Abstract
The peribacteroid membrane (PBM) forms the structural and functional interface between the legume plant and the rhizobia. The model legume Lotus japonicus was chosen to study the proteins present at the PBM by proteome analysis. PBM was purified from root nodules by an aqueous polymer two-phase system. Extracted proteins were subjected to a global trypsin digest. The peptides were separated by nanoscale liquid chromatography and analyzed by tandem mass spectrometry. Searching the nonredundant protein database and the green plant expressed sequence tag database using the tandem mass spectrometry data identified approximately 94 proteins, a number far exceeding the number of proteins reported for the PBM hitherto. In particular, a number of membrane proteins like transporters for sugars and sulfate; endomembrane-associated proteins such as GTP-binding proteins and vesicle receptors; and proteins involved in signaling, for example, receptor kinases, calmodulin, 14-3-3 proteins, and pathogen response-related proteins, including a so-called HIR protein, were detected. Several ATPases and aquaporins were present, indicating a more complex situation than previously thought. In addition, the unexpected presence of a number of proteins known to be located in other compartments was observed. Two characteristic protein complexes obtained from native gel electrophoresis of total PBM proteins were also analyzed. Together, the results identified specific proteins at the PBM involved in important physiological processes and localized proteins known from nodule-specific expressed sequence tag databases to the PBM.
The model legume Lotus japonicus forms nitrogen-fixing root nodules after infection by Mesorhizobium loti. The bacteria enter the plant cell by endocytosis, leading to the formation of a new compartment in the plant cell, the symbiosome. This compartment harbors the bacteroids and is surrounded by a peribacteroid membrane (PBM) formed from the plant plasma membrane (PM) during endocytosis of the bacteria (for review, see Verma and Hong, 1996; Whitehead and Day, 1997). The PBM develops further by receiving material from the endomembrane system (see Robertson et al., 1978; Verma and Hong, 1996) and finally forms the structural and functional interface between the symbionts (Robertson et al., 1978). The PBM plays a central role in the exchange of compounds between the organisms (see Udvardi and Day, 1997). To fulfill this role, the PBM is supplied with the specific components, like transporters and enzymes, necessary for the symbiotic exchange processes (see Verma and Hong, 1996; Whitehead and Day, 1997). Several of these activities have been characterized by biochemical and biophysical studies. For example, the activity of P-type H+-ATPase (P-ATPase) pumping protons from the cytoplasm to the peribacteroid space has been characterized (see Blumwald et al., 1985, and refs. therein). Dicarboxylates are supplied from the plant to the bacteroids, and a specific transporter present in the PBM has been characterized (see Udvardi and Day, 1997). The transport of the ammonia is probably mediated by a channel (Tyerman et al., 1995), but amino acid transport may also play a role in the delivery of the fixed nitrogen to the plant (see Day et al., 2001). However, only a few genes/proteins have been characterized by molecular genetic studies. This includes GmSAT1, a protein involved in the ammonium transport at the PBM (Kaiser et al., 1998), and ammonium transporters from nodules that might be involved in the transport across the PBM (see Day et al., 2001). Otherwise, nodulin 26, a major integral membrane protein of the PBM in soybean (Glycine max), has been cloned and characterized as a member of the major intrinsic protein family of proteins mainly functioning as water channels (Fortin et al., 1987).
Using transcriptional profiling with cDNA arrays from root nodules of L. japonicus, differences were detected in relative gene transcript abundance between nodules and uninfected roots (Colebatch et al., 2002). A number of transcripts were found to be up-regulated in nodules, indicating a specific role of the corresponding proteins in the nodule function.
Recently, proteome analysis has been used to understand more about the proteins involved in specific functions in model legumes. The use of two-dimensional electrophoresis (2-DE) and N-terminal sequencing led to the identification of some proteins attached to the PBM of soybean (Panter et al., 2000). A similar study using tandem mass spectrometry (MS/MS) revealed the presence of plant and bacteroid proteins in PBM preparations from pea (Pisum sativum; Saalbach et al., 2002). A root proteome reference map of Medicago truncatula was established using peptide mass fingerprinting and expressed sequence tag (EST) database searching (Mathesius et al., 2001). Similar tools were used to identify symbiosis-related proteins in M. truncatula (Bestel-Corre et al., 2002). Two special studies (Natera et al., 2000; Morris and Djordjevic, 2001) were devoted to the analysis of changes in protein expression in bacteroids compared with free bacteria, and in nodules compared with uninfected roots. Numerous changes were observed representing mostly abundant proteins of known function (like malate dehydrogenase [MDH], DNAK, GroEL, and leghemoglobin). After inoculation of roots with Rhizobium leguminosarum bacteria, 10 developmentally regulated proteins were identified including α-fucosidase, ethylene-induced proteins, tubulin, and chaperonin 21 (Morris and Djordjevic, 2001). The results of such studies have demonstrated that proteomics is a powerful tool to gain insight into the expression, abundance, and distribution of proteins in special tissues of model legumes.
In the present work, we have used global trypsin digestion combined with nanoscale liquid chromatography (nano-LC)/MS/MS for proteins extracted from PBM preparations in L. japonicus. This approach is very useful in identifying many proteins present in a complex mixture without previous separation of the proteins. For example, almost 1,500 proteins were identified in such a study in yeast (Saccharomyces cerevisiae; Washburn et al., 2001). The complex peptide mixture resulting from the digestion has to be separated by nano-LC, and the peptides are then directly analyzed by MS/MS. We have used this high-throughput approach and identified a large number of proteins related to the PBM. In particular, a number of membrane proteins such as transporters and kinases with potentially important functions at the PBM were detected.
RESULTS AND DISCUSSION
Isolation of PBM Vesicles and Proteins
PBM was prepared according to the well-established standard procedure (for example, see Price et al., 1987; Panter et al., 2000). To isolate the PBM, osmotic lysis and mechanical rupture were applied to the isolated symbiosomes. Thereafter, bacteroids were removed by a low-speed centrifugation, and the supernatant was subjected to an aqueous polymer two-phase partitioning to purify inside-out PBM vesicles from remaining contaminations. This step was included because it has been shown that a substantial amount of proteins from bacteroids and mitochondria can be present in PBM preparations (Panter et al., 2000; Saalbach et al., 2002). The polymer concentration was optimized by assaying for ATPase activity, a marker enzyme for PBM. The activity was highest (specific activity 7.4 μmol h−1 mg−1) at a polymer concentration of 6.1%. In the same fraction, the activities of cytochrome c oxidase and succinate-dependent cytochrome c reductase were zero (undetectable), indicating a very low level of contamination by mitochondrial and bacteroid membranes. Other membranes such as PM, tonoplast, and endoplasmatic reticulum (ER) are very unlikely to occur in PBM preparations because symbiosomes are pelleted at a low speed that does not sediment membranes. Such contaminations have never been detected by biochemical or electron microscopical examination of symbiosome or PBM preparations (Price et al., 1987; Panter et al., 2000). Together, these data indicate that PBM preparations of high quality were obtained for further analysis.
Identification of PBM Proteins
2-DE is a classical method in proteome analysis to separate and display proteins for further analysis. However, it is known to be problematic with membrane proteins. For example, no hydrophobic membrane proteins could be detected in two studies using 2-DE for PBM analysis (Panter et al., 2000; Saalbach et al., 2002). Therefore, it was avoided in the present study of PBM. Instead, total proteins obtained from the Triton X-114 fractionation were digested with trypsin (global trypsin digestion). The resulting peptide mixtures were analyzed by nano-LC/MS/MS. This approach identified a large number of proteins, including integral membrane proteins, in the PBM preparation. After searching the databases with the MS/MS data using the “Mascot” search program, a total of 160 significant matches were obtained. The significance is calculated by the “Mascot” program (see Perkins et al., 1999), and only significant matches according to “Mascot” are reported in Tables I to III.
Table I.
