Abstract
To understand further how pollination, seeds, auxin (4-chloroindole-3-acetic acid [4-Cl-IAA]), and gibberellins (GAs) regulate GA biosynthesis in pea (Pisum sativum) fruit, we studied expression of the gene PsGA3ox1 that codes for the enzyme that converts GA20 to biologically active GA1 using real-time reverse transcription-polymerase chain reaction analysis. PsGA3ox1 mRNA levels were minimally detectable in prepollinated pericarps and ovules (−2 d after anthesis [DAA]), increased dramatically after pollination (0 DAA), then decreased by 1 DAA. Seed PsGA3ox1 mRNA levels increased at 4 DAA and again 8 to 12 DAA, when seed development was rapid. Pericarp PsGA3ox1 mRNA levels peaked coincidentally with rapid pod diameter expansion (6–10 DAA) to accommodate the growing seeds. The effects of seeds and hormones on the expression of pericarp PsGA3ox1 were investigated over a 24-h treatment period. Pericarp PsGA3ox1 mRNA levels gradually increased from 2 to 3 DAA when seeds were present; however, when the seeds were removed, the pericarp transcript levels dramatically declined. When 2-DAA deseeded pericarps were treated with 4-Cl-IAA, PsGA3ox1 mRNA levels peaked 4 h after hormone treatment (270-fold increase), then decreased. PsGA3ox1 mRNA levels in deseeded pericarps treated with indole-3-acetic acid or GA3 were the same or lower than deseeded controls. These data show that PsGA3ox1 is expressed and developmentally regulated in pea pericarps and seeds. These data also show that pericarp PsGA3ox1 expression is hormonally regulated and suggest that the conversion of GA20 to GA1 occurs in the pericarp and is regulated by the presence of seeds and 4-Cl-IAA for fruit growth.
In pea (Pisum sativum), normal pericarp growth requires the presence of seeds (Eeuwens and Schwabe, 1975). Removal or destruction of the seeds 2 to 3 d after anthesis (DAA) results in the slowing of pericarp growth and subsequently abscission (Eeuwens and Schwabe, 1975; Ozga et al., 1992). Chemical signals originating from the seeds may be responsible for continued fruit development by maintaining hormone levels in the surrounding tissue (Eeuwens and Schwabe, 1975; Sponsel, 1982). Developing pea seeds and pericarps contain GAs (Garcia-Martinez et al., 1991; Rodrigo et al., 1997) and auxins (4-chloroindole-3-acetic acid [4-Cl-IAA] and indole-3-acetic acid (IAA); Marumo et al., 1968; Magnus et al., 1997). During early pericarp growth (2 DAA), application of the naturally occurring hormones, 4-Cl-IAA (Reinecke et al., 1995) and GA (Eeuwens and Schwabe, 1975; Ozga and Reinecke, 1999), to deseeded pericarp can substitute for seeds and stimulate pericarp growth. However, the other naturally occurring auxin in pea fruit, IAA, was ineffective in promoting growth (Reinecke et al., 1995).
Initial work comparing the growth promoting properties of 4-, 5-, 6-, and 7-Cl-IAAs and the corresponding F-IAA analogs demonstrated that a chlorine at the 4-position of the indole ring (4-Cl-IAA) was required for significant biological activity in pea pericarp growth (Reinecke et al., 1995). Studies comparing the growth-promoting response and the physicochemical properties of 4-Cl-IAA and 4-substituted analogs found that the 4-substituents' size and lipophilicity were associated with auxin's growth-promoting activity on pea pericarp (Reinecke et al., 1999). Pea pericarps respond in a qualitatively different fashion to two naturally occurring auxins that, in a variety of other auxin bioassays, showed only quantitative differences in activity (Reinecke, 1999). These data suggest unique ways of auxin action based on alternative molecular recognition mechanisms in this tissue.
Pea plants metabolize GAs by the early 13-hydroxylation pathway: GA12 → GA53 → GA44 → GA19 → GA20 → GA1 (Sponsel, 1995). Previous studies using the pea split-pericarp assay (test compounds are applied to the inner walls of split and deseeded 2-DAA pericarps) have shown that the presence of seeds or the application of 4-Cl-IAA to deseeded pea pericarp stimulated pericarp GA biosynthesis, specifically, the conversion of [14C]GA19 to [14C]GA20 (van Huizen et al., 1995), and expression of PsGA20ox1 (codes for enzyme that converts GA19 to GA20; van Huizen et al., 1997). IAA was ineffective in stimulating pericarp PsGA20ox1 expression and growth (Ngo et al., 2002).
