 10). Comparison of the partial TCP domains of amino acid sequences from group I and II showed that there were five synapomorphies, which suggests these clades are genuine (Fig. 11). These groupings were also supported by considerable differences in the variable region, such as presence or absence of motifs, which could not be included in the analysis.
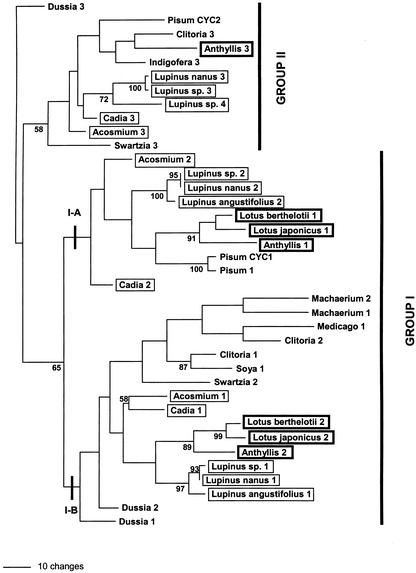
Figure 9.
Unrooted phylogram of single most parsimonious tree (748 steps, CI = 0.452, RI = 0.601) from the maximum parsimony analysis of 38 partial legume CYC-like sequences including some sequence data (292 characters, 153 parsimony informative) from the hypervariable region between the TCP and R domains. Bootstrap values (below in bold) are given for branches with >50% support. Major groups recovered in previous analyses (group I and group II) are shown. Clades containing Lupinus spp. and Lotus spp. sequences are highlighted (I-A and I-B) suggesting putative duplication events.
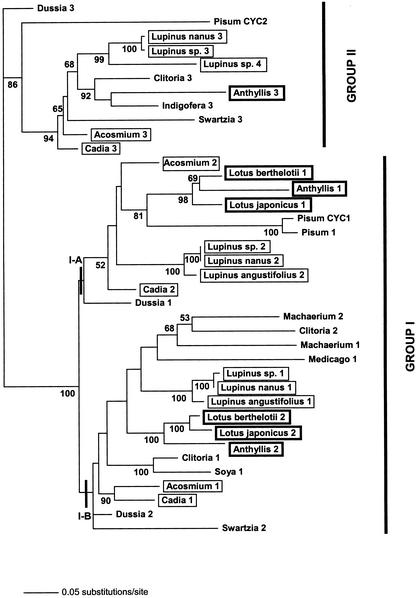
Figure 10.
Unrooted phylogram of the ML analysis (using the GTR + I + G model of nucleotide substitution) of partial legume CYC sequences. Support values at each node were obtained by Bayesian analysis of the data set and represent the frequency of each node in the MR consensus tree. The two main groups of LEGCYC (I and II) are highlighted, and one putative duplication event in group I is marked by A and B.
Figure 11.
Comparison of the partial TCP domain amino acid sequence from group I and II CYC-like sequences in legumes. Asterisk highlights group-specific changes; asterisks above and below bold sequences are amino acid differences found less frequently in these groups.
Within group I, two sequences from most taxa were found. These segregated into two clades (A and B, see Fig. 9), which for the most part contained one sequence per taxon, with a few exceptions (for example Machaerium 1 and 2). Clade A contained one LEGCYC sequence from representatives from both the genistoid (Lupinus spp., Cadia sp., and Acosmium spp.) and robinioid (Lotus spp. and Anthyllis sp.) clades, whereas clade B contained another LEGCYC sequence from these taxa. Although these clades have no bootstrap support in the parsimony analysis, they were found the ML tree and in most Bayesian trees. This suggests a putative orthology relationship between sequences within these clades (IA and IB) and a further conserved duplication in LEGCYC sequences (LEGCYC IA and IB) of possible functional significance.
DISCUSSION
Presence of TCP1/CYC Orthologs in Leguminosae
In the TCP gene family analyses, evidence from sequence similarity (PROTDIST) and evolution (ML and Bayesian analyses) strongly suggest that the legume CYC-like sequences examined here are homologous to the floral symmetry genes in snapdragon, CYC and DICH, and to the adaxially expressed floral gene TCP1 in Arabidopsis. Within this legume clade, a lower estimate of three CYC-like copies were found within the Papilionoideae, in species ranging from the basal-most clade (S. jorori) to higher papilionoids (e.g. the robinioid A. hermannia). Because of their apparent orthology with snapdragon CYC, these genes are candidates for floral developmental genes in the Leguminosae. However, these analyses, many of which lead to poorly resolved trees, highlight some of the difficulties in making detailed orthology statements within gene families and CYC-like genes in particular.