List of proteins identified by nano-LC/MS/MS in Triton X-114 fractions of PBM proteins from L. japonicus
| EST | Accession
No. |
Protein | Species | X114 |
|---|---|---|---|---|
| Membrane transporter | ||||
| MWM126g09_r | AV416407 | Aquaporin PIP1 | M. truncatula | b |
| LjNEST31d2r | BI417898 | Aquaporin PIP2 | Zea mays | b |
| MWM217d01_r | AV412267 | Tonoplast intrinsic protein, delta | G. hirsutum | b |
| EST508266 (Mt) | BG646647 | Aquaporin-like transmembrane channel | M. truncatula | b |
| T09260 | Aquaporin-like transmembrane channel | Arabidopsis | b | |
| AAF82790 | LIMP1: water-selective transport protein | L. japonicus | b | |
| LjNEST13e12r | AW720076 | Nitrate transporter (BAB19758), | Soybean | d |
| Peptide transporter (AAG21898) | O. sativa | |||
| LjNEST4f3r | AW719442 | Sulfate transporter-like protein | O. sativa | d |
| LjNEST58c12r | BI420529 | Putative sugar transporter (mannitol?) | Arabidopsis | d |
| LjNEST19b1r | AW720443 | Hexose transporter | Arabidopsis | d |
| Ljirnpest17-367-f7 | AW163951 | Suc transporter | Vitis vinifera | d |
| MWM205h12_r | AV411405 | Inorganic pyrophosphatase | Beta vulgaris | d |
| BAA36841 | Vacuolar H+-pyrophosphatase | Chara corallina | b | |
| T02083 | H+-transporting ATPase | Z. mays | b | |
| EST555109 (Le) | BI935220 | P-ATPase | Tomato | d |
| AAK31799 | Plasma membrane H+-ATPase | Lilium longiflorum | d | |
| NF085A12LF(Mt) | BG452216 | P-ATPase | Broad bean (Vicia faba) | d |
| bags17i02 (Hv) | AV916502 | V-ATPase | H. vulgare | d |
| AAC17840 | Vacuolar H+-ATPase catalytic subunit | G. hirsutum | d | |
| CAB70948 | Putative periplasmic-binding protein | Nostoc sp. | d | |
| NP_196853 | Adenosine nucleotide translocator | Arabidopsis | d | |
| NP_187470 | Adenylate translocator | Arabidopsis | d | |
| NP_196853 | Adenosine nucleotide translocator | Arabidopsis | d | |
| LjNEST30d5r | BI417536 | Adenine nucleotide translocator ANT1 | Z. mays | d |
| S05199 | ADP, ATP carrier protein G1 | Z. mays | d | |
| MWM163c10_r | AV418971 | Porin, anion channel VDAC | Pea | d |
| Signaling proteins | ||||
| LjNEST61b5r | BI420746 | Receptor protein kinase | Arabidopsis | d |
| LjNEST54h11r | BI420254 | Receptor protein kinase-like protein | Arabidopsis | d |
| ST55a7r | BI420272 | Receptor protein kinase | Arabidopsis | d |
| LjNEST43a8r | BI418550 | Ser/Thr kinase | Arabidopsis | a |
| AAA79977 | CDC2 (mitotic kinase) | Paramecium tetraur. | a | |
| Gm-c1009-3123 | AW306718 | Calmodulin | O. sativa | a |
| U09376 | GF14omega isoform | Arabidopsis | a | |
| BF153219 | 14-3-3 Protein homolog 30G | S. tuberosum | a | |
| AF121196 | 14-3-3 Protein | Populus × canescens | a | |
| U70534 | 14-3-3-Like protein B | Soybean | a | |
| X98866 | 14-3-3-Like protein 10 | Tomato | a | |
| U91727 | 14-3-3-Like protein F | Nicotiana tabacum | a | |
| Pathogen response-related proteins | ||||
| AAF68389 | Hypersensitive-induced response (HIR) protein | Z. mays | b | |
| LjNEST62h12r | BI420901 | HIR | O. sativa | b |
| HVSMEf0004I03f (Hv) | BF254610 | PR-4 protein, barwin protein | H. vulgare | d |
| LjNEST5d9r | AW719509 | PR-10 protein | M. truncatula | d |
| G077P02Y (Pc) | BI128498 | Beta-1,3-glucanase | Arabidopsis | a |
| BNLGHi10144 (Gh) | AI728205 | Putative beta-1,3-endoglucanase | Arabidopsis | a |
| Plasma- and endomembrane-related proteins | ||||
| AF000403 | Cytochrome P450 (71D11) | L. japonicus | d | |
| Gm-c1016-4435 | AW458588 | Cytochrome B5 | Arabidopsis | d |
| Ljirnpest33-648-e9 | BF177685 | Annexin | M. truncatula | a |
| Gm-c1063-2804 | BM731845 | Calreticulin | Ricinus communis | a |
| LjNEST44g6r | BI419312 | Protein disulfide-isomerase | Medicago sativa | a |
| Q16956 | Glc-regulated protein (BIP) | Aplysia californica | b | |
| AAK21920 | BiP-isoform D | Soybean | b | |
| GA__Ea0008J01f (Ga) | BG440513 | ADP-ribosylation factor (ARF) | Z. mays | d |
| JH0260 | ARF | Candida albicans | d | |
| CAA98170 | RAB7C | L. japonicus | d | |
| T03629 | RAB7D | N. tabacum | d | |
| MWL041g09_r | AV408457 | Rab7D | L. japonicus | d |
| BAB78669 | GTP-binding protein Rab7 family | O. sativa | d | |
| NP_175638 | RAB7D | Arabidopsis | d | |
| Gm-c1013-2333 | AW132296 | NSF attachment receptor | O. sativa | d |
| HWM006.H02 (Hv) | BE421201 | Membrane-associated protein | Arabidopsis | d |
| Gm-c1063-184 | BG047075 | Oligosaccharyltransferase like | Arabidopsis | b |
| Gm-c1040-2451 | BG352557 | Sterol-c-methyltransferase | Arabidopsis | d |
| BNLGHi13139 (Gh) | AI729343 | Glucanase homolog | Arabidopsis | a |
| LjNEST19b3r | AW720444 | Pectin methylesterase | Sesbania rostrata | b |
| Metabolic enzymes | ||||
| MWM218f08_r | AV412349 | Enolase | Arabidopsis | a |
| LjNEST19b8r | AW720446 | Enolase | Arabidopsis | a |
| CAC12826 | Malate dehydrogenase | N. tabacum | a | |
| Gm-c1061-1770 | BE805404 | Malate dehydrogenase | M. truncatula | a |
| NP_181788 | Phosphoenolpyruvate carboxylase (PEPC) | Arabidopsis | d | |
| NF120A05PL (Mt) | BI264706 | PEPC | G. hirsutum | d |
| CAB39757 | Suc synthase | L. japonicus | b | |
| CAB40794 | Suc synthase | M. truncatula | b | |
| BAA76430 | Fru-bisphosphate aldolase | Cicer arietinum | a | |
| MWL037h09_r | AV408222 | Fru-1,6-bisphosphatw aldolase | C. arietinum | a |
| P26520 | Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) | Petunia hybrida | a | |
| HVSMEb0004G08f (Hv) | BF627264 | Adenylate kinase A | O. sativa | a |
| Q08479 | Adenylate kinase A | O. sativa | a | |
| EST582638 (St) | BM408311 | Adenylate kinase B | O. sativa | a |
| Gm-c1065-2119 | BF070616 | Adenylate kinase B | O. sativa | a |
| NP_201145 | Adenylate kinase | Arabidopsis | a | |
| P19251 | Asn synthetase (ASN1) | Pea | b | |
| P49093 | Asn synthetase (ASN2) | L. japonicus | b | |
| CAA96526 | Asn synthetase | Broad bean | b | |
| LjNEST37f9r | BI418534 | Chorismate mutase (CM2) | Arabidopsis | a |
| LjNEST58b9r | BI420527 | Chorismate mutase | Arabidopsis | a |
| AAG24873 | Gln synthetase GSbeta1 | Soybean | a | |
| MWL071b11_r | AV410350 | Cys synthase (AtcysC1) | Arabidopsis | a |
| Q00834 | Cys synthase | Spinacia oleracea | a | |
| CAA11226 | Chalcone reductase | S. rostrata | a | |
| Gm-c1004-1546 | AI437675 | Quinone oxidase | C. arietinum | d |
| HVSMEf0022G06f (Hv) | BF260581 | Peroxidase | Z. mays | d |
| WHE1127_H02_P03 (Tae) | BE444079 | Acetoacetyl CoA thiolase | Z. mays | d |
| EST470535 (Le) | BG124889 | Protein phosphatase | Arabidopsis | a |
| Diverse proteins | ||||
| E007G04 (Os) | BI809008 | Heat shock protein (HSP70) | O. sativa | d |
| FM1_6_D07.b1_A003 (Sp) | BF421178 | Heat shock protein (HSP70) | O. sativa | d |