Although elongating pollinated pericarps (3 DAA) converted [14C]GA12 and [14C]GA19 to [14C]GA20, conversion to [14C]GA1 was not detected after a 24-h incubation period (Ozga et al., 1992; van Huizen et al., 1995). After a 48-h incubation period, Maki and Brenner (1991) reported metabolism of [2H]GA53 to [2H]GA1 in pollinated pericarp tissue; however, Rodrigo et al. (1997), using 5-DAA pollinated pericarps and [14C]GA12, obtained results similar to that of Ozga et al. (1992). In addition, Rodrigo et al. (1997) were unable to detect conversion of [14C]GA12 or [14C]GA20 to [14C]GA1 in 4-DAA pea seeds. A gene that codes for the enzyme that converts GA20 to the biologically active GA1 (PsGA3ox1; Mendel's LE gene that codes for a GA 3β-hydroxylase) has been cloned from pea (Lester et al., 1997; Martin et al., 1997). Using northern-blot analysis, Martin et al. (1997) were unable to detect PsGA3ox1 signal in unopened pea flowers or 1-cm young fruits, but low levels of seed PsGA3ox1 mRNA were detected at 14 DAA. Therefore, either GA 3β-hydroxylation of GA20 to GA1 does not occur in pea pericarp or seeds, or the methods used in the studies were not sensitive enough to detect pericarp PsGA3ox1 message level or GA 3β-hydroxylation activity (GA20 to GA1) in these tissues in two of the three reported studies.
Real-time reverse transcription (RT)-PCR is the method of choice for sensitive, specific, and reproducible quantification of mRNA (Bustin, 2000). The TaqMan (PE-Applied Biosystems, Foster City, CA) chemistry utilizes the 5′ → 3′ nuclease activity of Taq DNA polymerase to cleave a specific probe, labeled with a 5′ reporter and a 3′ quencher fluorescent dye, during strand elongation of the target sequence to generate a detectable fluorescent signal during amplification (Bustin, 2000). With the use of appropriate standard curves, absolute amounts of mRNA can be calculated. To determine if PsGA3ox1 message occurs in pea pericarp and further understand how seeds and hormones regulate GA biosynthesis in pea fruit, we studied the expression pattern of PsGA3ox1 in pea pericarps and seeds during pollination, development, and auxin- and/or GA-induced pericarp growth using real-time RT-PCR analysis.
RESULTS
Pericarp and Seed Growth
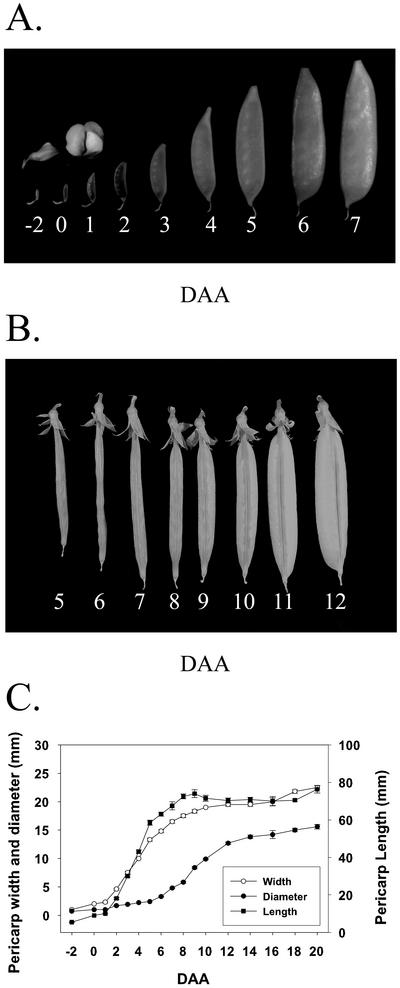
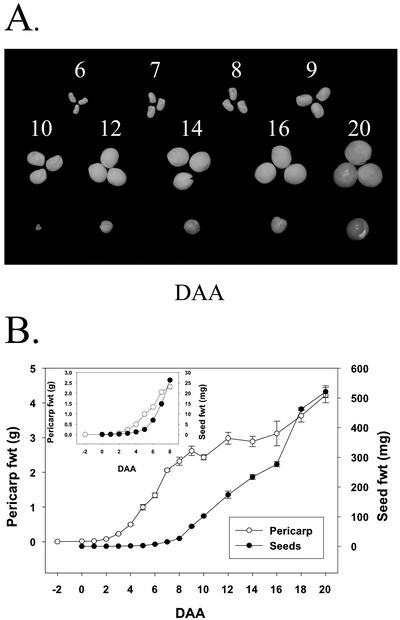
Pericarp growth in length and width was rapid from 2 to 5 DAA and was essentially complete by 8 DAA (Fig. 1, A and C). Subsequently, pericarp diameter rapidly increased from 6 to 12 DAA to accommodate the developing seeds (Fig. 1, B and C). Pericarp fresh weight increased rapidly from 3 to 9 DAA and continued to increase gradually to 20 DAA (Fig. 2B). After pollination, seed fresh weight increased rapidly from 8 to 20 DAA (Fig. 2, A and B). Pericarps from flowers emasculated at −2 DAA and harvested at the equivalent to −1, 0, 1, 2, and 3 DAA (pericarps from non-pollinated ovaries) ranged from 7 to 10 mm in length.