Complex Evolution of CYC-Like Genes in the Leguminosae
No simple pattern of gene evolution tracking organismal phylogeny within the legume CYC family was recovered in the phylogenetic analyses. Possible confounding factors such as intermediate levels of concerted evolution, variation in the rate of sequence evolution, and independent gene loss and duplication events, which render the interpretation of gene trees difficult (Doyle, 1994), cannot be ruled out here.
Different levels of variation in different parts of the sequences also made analysis difficult. The highly conserved TCP and R boxes were alignable but contained little phylogenetically informative information, whereas the variable region contained much variation but was difficult to align. Furthermore, the variation in the TCP and R domains was mainly at the synonymous third codon position and had a high degree of homoplastic variation (accounting for two-thirds of the steps required). High levels of homoplasy, possibly resulting in long-branch attraction and therefore artificial groupings, is suggested by the low support values of the trees from this analysis and the collapse of many nodes in the maximum parsimony strict consensus trees. Also, because the analysis includes clades between which functional differentiation may exist, particular amino acid positions may be subject to different selection pressure in different parts of the tree. This within-site rate variation, or heterotachy (Lopez et al., 2002), is also likely to make phylogenetic reconstruction more difficult.
Two Major Subgroups (I and II) of Legume CYC-Like Genes Represent a Probable Early Duplication
Despite the problematic nature of the data, certain patterns do emerge from the analyses. Results of the rooted Bayesian analysis suggests that LEGCYC genes can be divided into two main groups (referred to as I and II), which are characterized by different amino acid signatures. The results of the unrooted legume analyses of the extended dataset are also consistent with the two-group hypothesis, and these groups, although only moderately supported by maximum parsimony, are strongly supported by Bayesian inference. Taxa ranging from the basal-most papilionoids to highly derived species (from the “inverse repeat loss” clade, e.g. pea) have both groups of genes suggesting that these genes probably diverged after a duplication event before the evolution of the Papilionoideae. In addition to the putative amino acid synapomorphies in the TCP domain (Fig. 11), these groups are also distinguished by specific motifs in the otherwise variable region between the TCP and R domains.
Evidence for Two Subgroups (IA and IB) of Group I LEGCYC Sequences
Within group I, one other major duplication event appears to have occurred, giving rise to two subgroups IA and IB. We recovered genes belonging to both clades in a wide range of the species sampled here, implying that this duplication occurred at least early in the diversification of the papilionoids.
However, the relationships between sequences within these groups appear complex and require further investigation. Even though our sampling is fairly extensive compared with many studies of developmental gene phylogeny, further sampling may help resolve relationships within and between gene copies. However, these results are in agreement with a trend of independent duplications, and possible losses, with rapid gene evolution outside of the conserved TCP and R domains, previously documented in CYC-like genes families from other plant groups (e.g. Gesneriaceae; Citerne et al., 2000).
The Limitations and Potential of Phylogenomics
The lack of resolution resulting from problematic analyses (particularly using parsimony) highlights the limitations of phylogenomics, at least in rapidly evolving genes with high levels of homoplasy and in gene families where functional differentiation may lead to high levels of heterotachy (within-site rate variation). These problems may lead to difficulties in robust orthology estimation and hence functional prediction. In this study, Bayesian inference gives better resolution than parsimony; with the large amount of homoplasy in these data it is likely that model-based methods such as Bayesian inference will outperform parsimony.
The recognition of a major legume CYC-like (LEGCYC) group in this study does however suggest likely candidate genes for functional equivalents of CYC/TCP1. Furthermore, within this group of legume CYC candidates, further subgroups are recognized in this study (LEGCYC IA, IB, and II), inviting investigation of possible functional differences between these. Thus even where phylogenetic analyses are difficult, partial resolution may still enable hypotheses to be generated. Although we recognize the limitation of phylogenomics, we still regard this approach as extremely promising even with relatively intractable gene families.