| LjNEST15b10r | BI419761 | ENOD18 | Broad bean | b |
| LjNEST42b2r | BI419940 | Ethylene-regulated (ER6) protein | Arabidopsis | b |
| LjNEST1f4r | AW719244 | Leghemoglobin | L. japonicus | b |
| BAB18108 | Leghemoglobin | L. japonicus | b | |
| MtBC10A03F1 (Mt) | AL382789 | Ubiquitin | Z. mays | d |
| AB04E05 | Similar to fibrillin | Arabidopsis | a | |
| BAB10497 | Nuclear pore protein like | Arabidopsis | d | |
| BAA03710 | Cytokinin-binding protein (CBP57) | Nicotiana sylvestris | d | |
| LjNEST24c11r | BI418071 | Chloroplast nucleoid DNA-binding protein | Arabidopsis | d |
| Bacteroid proteins | ||||
| NC_002678 | Hypothetical protein | Mesorhizobium loti | a | |
| NC_002678 | Unknown protein | M. loti | d | |
| NC_002678 | Outer membrane protein | M. loti | d | |
| NC_002678 | Cytochrome-c oxidase fixP chain | M. loti | d | |
| NC_002678 | Unknown protein | M. loti | d | |
| NP_436666 | Putative sugar ABC transporter protein | Sinorhizobium meliloti | d | |
| AAA21773 | Nitrogenase iron protein | R. leguminosarum | a | |
| P54978 | Phytoene dehydrogenase | Agrobacterium sp. | d | |
| NP_578667 | 3-Isopropylmalate dehydratase | Pyrococcus furiosus | a | |
| NC_000913 | Putative asparaginase | Escherichia coli | d | |
| NC_003030 | ABC transporter, ATP-binding protein | Clostridium acetobutylicum | d | |
| AAA99874 | Heat shock protein (hsp70) | Euplotes eurystomus | d | |
| AAD03835 | 4-Oxalocrotonate decarboxylase | Sphingomonas chungbuken. | a | |
| NP_269026 | Putative chorismate synthase | Streptococcus pyogenes | d | |
| T35824 | Probable oxidoreductase | Streptomyces coelicolor | a |
Proteins are grouped according to functional similarity. The partitioning into the Triton X-114 phases is indicated in column 5 (X114): a, aqueous phase; d, detergent phase; and b, in both phases. A match in an EST is indicated by the corresponding EST name in column 1. An indication of the species for the EST in column 1 is only given if the EST is not from L. japonicus and is also omitted from soybean ESTs, which are characterized by Gm in the EST name. The following abbreviations of species names are used in column 1: Ga, Gossypium arboreum; Gh, Gossypium hirsutum; Hv, Hordeum vulgare; Le, tomato (Lycopersicon esculentum); Mt, M. truncatula,Os, Oryza sativa, Pc, Populus cambium; Sp, Sorghum propinquum;St, Solanum tuberosum; and Tae, Triticum aestivum. If there is no entry in column 1, the match is directly in the nr database. In column 2 the accession nos. of the ESTs or proteins are given. Column 3 contains the name of the protein identified in the nr database either by a direct match or after a BLAST search with the corresponding EST. The BLAST searches mostly yielded a no. of hits in similar proteins, but only the best one is given. The name of the species corresponding to the identified protein is given in column 4.
Table III.
List of the identified ESTs with no similarity to other proteins (see also legend to Table I)
| EST | Accession No. | Species | Origin | X114 |
|---|---|---|---|---|
| LjNEST22c12rc | AW720598 | L. japonicus | Nodules | d |
| LjNEST3g1r | AW719382 | L. japonicus | Nodules | d |
| LjNEST11e1rc | AW720618 | L. japonicus | Nodules | d |
| LjNEST3a10r | AW719341 | L. japonicus | Nodules | d |
| MWM245c05_r | AV414507 | L. japonicus | Young plants | a |
| MWM178f04_r | AV420140 | L. japonicus | Young plants | a |
| MWL025f02_r | AV407519 | L. japonicus | Young plants | d |
| MWM171h12_r | AV419560 | L. japonicus | Young plants | d |
| Gm-c1020-1157 | AI973715 | Soybean | Nodules | d |
| Gm-c1036-1221 | AW756075 | Soybean | Somatic embryos | d |
| Gm-c1027-5614 | AW760211 | Soybean | Cotyledons | d |
| Gm-r1070-6097 | BE821765 | Soybean | Mixed | a |
| Gm-r1070-5509 | BE821529 | Soybean | Mixed | a |
| pHOGA-7L14 | BG646466 | M. truncatula | Seedling roots | d |
| NF088H12IN | BI265149 | M. truncatula | Leaves fed upon by Spodoptera exigua | d |
| NF078B05EC | BF646608 | M. truncatula | Elicited cell culture | a |
| cLED17E2 | AI489301 | Tomato | Carpel | d |
| cLEG31D1 | BE460472 | Tomato | Pericarp, breaker stage | d |
| cTOB30B18 | BI929926 | Tomato | Flower buds | d |
| HVSMEf0011H11f | BF256953 | H. vulgare | Seedling root | d |
| BCD178_WHE01B07 | BE438623 | H. vulgare | Leaf | d |
| PIC1_7_C02.g1_A002 | BM327529 | Sorghum bicolor | Seedlings infected with Colletotrichum raminicola | d |
| C11289_1A | AU067946 | O. sativa | Callus | d |
| PPN281517 | BI740967 | Physcomitrella patens | Protonemata: 7-d-old tissue, auxin treated | d |
Most of the matches were significant single peptide matches, and in a number of cases several single matches identified similar, homologous proteins of the same protein type. This was especially the case with ATPases, aquaporins, and 14-3-3 proteins. Seventy of the matches were directly obtained in the nonredundant protein database (National Center for Biotechnology Information), which identified 52 proteins, whereas 90 of the matches were in the EST database (green plant). Subsequently, these EST sequences were searched against the nonredundant database by BLAST. In this way, another 33 proteins were identified. In addition, eight hypothetical proteins were found, and 24 ESTs had no homology to any known proteins.
The matches and identified proteins are grouped in Table I according to similarity of function. Proteins related to membrane transport, signaling, pathogen response, and the endomembrane system were identified. A number of soluble metabolic enzymes and other diverse proteins were also present. The following discussion is based on this grouping.
Membrane Transporters
Aquaporins of the nodulin 26 family are known to occur at the PBM in soybean (Fortin et al., 1987). In L. japonicus, LIMP2 is considered the nodulin 26 homolog (Guenther and Roberts, 2000). Our search did not identify this protein. However, several other aquaporins were found. First, LIMP1, known to be expressed in both roots and nodules in L. japonicus (Guenther and Roberts, 2000), was identified in the PBM preparation. It was also detected as a member of a PBM protein complex (see below) and, furthermore, was present in PM preparations from uninfected roots (data not shown). In addition, three other matches in L. japonicus ESTs led to the identification of aquaporins related to PIP1, PIP2, and δ-TIP. All three matches were based on specific peptides with no similarity to the L. japonicus aquaporins LIMP1 and LIMP2 (data not shown), indicating the existence of several types of aquaporins at the PBM of L. japonicus.