Figure 1.
Pea pericarp development. Development of pericarps during the rapid phases of growth in length, width (−2, 0, and 1–7 DAA; flower bud and flower at anthesis are at −2 and 0 DAA, respectively; A) and diameter (5–12 DAA; B). Growth of pericarp in length, width, and diameter from −2 to 20 DAA (C). Data are means ± se, n = 4 to 12.
Figure 2.
Pericarp and seed growth. A, Seed development from 6 DAA to cotyledon contact point (20 DAA; from left to right, row 1, 6–9-DAA seeds; row 2, 10-, 12-, 14-, 16-, and 20-DAA seeds; and row 3, embryos dissected from 10-, 12-, 14-, 16-, and 20-DAA seeds). B, Growth in fresh weight of pericarps and seeds from −2 DAA to 20 DAA. Data are means ± se, n = 4 to 12 for pericarp fresh weight; n = 2 to 4 for seed fresh weight.
PsGA3ox1 mRNA Quantitation
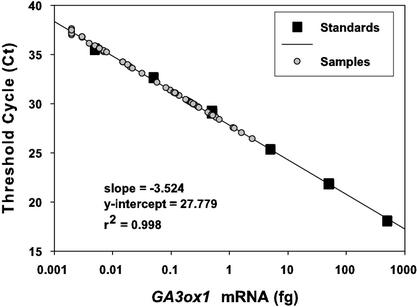
Pea GA3ox87 forward and reverse primers produced a single product of the expected size (87 bp) when completed TaqMan RT-PCR reactions (40 cycles) were analyzed by agarose gel electrophoresis for end-point detection (data not shown). The PsGA3ox1 target sequence (amplicon) was amplified clearly and reproducibly by real-time RT-PCR. Quantification of starting template (plant tissue PsGA3ox1) concentration in real-time RT-PCR assays was achieved by comparison against serially diluted plasmid-generated PsGA3ox1 mRNAs from 5 × 10−3 to 5 × 102 femtogram (fg) at one-log unit intervals (Fig. 3). The dilution series had a linear correlation coefficient (r2) of 0.998 that enabled template quantification over at least six orders of magnitude with cycle threshold (Ct) values between 17 and 37 PCR cycles.
Figure 3.
Standard curve for PsGA3ox1. In vitro-transcribed PsGA3ox1 mRNAs were serially diluted from 5 × 10−3 to 5 × 102 fg at one-log unit intervals. To illustrate the data spread over the standard curve dilution series, samples from Fig. 6B were plotted on the standard curve used to derive the starting template concentration of these samples.
Pea 18S small subunit nuclear ribosomal RNA forward and reverse primers produced a single product of the expected size (62 bp) when completed TaqMan RT-PCR reactions (40 cycles) were analyzed by agarose gel electrophoresis for end-point detection (data not shown). The 18S amplicon was used as a loading control to estimate the variation in input total RNA for one complete replication of the pericarp and seed developmental study (−2 to 20 DAA) and one complete replication of the hormonal regulation study. The average Ct value (±sd) for the 18S ribosomal amplicon in pericarp samples from −2 to 20 DAA was 19.1 ± 0.25 (n = 28; coefficient of variation [CV] = 1.3%); in seed samples from −2 to 20 DAA, Ct was 19.0 ± 0.25 (n = 26; CV = 1.3%); in pericarps treated with IAA, 4-Cl-IAA, GA3, or GA3 + 4-Cl-IAA, and split pod no seeds (SPNS), split pod (SP), and intact pericarp controls, Ct was 20.32 ± 0.21 (n = 56; CV = 1.0%). Because the coefficient of variation of the 18S ribosomal amplicon Ct values was extremely low among all the samples assayed (1%–1.3% CV), PsGA3ox1 mRNA values were not normalized to the 18S signal.