MATERIALS AND METHODS
Molecular Methods: DNA Extraction, PCR, Cloning, and Sequencing
For each species, genomic DNA was extracted from either fresh or silica dried leaf material following a modification of the cetyl-trimethyl-ammonium bromide procedure of Doyle and Doyle (1987). Previously extracted DNA was available for Dialium guinanense (R.T. Pennington, Royal Botanic Garden Edinburgh), pea (line 399; J. Hofer, John Innes Centre), and Lupinus angustifolius cv Merrit (S. Barker, University of Western Australia, Perth).
The region delimited by the conserved TCP and R domains was amplified using primers LEGCYC/F1, 5′-TCA GGG SYT GAG GGA CCG-3′, and LEGCYC/R1, 5′-TCC CTT GCT CTT GCT CTT GC-3′. These primers were designed based on available sequences of CYC-like genes from Lotus japonicus and soybean (Glycine max; D. Luo, unpublished data), compared with nucleotide sequences of the TCP and R domains from snapdragon (Antirrhinum majus; CYC, Y16313; and DICH, AF199465), Arabidopsis (TCP1, AC002130; TCP12, AC011914; and TCP18, AP001303) and maize (Zea mays subsp. mays; TB1, AF340199). PCR amplifications were carried out using Taq and reagents (Bioline, London) in a 50-μL mix containing 2.5 μL of 50 mm MgCl2, 5 μL of a 2 mm dNTP mix, 2.5 μL of each primer (10 μm; MWG Biotech, Gersberg, Germany), 1 unit of BIOTAQ, and 10 to 20 ng of DNA. Conditions consisted of an initial denaturation step at 94°C (3 min), followed by 30 cycles of denaturation at 94°C (1 min), annealing at 50°C to 55°C (30 s), and extension at 72°C (30 s), followed by a final extension step at 72°C (5 min). PCR products were purified using the QIAquick PCR Purification Kit (Qiagen Ltd, Dorking, Surrey, UK) and then cloned using TOPO-TA Cloning Kit for Sequencing (Invitrogen, Carlsbad, CA). Dye-terminator cycle sequencing was carried out using Thermosequenase II (Amersham Biosciences UK, Little Chalfont, Buckinghamshire, UK). Samples were analyzed on an ABI 377 Prism Automatic DNA Sequencer (Applied Biosystems, Foster City, CA). In taxa of particular interest (Cadia purpurea and Lupinus nanus), 36 to 39 clones were sequenced, respectively. In addition, the entire open reading frame of two gene pairs in C. purpurea and L. nanus was sequenced by genome walking (modified from Siebert et al., 1995).
Phylogenetic Analysis: Taxon and Sequence Selection
CYC-like genes from legumes were placed in the context of the TCP gene family, represented by certain key sequences from L. japonicus and C. purpurea (Lotus japonicus 1 and 2, Cadia 1–4; Table I). To simplify the analysis, certain Arabidopsis TCP genes belonging to the PCF group (Cubas, 2002) were excluded (TCP7, TCP8, TCP14, TCP15, TCP20, TCP21, and TCP22 following the nomenclature of Cubas [2002]), whereas other sequences of particular interest were added: Gossypium hirsutum AUXIN (AF165924), Lupinus albus ‘TCP1 ’ (AJ426419), Linaria vulgaris LCYC (AF161252), and snapdragon DICH (AF199465). The 58 amino acids of the TCP domain were aligned manually. The matrix of 31 sequences was analyzed using not only protein distance methods similar to that of Cubas (2002), but also maximum parsimony, ML, and Bayesian methods (see below).
Results from these analyses guided the choice of sequences sampled to investigate the evolution of CYC-like genes in the legume family, using nucleotides of the TCP and R domains, with CYC, DICH, and TCP1 as outgroups. Twenty-nine taxa were sampled to represent the phylogenetic range of the papilionoids.