Recent biochemical and immunological studies (Blumwald et al., 1985; Fedorova et al., 1999) indicated that the major H+-ATPase of symbiosomes is a P-ATPase. In accordance, four matches led to the identification of P-ATPases in our study. The occurrence of V-type H+-ATPase (V-ATPase) has not been demonstrated unequivocally so far (see Fedorova et al., 1999, and refs. therein), but it was detected in a proteome study of pea PBM (Saalbach et al., 2002). The finding of V-ATPase in the present study supports the presence of this type at the PBM. The V-ATPase was also found in a protein complex isolated from PBM of L. japonicus and soybean (see below). Furthermore, two matches identified a vacuolar H+-pyrophosphatase in the PBM preparations, an enzyme working in parallel to the V-ATPase at the vacuolar membrane. Taken together, our data indicate a more complex situation regarding both aquaporins and ATPases at the PBM than previously thought.
Other transporters have not been well characterized at the PBM. Our search identified several sugar transporters and also transporters for sulfate and nitrate. All matches were in ESTs from a L. japonicus nodule library and, thus, localized the translation products of these nodule transcripts to the PBM. The sulfate transporter transcript was shown to be highly up-regulated in nodules compared with uninfected roots (Trevaskis et al., 2002). The translation product is also abundant because in addition to the identification in global PBM digests, it could also be identified as a band on a one-dimensional SDS gel (data not shown). The identified nitrate transporter has very high similarity to peptide transporters. Therefore, its function remains unclear. The identified transporters for sugars like Suc and hexoses might play an important role in the plant's supply of carbohydrate to the bacteroids.
A porin (anion channel) and a nucleotide translocator (five matches) represent the only mitochondrial proteins detected in this study. The porin was also found in a proteome study of pea PBM (Saalbach et al., 2002). Other mitochondrial proteins were reported in the same paper and in a proteome study of soybean PBM (Panter et al., 2000). Despite the absence of a mitochondrial marker enzyme in the PBM preparations (see above), these proteins might represent a contamination. On the other hand, mitochondrial proteins were also observed in a proteome study of phagosomes, the key organelles of macrophages (Garin et al., 2001). These organelles of animal cells are formed by endocytosis similar to the formation of the symbiosomes in the plant system. Furthermore, the occurrence of mitochondrial proteins (like HSP60) at other sites has been reported earlier for a number of proteins (for review, see Soltys and Gupta, 1999). Therefore, other approaches such as immunolocalization have to be used to confirm the possible location of the specific mitochondrial proteins at the PBM.
Proteins Involved in Signaling
Several proteins that may play a role in signaling were found in the L. japonicus PBM preparations. In particular, three matches were obtained in ESTs from L. japonicus nodules encoding proteins homologous to a receptor protein kinase in Arabidopsis. These sequences show no significant similarity to the recently described SYMRK (Stracke et al., 2002) and NORK (Endre et al., 2002) receptor kinases that are involved in the Nod-factor perception/transduction system, a symbiotic signal transduction pathway leading from the perception of microbial signal molecules to rapid symbiosis-related gene activation. Therefore, the receptor kinase identified in our study has a different and so far unknown signaling function at the PBM.
Finally, several isoforms of the plant 14-3-3 protein family were matched in our search. The presence of 14-3-3 proteins at the PBM is not surprising because many diverse functions in enzyme regulation and signal transduction at different cellular locations have been attributed to the 14-3-3 protein family (for recent review, see Aducci et al., 2002; Sehnke et al., 2002). One specific function is the regulation of the PM H+-ATPase (Baunsgaard et al., 1998). This ATPase is also present at the PBM (see above), and it could be suggested that one function of the 14-3-3 proteins at the PBM is the regulation of this ATPase. Other functions, such as the regulation of kinases, are likely.
Pathogen Response-Related Proteins
The plant pathogen responses include the induction of pathogenesis-related (PR) proteins. This protein family can be subdivided into several groups, which are partly also induced by other stress. PR proteins are secretory proteins and are known to be located extracellularly (for review, see Kitajima and Sato, 1999). In our study, we have identified homologs of three members of this family, PR-2, PR-4, and PR-10. Because they are secretory proteins, it could be suggested that they are located at the inner side of the PBM in the space between the PBM and the bacteroids. The PR-2 proteins are endo-β-1,3-glucanases that are induced by viral, bacterial, and fungal pathogens (Beffa and Meins, 1996). At the PBM, they might play a role in the modification of the bacteroid cell wall. The PR-4 family is formed by small proteins with a signal peptide for extracellular targeting and with similarity to wound-inducible proteins (win; Linthorst et al., 1991). The PR-10 proteins have ribonuclease activity (Bantignies et al., 2000) and have also been localized extracellularly (Pinto and Ricardo, 1995). The observed PR-10 homolog is similar to MtPR10-1 that is constitutively expressed in roots of M. truncatula and does not respond during nodulation, but is pathogen inducible in leaves (Gamas et al., 1998). Another PR-10 protein in M. truncatula, MtN13, is exclusively expressed in response to infection with rhizobia (Gamas et al., 1998).
Furthermore, a homolog of another potential plant defense protein family, HIR, was detected in the PBM preparations. There is little information on the function of these proteins available, but it is thought that they are involved in the hypersensitive reactions leading to cell death and pathogen resistance (Nadimpalli et al., 2000). This is also concluded from their structural similarity to prohibitins and stomatins (Nadimpalli et al., 2000), both representing families of membrane proteins. HIR proteins have not been detected in nodules so far. However, when the PBM preparations were separated on one-dimensional SDS gels, the HIR protein could also be identified in a clearly visible band (data not shown). This, together with the frequent peptide matches observed in our database searches (data not shown), indicate that it is an abundant protein in nodule cells. The EST matched in our search (LjNEST62h12r) represents a full-length cDNA clone isolated from nodules of L. japonicus. Interestingly, the HIR homolog was also detected in a protein complex when PBM preparations were resolved on a BN gel (see below).
Endomembrane and PM Proteins
Similar to studies on soybean (Panter et al., 2000) and pea (Saalbach et al., 2002) PBM, proteins characteristic for the endomembrane system have been found in the present study of L. japonicus PBM. As discussed previously (Saalbach et al., 2002), such proteins can be expected in the symbiosomes and at the PBM because other studies have shown the involvement of endomembranes in the formation of symbiosomes (for review, see Whitehead and Day, 1997). Interestingly, many ER and Golgi proteins were also detected in the proteome study of phagosomes, the key organelles of macrophages (Garin et al., 2001). These organelles of animal cells are formed by endocytosis similar to the formation of the symbiosomes in the plant system. Together, the results point to a similar mechanism of formation of these organelles and support the known connection between the endomembrane system and symbiosome biogenesis.
In the present study, a number of novel proteins including secreted and vesicle proteins not yet described for the PBM were detected. For example, an annexin homolog was present in the PBM preparations. Annexins have diverse cellular roles including functions in vesicle transport and secretion (for review, see Delmer and Potikha, 1997), and in the presence of calcium, annexins can be membrane associated (for review, see König and Gerke, 2000). In M. truncatula, an annexin was expressed during the early symbiotic response indicating a role in Nod-factor signaling pathway(s) (Niebel et al., 1998).
Cytochrome P450, characteristic for the ER and found to be up-regulated in nodules (Colebatch et al., 2002) and cytochrome b5, normally located at the PM, were found in the PBM preparations. Otherwise, homologs of the small GTP-binding proteins ARF and Rab7 involved in vesicle traffic were present. Rab7 in particular has been shown to be expressed in nodules of L. japonicus (Borg et al., 1997) and to be essential for the biogenesis of symbiosomes and the PBM (Cheon et al., 1993).