Pea GA3ox126 forward and reverse primers produced a single product of the expected size (126 bp) when completed TaqMan RT-PCR reactions (40) cycles were analyzed by agarose gel electrophoresis for end-point detection (data not shown). The trend in Ct values was similar when 2-, 6-, 10-, and 14-DAA pericarp samples were assayed using GA3ox126 forward and reverse primers (Ct = 29.9, 22.7, 21.6, 22.5, respectively) and GA3ox87 forward and reverse primers (Ct = 32.3, 24.7, 24.0, and 24.2, respectively) with the same fluorescent probe.
A BLAST search of all GenBank, EMBL, DNA Data Bank of Japan, and Protein Data Base database sequences (excluding expressed sequence tag [EST], sequence tagged sites, genome survey sequences, and phase 0, 1, or 2 high-throughput genomic sequences) found significant sequence alignment only between the 86- and 126-bp PCR amplicon sequences and pea GA 3β-hydroxylase (PsGA3ox1) sequences (E values 4 × 10−64 to 4 × 10−39). We did not pick up any significant homology to GA 3β-hydroxylases from other plant species or other GA 2-oxoglutarate-dependent dioxygenases in pea. In a BLAST search of ESTs in GenBank, EMBL, and DNA Data Bank of Japan, no pea ESTs were found to have significant sequence alignment to the 86- and 126-bp PCR amplicon sequences. However, two EST sequences from Medicago truncatula (E values ranged from 5 × 10−26 to 1 × 10−10) and one from Glycine max (E values ranged from 3 × 10−6 to 2 × 10−3) showed significant homology to both amplicons.
Effect of Pollination on PsGA3ox1 mRNA Levels
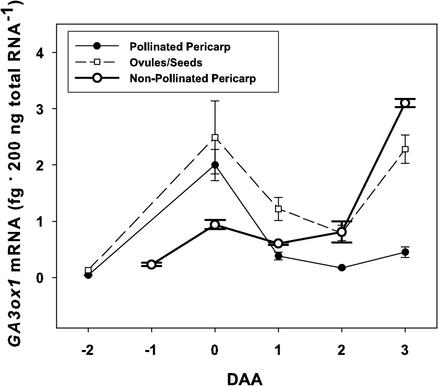
PsGA3ox1 mRNA levels were minimal in both pericarps (0.04 fg 200 ng of total RNA−1) and ovules (0.13 fg 200 ng of total RNA−1) before pollination (−2 DAA; Fig. 4). After pollination (0 DAA), PsGA3ox1 mRNA levels increased dramatically in both tissues (50 times in pericarps and 19 times in ovules), then decreased by 1 DAA to levels approximately 10 and 9 times of that of prepollinated tissues (−2 DAA pericarps and ovules, respectively; Fig. 4). Pericarps from non-pollinated ovaries did not exhibit the dramatic increase in PsGA3ox1 mRNA at the equivalent to anthesis (0 DAA) as seen in pericarps from pollinated ovaries. Furthermore, PsGA3ox1 expression in non-pollinated pericarps dramatically increased at the equivalent to 3 DAA, just before pericarp senescence (Fig. 4).
Figure 4.
Effect of pollination on PsGA3ox1 mRNA level in pea fruit. PsGA3ox1 mRNA (femtogram 200 ng of total RNA−1) in pericarps from pollinated and non-pollinated ovaries, and ovules or seeds at −2 to 3 DAA. Non-pollinated pericarps were green and turgid through the equivalent to 3 DAA, but did not exceed 10 mm in length. Data are means ± se, n = 3 to 5 for pollinated pericarps; n = 2 for non-pollinated pericarps; n = 3 to 4 for ovules/seeds.