For the detailed analysis within the legumes including the nucleotide region between the TCP and R domains, a larger number of species was used, with representatives from the three subfamilies Caesalpinioideae, Mimosoideae, and Papilionoideae (Table II). Particular emphasis was placed on sampling representatives from all major papilionoid groups defined by current molecular phylogenetic evidence (Doyle et al., 1997; Hu et al., 2000; Kajita et al., 2001; Lavin et al., 2001; Pennington et al., 2001; M. Wojciechowski, M. Lavin, and M. Sanderson, unpublished data; Fig. 3, names of groups follow [Pennington et al., 2001]). All legume sequences obtained with primers LEGCYC/F1-R1, with the exception of Cadia 4, were selected as the ingroup. Additional legume sequences from separate studies were included in this analysis: L. japonicus (Lotus japonicus 1, Lotus japonicus 2), soybean (Soya 1), pea (Pisum CYC1, Pisum CYC2; D. Luo, personal communication), and Medicago truncatula (Medicago 1, BG455508). Snapdragon CYC and DICH and Arabidopsis TCP1 were chosen as outgroups in the partial TCP and R domains nucleotide sequence analysis.
Table II.
Species used in survey of CYC-like genes using primers LEGCYC-F1 and R1
| Subfamily | Clade | Taxon | Sourcea | Location |
|---|---|---|---|---|
| Caesalpinioideae | Ceratonia oroethauma (Hillc.) Lewis & Verdc. | 1996 0942A | Oman | |
| Dialium guianense (Aubl.) Sandw. | R.T. Pennington 639 | Napo, Ecuador | ||
| Mimosoideae | Zapoteca tetragona (Willd.) H.M. Hernandez | 1999 1149 | Guatemala | |
| Papilionoideae | Inverse Repeat Loss clade | Pea (Pisum sativum) line 399 | – | cultivated, John Innes Centre, Norwich, UK |
| Robinioid clade | Anthyllis hermanniae L. | 1975 1501 | Mediterranean | |
| Lotus berthelotii Masf. | 1978 0702B | Canary Islands | ||
| Old WorldTropical clade | Indigofera pendula Franch. | 1991 0547A | China | |
| Clitoria sp. | R.T. Pennington 990 | San Martín, Peru | ||
| Genistoid clade | Cadia purpurea (Picc.) Aiton | 1994 2001A | Yemen | |
| Acosmium subelegans (Mohl.) Yakovlev | Bridgewater 358 | Mato Grosso do Sul, Brazil | ||
| Lupinus sp. | R.T. Pennington 815 | Piura, Peru | ||
| L. nanus Doug. ex Benth. | – | commercial seed (Sutton Seeds, Paignton, Devon, UK) | ||
| Lupinus angustifolius L. cv Merrit | – | cultivated, University of Western Australia, Perth | ||
| Dalbergioid clade | Machaerium scleroxylon Tul. | 1999 0888A | Brazil | |
| Amicia glandulosa Kunth | R.T. Pennington 654 | Loja, Ecuador | ||
| Basal Papilionoideae | Dussia macroprophyllata Harms | 1995 1539A | Heredia, Costa Rica | |
| Swartzia jorori Harms | R.T. Pennington 938 | Santa Cruz, Bolivia |
Relationship of major Papilionoid clades (from Doyle et al., 1997; Hu et al., 2000; Kajita et al., 2001; Pennington et al., 2001) given in figure 3. –, XXX.
Source number refers to either RBGE living collection number (e.g. 1996 0942A) or collector's voucher number from wild collections (e.g. R.T. Pennington 639). All herbanium vouchers at RGBE.
DNA Sequence Alignment
Unambiguous alignment of all 54 legume CYC-like DNA sequences from 25 taxa was only possible in the TCP and R domains and reduced the matrix to 145 nucleotide characters. However, by excluding certain problematic sequences, it was possible to align certain parts of the variable region between these two conserved domains as protein sequences that were then analyzed as nucleotide sequences. Protein sequences were aligned using ClustalX (Thompson et al., 1997), followed by manual adjustments taking both amino acids and nucleotides into consideration.