Several proteins that are normally secreted such as PR proteins (see above) and pectin methylesterase were also constituents of the PBM preparations. For these and other detected proteins, the question arises as to how they are located to the PBM and which specific functions they fulfill there.
Metabolic Enzymes
Surprisingly, a number of soluble metabolic enzymes were identified in the PBM fraction. According to their solubility, most of them were separated into the aqueous phase of the Triton X-114 partitioning. These proteins might represent a contamination. Cytoplasmic proteins could stick nonspecifically to the membrane or could be caught in membrane vesicles during the disruption and partitioning of the PBM, and abundant proteins like the observed enzymes could then be detected in the analysis. However, it should be noted that, with the used methodological approach, quantification is not possible. This means that no conclusion can be made as to the level of the possible contamination.
On the other hand, proteome studies often localize proteins at unexpected locations, and specific experiments have localized proteins to different compartments. This was also the case in a proteome analysis of the Arabidopsis cell wall (Chivasa et al., 2002), where five proteins that are normally not considered to be secreted were detected in cell wall extracts. This included the mitochondrial enzyme citrate synthase and the soluble enzymes enolase and GAPDH. Enolase and GAPDH, which were also present in our PBM preparations, were immunologically localized to the cell wall of yeast (Gozalbo et al., 1998; Edwards et al., 1999).
Several of the enzymes detected in our study are known to be important for the nodule metabolism and to be up-regulated in nodules. Up-regulation compared with uninfected roots was shown for Suc synthase, MDH, PEPC, GAPDH, Asn synthase, Gln synthase, and chorismate mutase (Colebatch et al., 2002). In the case of Suc synthase (nodulin 100), it is known that it can be bound to the PM (Komina et al., 2002). This enzyme was also found in a special PBM protein complex separated on a native gel (see below). These results indicate that Suc synthase is not only bound to the PM but also to the PBM. The other enzymes related to glycolysis are especially important for the supply of carbohydrate to the bacteroids. Carbon dioxide fixation by PEPC leads to the formation of dicarboxylic acids, which are reduced to malate via MDH. PEP formation also involves the action of enolase that was present in the PBM preparations as well. Malate serves as the primary carbon source to support the respiratory needs of the bacteroids. Furthermore, enzymes such as the Asn and Gln synthases are key enzymes of the assimilation of the ammonia released by the bacteroids. On the basis of these functions and our findings, a model could be suggested as an alternative to contamination, where cytosolic functional protein complexes closely surround the symbiosomes and can be bound to the PBM. For proteins normally secreted (see PM proteins above) and for proteins that might be secreted (like enolase and GAPDH; see above), a localization inside the symbiosomes, i.e. in the peribacteroid space between the PBM and the outer bacteroid membrane, also has to be considered.
Possible PBM Protein Complexes
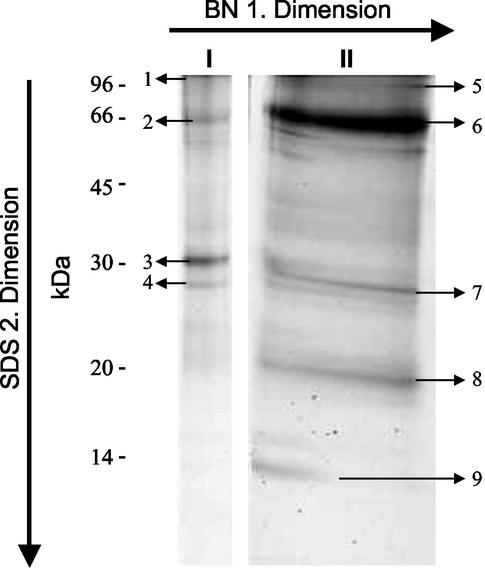
Recent proteome studies have shown that most proteins occur in complexes with other proteins (e.g. Ho et al., 2002). To get a first insight into possible protein interactions in the PBM, proteins were extracted under native conditions and separated with a BN gel. On such gels only two abundant and distinct protein complexes could be detected. This pattern was very similar with PBM from L. japonicus and soybean (data not shown). These complexes were cut out from the gel and loaded for a second-dimension separation onto an SDS gel (Fig. 1). Several abundant bands (see Fig. 1) were obtained from both complexes and were analyzed by MS/MS. The identified proteins are listed in Table II. The occurrence of these special proteins in a complex is surprising, and the results are difficult to interpret. Complex I contained P-ATPase as the only integral membrane protein. Otherwise, the HIR protein discussed above and remorin were found in this complex. Remorin is a small hydrophilic protein tightly associated to the PM. It binds uronides and is thought to be a plasmodesmata-associated protein involved in cell-to-cell signaling or transport (Reymond et al., 1996). Interestingly, the enzyme isocitrate lyase normally located in glyoxysomes was also found as a component of complex I. This is comparable with the finding of the mitochondrial enzyme citrate synthase in cell wall extracts in Arabidopsis (Chivasa et al., 2002). The latter can also occur in the cytoplasm, where it forms filaments binding to the eukaryotic elongation factor eEF1α (Numata et al., 1991), and thus has been found at three different locations. Similarly, isocitrate lyase might not only be targeted to glyoxysomes but (possibly on the basis of posttranslational modifications) also to symbiosomes to fulfill special functions (see also discussion above).
Figure 1.
Results of a two-dimensional separation of total L. japonicus PBM proteins with Blue Native (BN) gel electrophoresis as the first dimension and SDS gel electrophoresis as the second dimension. Lanes I and II represent the lanes from the SDS gel after separation of the complexes I and II from the BN gel. Arrows indicate the bands analyzed by MS/MS as described in “Materials and Methods.”
Table II.
Proteins identified by MS/MS in bands obtained after a two-dimensional separation of total L. japonicus PBM proteins with Blue Native (BN) gel electrophoresis as the first dimension and SDS-gel electrophoresis as the second dimension (see Fig. 1)
| EST | Accession No. | Protein Name | Species | Band No. | Complex |
|---|---|---|---|---|---|
| AJ310523 | P-ATPase | Broad bean | 1 | I | |
| P15479 | Isocitrate lyase | R. communis | 2 | I | |
| AF236373 | HIR | Z. mays | 3 | I | |
| LjNEST60e3r | U72489 | Remorin | S. tuberosum | 4 | I |
| AJ131964 | Suc synthase | M. truncatula | 5 | II | |
| AY037143 | V-ATPase | Pea | 6 | II | |
| LjNEST3g1r | AW719382 | No match | – | 7 | II |
| AF275315 | LIMP1 | L. japonicus | 8 | II | |
| AY063865 | Unknown | Arabidopsis | 9 | II |
Complex II contained the membrane proteins V-ATPase and LIMP1. LIMP1 represents an aquaporin know to occur in roots and nodules (Guenther and Roberts, 2000; see also discussion above). Like in the global PBM analysis, LIMP2, the possible homolog of soybean nodulin 26 (Guenther and Roberts, 2000), could not be detected in the abundant bands. However, when soybean PBM was analyzed, nodulin 26 was clearly present in complex II (data not shown). Otherwise, the enzyme Suc synthase was detected in complex II from L. japonicus PBM. Suc synthase can be bound to the PM and was also found in the global PBM preparations (see Table I and discussion above). Finally, two unknown proteins were present. One of them, represented by EST LjNEST3g1 from a L. japonicus nodule cDNA library, was also found in the general analysis of the PBM (see Table III). Because it was clearly detected in a band from complex II, it might represent a novel important PBM protein.
The identified proteins were found in clear and abundant bands, and it can be concluded that they represent specific components of the complexes. However, there is no known functional relationship among any of the identified proteins, and no specific functional interactions can be deduced from the results. The results might also point to a special structural protein arrangement in the PBM. It can be expected that both complexes contain more proteins than the identified ones, and further analysis might lead to a better understanding of the importance of these complexes.
Diverse Proteins
In addition to the protein groups described above, diverse proteins from plants and the bacteroids were detected (see Table I). In some cases, the matches were in non-plant and non-Rhizobium spp., and an assignment of origin cannot be made. Of the plant proteins, leghemoglobin was also detected in the proteome study of pea PBM (Saalbach et al., 2002), and its binding to the PBM was discussed therein. Enod18 is an early nodulin and, therefore, most likely involved in early events of the infection process. It belongs to a novel family of ATP-binding proteins (Hohnjec et al., 2000), and immunological studies demonstrated that it was localized to the cytoplasm of infected cells in the nitrogen-fixing zone of broad bean nodules (Becker et al., 2001). Its function is unknown, and similar to other proteins found in our study, the specificity and importance of the binding to the PBM have to be investigated.
The ethylene-regulated protein ER6 found in our study was also found to be up-regulated in L. japonicus nodules (Colebatch et al., 2002). It is a small soluble protein of unknown function that was found to be induced by ethylene in tomato fruits, but not in leaves and roots (Zegzouti et al., 1999).
Fifteen matches were in proteins from Rhizobium spp. and other bacteria (see Table I). Most likely, these proteins represent a contamination of the PBM preparation by bacteroid proteins, although this contamination would be relatively low compared with the total number of detected proteins. A much higher contamination by bacteroid proteins was observed and discussed in a recent study of pea PBM (Saalbach et al., 2002). This difference might be due to the different structure of symbiosomes from pea and L. japonicus and to the different isolation procedures (see also Saalbach et al., 2002). A number of matches, mostly in ESTs, led to hypothetical proteins (mostly of Arabidopsis) with unknown function (see Table I). Furthermore, 24 matches were obtained in ESTs, the translation products of which showed no homology to known proteins (Table III). Eight of those were in ESTs from L. japonicus, and another eight in other legume ESTs. Two of the L. japonicus ESTs (LjNEST11e1 and LjNEST3a10) were found to be up-regulated in nodules, the latter possibly even nodule specific (Colebatch et al., 2002). The ESTs the products of which show no homology to known proteins potentially encode novel PBM proteins, and further studies will reveal their nature and their function.
CONCLUSIONS
In this proteome study of the PBM, we have used global trypsin digestion and nano-LC/MS/MS for protein analysis. This approach led to the identification of a large number of proteins: 94 known proteins and 24 ESTs of unknown identity were detected. This number by far exceeds the number of proteins hitherto reported for the PBM. A similar study on soybean PBM reported 17 proteins (Panter et al., 2000), and in a study on pea, only very few PBM proteins could be detected because of the special structure of pea symbiosomes (Saalbach et al., 2002). In the present study, a number of hydrophobic membrane proteins such as transporters for sugars, peptides, and sulfate were detected for the first time, to our knowledge. In special cases, such as the occurrence of aquaporins, our results indicate a more complex situation than previously thought. These results provide a new valuable basis for specific molecular functional studies on metabolite exchange through the PBM.
Many of the proteins were identified via matches in the EST database (different cDNA libraries). In this way, translation products of nodule-specific transcripts were localized to the PBM. In addition, 24 ESTs of unknown identity (and function) were identified, which provide a basis for new studies on unknown proteins and functions of the PBM.
A receptor protein kinase different to two recently described receptor kinases involved in nodule formation (Endre et al., 2002; Stracke et al., 2002) was found. This protein seems to be of special interest for further studies on signal transduction at the PBM. Furthermore, a potential plant defense protein (HIR protein) was for the first time detected in the PBM preparations. The function of this protein is unknown, but it is thought to be involved in hypersensitive reactions leading to cell death and pathogen resistance (Nadimpalli et al., 2000). It seems to be abundant at the PBM and represents an interesting novel candidate for the investigation of the function of a pathogen-related protein at the PBM. Work is in progress to study the specific localization and possible interactions with other proteins by using gene fusion constructs.
Similar to other studies on the PBM, endomembrane proteins were detected in our proteome analysis. These results support the involvement of the endomembrane system in symbiosome biogenesis, which is consistent with results obtained in a proteome study of phagosomes (Garin et al., 2001). The latter study led to new conclusions as to the extent and importance of the endomembrane system in the biogenesis of these endocytotic organelles. This and our finding of PM and extracellular proteins should stimulate future studies on intracellular protein targeting to demonstrate whether specific transport pathways and signals exist for the delivery of such proteins to the symbiosomes.
Like in many other proteome studies, a number of unexpected proteins was detected in the PBM preparations. Most of them are soluble cytoplasmic and extracellular proteins. Such proteins might represent a contamination caused by unspecific binding to the PBM. However, no conclusion can be made as to the level of the possible contamination. On the other hand, soluble proteins could also specifically bind and form functional complexes at the PBM. Evidence for this localization has to come from further studies using immunolocalization and gene fusion constructs, as mentioned above for the HIR protein.
Taken together, our results provide new information and, in particular, a valuable basis for further studies to gain insights into the structure and function of the PBM.
MATERIALS AND METHODS
Plant Material
Lotus japonicus GIFU (B-129) seeds were germinated and grown in hydroponic culture with vermiculite in controlled environment (16-h day/8-h night, 21°C/17°C day/night temperature, and 60% relative humidity). Plants were inoculated when 7 d old with Mesorhizobium loti strain R7A and were provided with 1 mm KNO3 for the first 2 weeks of growth.
PBM and PM Vesicle Isolation
Nodules were harvested from 7-week-old plants, and a raw symbiosome pellet was prepared as described previously (Saalbach et al., 2002). After the first pelleting of the symbiosomes, the pellet was immediately resuspended in S buffer (0.33 m Suc, 10 mm dithiothreitol, 0.1 mm Na2EDTA, and 5 mm Tris-HCl [pH 7.8]) in a glass homogenizer to disrupt the symbiosomes. The homogenate was centrifuged at 7,000g for 10 min to pellet the bacteroids. The supernatant was centrifuged at 100,000g for 1 h to pellet membranes. This pellet was resuspended in 4 mL of S buffer in a glass homogenizer. The PBM was purified from the homogenate by using an aqueous polymer two-phase system according to Larsson et al. (1987). A 16-g system prepared in 5 mm potassium phosphate buffer (pH 7.8; 0.33 m Suc, 1 mm dithiothreitol, and 0.1 mm Na2EDTA) was used for material from 20- to 40-g nodules. The final upper aqueous phase was centrifuged at 100,000g for 1 h and the pellet representing the PBM fraction was stored at −80°C. The polymer concentration was optimized for PBM purification by assaying the marker enzyme activities of ATPase and cytochrome c oxidase (Christiansen et al., 1995) and the succinate-dependent cytochrome c reductase (Douce et al., 1972). The protein content was estimated according to Bradford (1976).
Protein Extraction, BN PAGE, and Trypsin Digestion
For global trypsin digests, the proteins were extracted from the PBM by using Triton X-114 according to Vachon et al. (1991). After Triton X-114 separation, the samples were precipitated with acetone and redissolved in 25 mm NH4HCO3, 5% (v/v) acetonitrile, and 0.5 mm CaCl2 and digested with trypsin (1 μg/100 μg protein) at 37°C overnight. For BN gels, a PBM pellet (1 mg of protein) was extracted as described by Brookes et al. (2002), except that the samples were stored on ice for about 1 h before gel loading. BN gel electrophoresis was performed as described by Schägger and von Jagow (1991).
Bands were cut from the BN gel and loaded onto an SDS gel to separate the individual proteins of the complexes. The SDS gel was stained with Coomassie Brilliant Blue G250, and bands were cut and treated with trypsin and analyzed by MS/MS as described previously (Saalbach et al., 2002).