Phylogenetic Analysis
Protein Methods
Protein distance analysis was carried out using program from the PHYLIP package (Felsenstein, 1993). One hundred half-deletion jackknife data sets were obtained with SEQBOOT, distance matrices were calculated with PROTDIST using the PAM-Dayhoff model of amino acid substitution, neighbor-joining trees were obtained with NEIGHBOR, and a consensus tree was produced by CONSENSE. Branches with <50% support were collapsed. Protein ML analysis was also carried out using PHYLIP. The most parsimonious trees were calculated with PROTPARS (Felsenstein, 1993), with support values obtained by 100 half-deletion jackknife replicates as described above. A 50% MR consensus tree was obtained with CONSENSE, collapsing branches with <50% jackknife support. Protein ML analysis was carried out using TREEPUZZLE v5 (Schmidt et al., 2002) with the BLOSUM 62 model of substitution (Henikoff and Henikoff, 1992) allowing for two rates of heterogeneity (1 invariable + 1 variable). To provide support values, Bayesian analysis was carried out using MrBayes v2.01 (Huelsenbeck and Ronquist, 2001), using the PAM-Dayhoff amino acid substitution model with one million generations sampled every 100 generations with a burn-in of 100,000 generations.
DNA Methods
Maximum parsimony analysis was carried out using PAUP* 4.0b7 (Phylogenetic Analysis Using Parsimony, version 4.0b7, Sinaur Associates, Sunderland, MA). Heuristic searches with 1,000 random addition replicates (to avoid local optima) and tree bisection reconnection (TBR) branch swapping were conducted with steepest descent and multrees options selected. A maximum of 10 minimal length trees was retained per replicate, and a further heuristic search by TBR was carried out on the shortest trees. Branch support values were calculated by 1,000 bootstrap replicates with simple sequence addition and a maximum of 10 minimal length trees retained per replicate. This search method was carried out both for the TCP and R nucleotide matrices, as well as the matrix incorporating certain variable regions. Bayesian phylogenetic analysis of the TCP plus R data set was carried out using MrBayes v2.01 (Huelsenbeck and Ronquist, 2001), using a general time reversible (GTR) model and site-specific rates partitioned by codon. Chains were run for 600,000 generations (burn-in of 100,000 generations) sampled every 100 generations. Resultant trees were used to generate a 50% MR consensus tree in PAUP* v4.0b7.
ML analyses were carried out for the matrix incorporating the more variable regions. The best-fit model was GTR + I + G (GTR model estimating the proportion of invariable sites and γ-distribution; Rodriguez et al., 1990), selected as the best-fit model of nucleotide substitution by the Akaike Information Criterion using Modeltest v3.06 (Posada and Crandall, 1998). A heuristic ML analysis with TBR branch swapping was carried out using PAUP* v4.0b7 with the parameters defined from above.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank the Royal Botanic Garden Edinburgh for use of laboratory and glasshouse facilities, the horticultural staff, and the laboratory staff (particularly Michelle Hollingsworth and Alex Ponge) for assistance. Julie Hofer (John Innes Centre) and Susan Barker (University of Western Australia) kindly made available DNA samples. We thank Debbie White (RGBE) for the photographs.
Footnotes
This work was supported by The Carnegie Trust for the Universities of Scotland and by the Systematics Association.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.016311.
LITERATURE CITED
- Almeida J, Rocheta M, Galego L. Genetic control of flower shape in Antirrhinum. Development. 1997;124:1387–1392. doi: 10.1242/dev.124.7.1387. [DOI] [PubMed] [Google Scholar]
- Baum DA. The evolution of plant development. Curr Opin Plant Biol. 1998;1:79–86. doi: 10.1016/s1369-5266(98)80132-5. [DOI] [PubMed] [Google Scholar]
- Bruneau A, Forest F, Herendeen PS, Klitgaard BB, Lewis GP. Phylogenetic relationships in the Caesalpinioideae (Leguminosae) as inferred from chloroplast trnL intron sequences. Syst Bot. 2001;26:487–514. [Google Scholar]
- Citerne HL, Moller M, Cronk QCB. Diversity of cycloidea-like genes in Gesneriaceae in relation to floral symmetry. Ann Bot. 2000;86:167–176. [Google Scholar]
- Cronk QCB. Plant evolution and development in a post-genomic context. Nat Rev Genet. 2001;2:607–619. doi: 10.1038/35084556. [DOI] [PubMed] [Google Scholar]