Nano-LC/MS/MS of Triton-X114-Separated PBM Proteins
Trypsin-digested samples were lyophilized and redissolved in 5% (w/v) acetonitrile and 0.1% (v/v) formic acid in water. Aliquots corresponding to approximately 5 μg of protein were loaded onto a nano-reversed phase column (75-μm × 15-cm i.d., 3-μm C18, 100 Å; LC-Packings, Amsterdam). Peptides were eluted from the column with a gradient of 5% to 80% (v/v) acetonitrile for 2.75 h at a flow rate of 130 nL min−1 using an UltiMate nano LC system (LC-Packings). Eluting peptides were continuously analyzed by MS/MS with an Ultima Q-Tof mass spectrometer (Micromass, Manchester, UK) with two dependent MS/MS scans per full MS. The resulting PKL file was used to search the National Center for Biotechnology Information nonredundant and dbEST databases with the MS/MS ion search option of the Mascot search program (www.matrixscience.com).
ACKNOWLEDGMENTS
We thank Allan Stensballe (University of Southern Denmark, Odense, Denmark) for expert assistance in nano-LC/MS/MS performance, Helge Egsgaard (Risø, Roskilde, Denmark) for general support in MS, and Natalia Bykova (Risø) for providing expertise and technical supply in running BN electrophoresis. We are grateful to Ina Hansen and Gertrud Kock for their excellent technical assistance and to Hans Thordal-Christensen, Julia Kinane, and Solveig Christiansen (Risø) for helpful discussions and critical reading of the manuscript.
Footnotes
This work was supported by a 2-year trainee program of the European Union research training network Lotus (to S.W.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.015362.
LITERATURE CITED
- Aducci P, Camoni L, Marra M, Visconti S. From cytosol to organelles: 14-3-3 proteins as multifunctional regulators of plant cell. IUBMB Life. 2002;53:49–55. doi: 10.1080/15216540210813. [DOI] [PubMed] [Google Scholar]
- Bantignies B, Seguin J, Muzac I, Dedaldechamp F, Gulick P, Ibrahim R. Direct evidence for ribonucleolytic activity of a PR-10-like protein from white lupin roots. Plant Mol Biol. 2000;42:871–881. doi: 10.1023/a:1006475303115. [DOI] [PubMed] [Google Scholar]
- Baunsgaard L, Fuglsang AT, Jahn T, Korthout HA, de Boer AH, Palmgren MG. The 14-3-3 proteins associate with the plant plasma membrane H(+)-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J. 1998;13:661–671. doi: 10.1046/j.1365-313x.1998.00083.x. [DOI] [PubMed] [Google Scholar]
- Becker JD, Moreira LM, Kapp D, Frosch SC, Pühler A, Perlic AM. The nodulin vfENOD18 is an ATP-binding protein in infected cells of Vicia fabaL. nodules. Plant Mol Biol. 2001;47:749–759. doi: 10.1023/a:1013664311052. [DOI] [PubMed] [Google Scholar]
- Beffa R, Meins F., Jr Pathogenesis-related functions of plant beta-1,3-glucanases investigated by antisense transformation: a review. Gene. 1996;179:97–103. doi: 10.1016/s0378-1119(96)00421-0. [DOI] [PubMed] [Google Scholar]
- Bestel-Corre G, Dumas-Gaudot E, Poinsot V, Dieu M, Dierick JF, van Tuinen D, Remacle J, Gianinazzi-Pearson V, Gianinazzi S. Proteome analysis and identification of symbiosis-related proteins from Medicago truncatulaGaertn. by two-dimensional electrophoresis and mass spectrometry. Electrophoresis. 2002;23:122–137. doi: 10.1002/1522-2683(200201)23:1<122::AID-ELPS122>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Blumwald E, Fortin MG, Rea PA, Verma DPS, Poole RJ. Presence of host-plasma membrane type H+-ATPase in the membrane envelope enclosing the bacteroids in soybean root nodules. Plant Physiol. 1985;78:665–672. doi: 10.1104/pp.78.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg S, Brandstrup B, Jensen TJ, Poulsen C. Identification of new protein species among 33 different small GTP-binding proteins encoded by cDNAs from Lotus japonicus, and expression of corresponding mRNAs in developing root nodules. Plant J. 1997;11:237–250. doi: 10.1046/j.1365-313x.1997.11020237.x. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brookes PS, Pinner A, Ramachandran A, Coward L, Barnes S, Kim H, Darley-Usmar VM. High throughput two-dimensional blue-native electrophoresis: a tool for functional proteomics of mitochondria and signaling complexes. Proteomics. 2002;2:969–977. doi: 10.1002/1615-9861(200208)2:8<969::AID-PROT969>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Cheon CI, Lee NG, Siddique ABM, Bal AK, Verma DPS. Roles of plant homologs of Rab1p and Rab7p in the biogenesis of the peribacteroid membrane, a subcellular compartment formed de-novo during root-nodule symbiosis. EMBO J. 1993;12:4125–4135. doi: 10.1002/j.1460-2075.1993.tb06096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivasa S, Ndimba BK, Simon WJ, Robertson D, Yu XL, Knox JP, Bolwell P, Slabas AR. Proteomic analysis of the Arabidopsis thalianacell wall. Electrophoresis. 2002;23:1754–1765. doi: 10.1002/1522-2683(200206)23:11<1754::AID-ELPS1754>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Christiansen JH, Rosendahl L, Widell S. Preparation and characterization of sealed inside-out peribacteroid membrane vesicles from Pisum sativum L. and Glycine maxL. root nodules by aqueous polymer two-phase partitioning. J Plant Physiol. 1995;147:175–181. [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK. Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Mol Plant-Microbe Interact. 2002;15:411–420. doi: 10.1094/MPMI.2002.15.5.411. [DOI] [PubMed] [Google Scholar]
- Day DA, Poole PS, Tyerman SD, Rosendahl L. Ammonia and amino acid transport across symbiotic membranes in nitrogen-fixing legume nodules. Cell Mol Life Sci. 2001;58:61–71. doi: 10.1007/PL00000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP, Potikha TS. Structures and functions of annexins in plants. Cell Mol Life Sci. 1997;53:546–553. doi: 10.1007/s000180050070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R, Christensen EL, Bonner WD., Jr Preparation of intact plant mitochondria. Biochim Biophys Acta. 1972;275:148–160. doi: 10.1016/0005-2728(72)90035-7. [DOI] [PubMed] [Google Scholar]
- Edwards SR, Braley R, Chaffin WL. Enolase is present in the cell wall of Saccharomyces cerevisiae. FEMS Microbiol Lett. 1999;177:211–216. doi: 10.1111/j.1574-6968.1999.tb13734.x. [DOI] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB. A receptor kinase gene regulating symbiotic nodule development. Nature. 2002;417:962–966. doi: 10.1038/nature00842. [DOI] [PubMed] [Google Scholar]
- Fedorova E, Thomson R, Whitehead LF, Maudoux O, Udvardi MK, Day DA. Localization of H+-ATPase in soybean root nodules. Planta. 1999;209:25–32. doi: 10.1007/s004250050603. [DOI] [PubMed] [Google Scholar]
- Fortin MG, Morrison NA, Verma DP. Nodulin-26, a peribacteroid membrane nodulin is expressed independently of the development of the peribacteroid compartment. Nucleic Acids Res. 1987;15:813–824. doi: 10.1093/nar/15.2.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P, de Billy F, Truchet G. Symbiosis-specific expression of two Medicago truncatulanodulin genes, MtN1 and MtN13, encoding products homologous to plant defence proteins. Mol Plant-Microbe Interact. 1998;11:393–403. doi: 10.1094/MPMI.1998.11.5.393. [DOI] [PubMed] [Google Scholar]
- Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M. The phagosome proteome: insight into phagosome functions. J Cell Biol. 2001;152:165–180. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozalbo D, Gil-Navarro I, Azorin I, Renau-Piqueras J, Martinez JP, Gil ML. The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicansis also a fibronectin and laminin binding protein. Infect Immunol. 1998;66:2052–2059. doi: 10.1128/iai.66.5.2052-2059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther JF, Roberts DM. Water-selective and multifunctional aquaporins from Lotus japonicusnodules. Planta. 2000;210:741–748. doi: 10.1007/s004250050675. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K et al. Systematic identification of protein complexes in Saccharomyces cerevisiaeby mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Küster H, Albus U, Frosch SC, Becker JD, Pühler A, Perlick AM, Frühling M. The broad bean nodulin VfENOD18 is a member of a novel family of plant proteins with homologies to the bacterial MJ0577 superfamily. Mol Gen Genet. 2000;264:241–250. doi: 10.1007/s004380000292. [DOI] [PubMed] [Google Scholar]
- Kaiser BN, Finnegan PM, Tyerman SD, Whitehead LF, Bergersen FJ, Day DA, Udvardi MK. Characterization of an ammonium transport protein from the peribacteroid membrane of soybean nodules. Science. 1998;281:1202–1206. doi: 10.1126/science.281.5380.1202. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Sato F. Plant pathogenesis-related proteins: molecular mechanisms of gene expression and protein function. J Biochem. 1999;125:1–8. doi: 10.1093/oxfordjournals.jbchem.a022244. [DOI] [PubMed] [Google Scholar]
- Komina O, Zhou Y, Sarath G, Chollet R. In vivo and in vitro phosphorylation of membrane and soluble forms of soybean nodule sucrose synthase. Plant Physiol. 2002;129:1664–1673. doi: 10.1104/pp.002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Gerke V. Modes of annexin-membrane interactions analyzed by employing chimeric annexin proteins. Biochim Biophys Acta. 2000;1498:174–180. doi: 10.1016/s0167-4889(00)00094-x. [DOI] [PubMed] [Google Scholar]
- Larsson C, Widell S, Kjellbom P. Preparation of high purity plasma membranes. Methods Enzymol. 1987;148:558–568. [Google Scholar]
- Linthorst HJ, Danhash N, Brederode FT, Van Kan JA, De Wit PJ, Bol JF. Tobacco and tomato PR proteins homologous to win and pro-hevein lack the “hevein” domain. Mol Plant-Microbe Interact. 1991;4:586–592. doi: 10.1094/mpmi-4-586. [DOI] [PubMed] [Google Scholar]
- Mathesius U, Keijzers G, Natera SH, Weinman JJ, Djordjevic MA, Rolfe BG. Establishment of a root proteome reference map for the model legume Medicago truncatulausing the expressed sequence tag database for peptide mass fingerprinting. Proteomics. 2001;1:1424–1440. doi: 10.1002/1615-9861(200111)1:11<1424::AID-PROT1424>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Morris AC, Djordjevic MA. Proteome analysis of cultivar-specific interactions between Rhizobium leguminosarumbiovar trifolii and subterranean clover cultivar Woogenellup. Electrophoresis. 2001;22:586–598. doi: 10.1002/1522-2683(200102)22:3<586::AID-ELPS586>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Nadimpalli R, Yalpani N, Johal GS, Simmons CR. Prohibitins, stomatins, and plant disease response genes compose a protein superfamily that controls cell proliferation, ion channel regulation, and death. J Biol Chem. 2000;275:29579–29586. doi: 10.1074/jbc.M002339200. [DOI] [PubMed] [Google Scholar]
- Natera SHA, Guerreiro N, Djordjevic NA. Proteome analysis of differentially displayed proteins as a tool for the investigation of symbiosis. Mol Plant-Microbe Interact. 2000;13:995–1009. doi: 10.1094/MPMI.2000.13.9.995. [DOI] [PubMed] [Google Scholar]
- Niebel FC, Lescure N, Cullimore JV, Gamas P. The Medicago truncatula MtAnn1 gene encoding an annexin is induced by Nod factors and during the symbiotic interaction with Rhizobium meliloti. Mol Plant-Microbe Interact. 1998;11:504–513. doi: 10.1094/MPMI.1998.11.6.504. [DOI] [PubMed] [Google Scholar]
- Numata O, Takemasa T, Takagi I, Hirono M, Hirano H, Chiba J, Watanabe Y. Tetrahymena 14-nm filament-forming protein has citrate synthase activity. Biochem Biophys Res Commun. 1991;174:1028–1034. doi: 10.1016/0006-291x(91)91522-e. [DOI] [PubMed] [Google Scholar]
- Panter S, Thomson R, de Bruxelles G, Laver D, Trevaskis B, Udvardi M. Identification with proteomics of novel proteins associated with the peribacteroid membrane of soybean root nodules. Mol Plant-Microbe Interact. 2000;13:325–333. doi: 10.1094/MPMI.2000.13.3.325. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Pinto MP, Ricardo CP. Lupinus albusL. pathogenesis-related proteins that show similarity to PR-10 proteins. Plant Physiol. 1995;109:1345–1351. doi: 10.1104/pp.109.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Day DA, Gresshoff PM. Rapid isolation of intact peribacteroid envelopes from soybean nodules and demonstration of selective permeability to metabolites. J Plant Physiol. 1987;130:157–164. [Google Scholar]
- Reymond P, Kunz B, Paul-Pletzer K, Grimm R, Eckerskorn C, Farmer EE. Cloning of a cDNA encoding a plasma membrane-associated, uronide binding phosphoprotein with physical properties similar to viral movement proteins. Plant Cell. 1996;8:2265–2276. doi: 10.1105/tpc.8.12.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JG, Lyttleton P, Bullivant S, Grayston GF. Membranes in lupin root nodules: I. The role of Golgi bodies in the biogenesis of infection threads and peribacteroid membranes. J Cell Sci. 1978;30:129–149. doi: 10.1242/jcs.30.1.129. [DOI] [PubMed] [Google Scholar]
- Saalbach G, Erik P, Wienkoop S. Characterisation by proteomics of peribacteroid space and peribacteroid membrane preparations from pea (Pisum sativum) symbiosomes. Proteomics. 2002;2:325–337. doi: 10.1002/1615-9861(200203)2:3<325::aid-prot325>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Sehnke PC, DeLille JM, Ferl RJ. Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell Suppl. 2002;14:S339–S354. doi: 10.1105/tpc.010430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltys BJ, Gupta RS. Mitochondrial-matrix proteins at unexpected locations: Are they exported? Trends Biochem Sci. 1999;24:174–177. doi: 10.1016/s0968-0004(99)01390-0. [DOI] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Colebatch G, Desbrosses G, Wanrey M, Wienkoop S, Saalbach G, Udvardi M. Differentiation of plant cells during symbiotic nitrogen fixation. Comp Funct Genomics. 2002;3:151–157. doi: 10.1002/cfg.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyerman SD, Whitehead LF, Day DA. A channel-like transporter for NH4+ on the symbiotic interface of N2-fixing plants. Nature. 1995;378:629–632. [Google Scholar]
- Udvardi MK, Day DA. Metabolite transport across symbiotic membranes of legume nodules. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:493–523. doi: 10.1146/annurev.arplant.48.1.493. [DOI] [PubMed] [Google Scholar]
- Vachon V, Pouliot JF, Laprade R, Beliveau R. Fractionation of renal brush border membrane proteins with Triton X-114 phase partitioning. Biochem Cell Biol. 1991;69:206–211. doi: 10.1139/o91-031. [DOI] [PubMed] [Google Scholar]
- Verma DPS, Hong ZL. Biogenesis of the peribacteroid membrane in root nodules. Trends Microbiol. 1996;4:364–368. doi: 10.1016/0966-842x(96)10053-6. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Whitehead LF, Day DA. The peribacteroid membrane. Physiol Plant. 1997;100:30–44. [Google Scholar]
- Zegzouti H, Jones B, Frasse P, Marty C, Maitre B, Latch A, Pech JC, Bouzayen M. Ethylene-regulated gene expression in tomato fruit: characterization of novel ethylene-responsive and ripening-related genes isolated by differential display. Plant J. 1999;18:589–600. doi: 10.1046/j.1365-313x.1999.00483.x. [DOI] [PubMed] [Google Scholar]