- Cubas P. Role of TCP genes in the evolution of morphological characters in angiosperms. In: Cronk QCB, Bateman RM, Hawkins JA, editors. Developmental Genetics and Plant Evolution. London: Taylor and Francis; 2002. pp. 247–266. [Google Scholar]
- Cubas P, Coen E, Zapater JMM. Ancient asymmetries in the evolution of flowers. Curr Biol. 2001;11:1050–1052. doi: 10.1016/s0960-9822(01)00295-0. [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 1999a;18:2115–2222. doi: 10.1046/j.1365-313x.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999b;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- Doebley J, Lukens L. Transcriptional regulators and the evolution of plant form. Plant Cell. 1998;10:1075–1082. doi: 10.1105/tpc.10.7.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue MJ, Ree RH, Baum DA. Phylogeny and the evolution of flower symmetry in the Asteridae. Trends Plant Sci. 1998;3:311–317. [Google Scholar]
- Doyle JJ. Evolution of a plant homeotic multigene family: towards connecting molecular systematics and molecular developmental genetics. Syst Biol. 1994;43:307–328. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Doyle JJ, Doyle JL, Ballenger JA, Dickson EE, Kajita T, Ohashi H. A phylogeny of the chloroplast gene rbcL in the Leguminosae: the taxonomic correlations and insights into the evolution of nodulation. Amer J Bot. 1997;84:541–554. [PubMed] [Google Scholar]
- Eisen JA. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. Washington: Department of Genetics, University of Seattle; 1993. [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J-M, Lavin M, Wojciechowski M, Sanderson MJ. Phylogenetic systematics of the tribe Millettieae (Leguminosae) based on chloroplast trnK/matK sequences, and its implications for evolutionary patterns in Papilionoideae. Am J Bot. 2000;87:418–430. [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kajita T, Ohashi H, Tateishi Y, Bailey D, Doyle JJ. rbcL legume phylogeny, with particular reference to Phaseoleae, Millitteae, and allies. Syst Bot. 2001;26:515–536. [Google Scholar]
- Kramer EM, Irish VF. Evolution of genetic mechanisms controlling petal development. Nature. 1999;399:144–148. doi: 10.1038/20172. [DOI] [PubMed] [Google Scholar]
- Lavin M, Pennington RT, Klitgaard BB, Sprent JI, de Lima HC, Gasson P. The dalbergioid legumes (Fabaceae): delimitation of a pantropical monophyletic clade. Am J Bot. 2001;88:503–533. [PubMed] [Google Scholar]
- Lopez P, Casane D, Philippe H. Heterotachy, an important process of protein evolution. Mol Biol Evol. 2002;19:1–7. doi: 10.1093/oxfordjournals.molbev.a003973. [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen E. Control of organ asymmetry in flowers of Antirrhinum. Cell. 1999;99:367–376. doi: 10.1016/s0092-8674(00)81523-8. [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383:794–799. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- McSteen P, Hake S. Genetic control of plant development. Curr Opin Biotechnol. 1998;9:189–195. [Google Scholar]
- Pennington RT, Klitgaard BB, Ireland H, Lavin M. New insights into floral evolution of basal Papilionoideae from molecular phylogenies. In: Herendeen PS, Bruneau A, editors. Advances in Legume Systematics. Vol. 9. Kew, UK: Royal Botanic Gardens; 2000. pp. 233–248. [Google Scholar]
- Pennington RT, Lavin M, Ireland H, Klitgaard B, Preston J, Hu J-M. Phylogenetic relationships of basal papilionoid legumes based upon sequences of the chloroplast trnL intron. Syst Bot. 2001;26:537–556. [Google Scholar]
- Polhill RM. Papilionoideae. In: Polhill RM, Raven PH, editors. Advances in Legume Systematics. Vol. 1. Kew, UK: Royal Botanic Gardens; 1981. pp. 191–208. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rodriguez F, Oliver JF, Marin A, Medina JR. The general stochastic model of nucleotide substitutions. J Theor Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Shepard KA, Purugganan MD. The genetics of plant morphological evolution. Curr Opin Plant Biol. 2002;5:49–55. doi: 10.1016/s1369-5266(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Siebert PD, Chenchick A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. Flowering Plants: Evolution above the Species Level. Harvard University Press; 1974. [Google Scholar]
- Thompson JF, Gibson F, Plewmiak F, Jenamougin F, Higgins DG. The ClustalX window interface: flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]