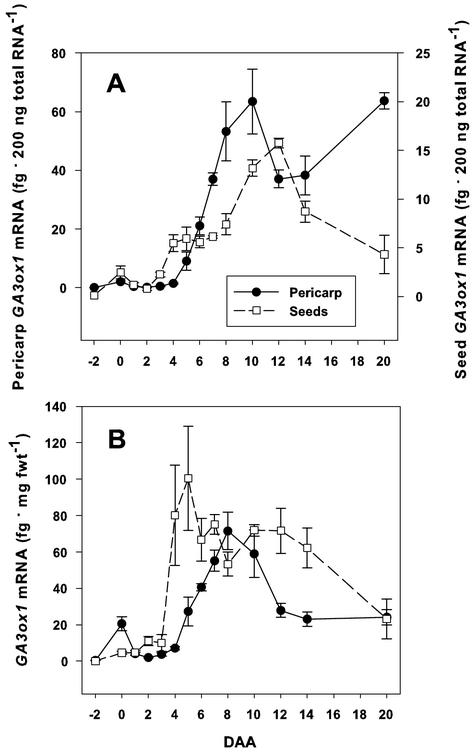
Developmental Profile of PsGA3ox1 mRNA Levels
After pollination, pericarp PsGA3ox1 mRNA levels increased linearly from 6 to 10 DAA (Fig. 5A), coincident with rapid pericarp diameter expansion (6–12 DAA), to accommodate the growing seeds. PsGA3ox1 mRNA levels in seeds (Fig. 5A) increased from 2 to 4 DAA and remained at this level until a further increase from 8 to 12 DAA, when seed development was rapid (Fig. 2A). In general, the seed and pericarp PsGA3ox1 developmental profile was similar when expressed as femtogram 200 ng of total RNA−1 or as femtogram milligram fresh weight−1, with two exceptions (Fig. 5, A and B). The increase in seed PsGA3ox1 transcript level at 4 DAA was magnified when expressed as femtogram milligram fresh weight−1, and pericarp PsGA3ox1 transcript level increased 20 DAA when expressed as femtogram 200 ng of total RNA−1, but was not when expressed as femtogram milligram fresh weight−1. In addition, when comparing the PsGA3ox1 developmental profiles in seeds and pericarps, seeds had lower mRNA levels per total RNA than pericarps, but similar levels when expressed per milligram fresh weight (Fig. 5, A and B). The differences between seeds and pericarps were due to seeds, in general, having higher levels of total RNA per gram of fresh weight than pericarps (data not shown).
Figure 5.
Developmental regulation of PsGA3ox1 mRNA levels in pea pericarps and seeds. PsGA3ox1 mRNA levels from −2 to 20 DAA in pericarps and ovules or seeds expressed as femtogram 200 ng of total RNA−1 (A) and femtogram milligram fresh weight−1 (B). Data are means ±se, n = 3 to 6 for A; and n = 2 to 6 for B, with one exception, −2 DAA ovule, n = 1 for A and B.
Hormonal Regulation of PsGA3ox1 mRNA Levels
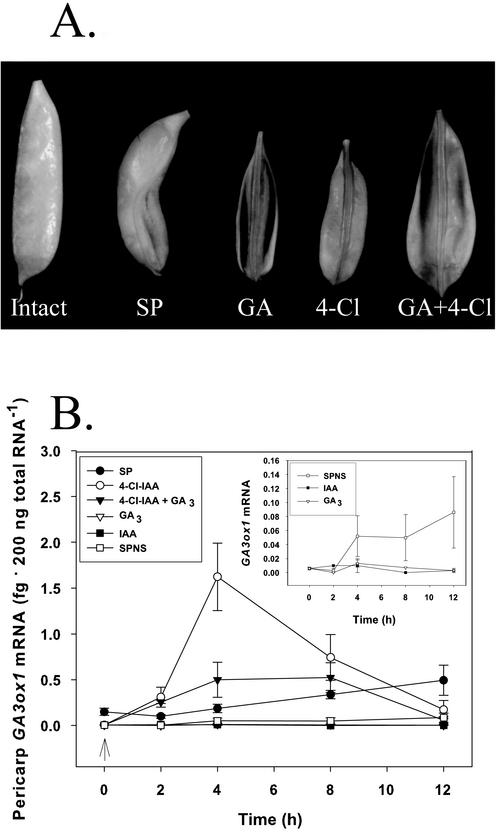
The effect of pericarp splitting and seed removal plus hormone application (at 2 DAA) on pericarp growth 5 d after treatment is shown in Figure 6A. Pericarps continued to grow after splitting of the pericarp 2 DAA without disturbing the seeds (SP; Fig. 6A); however, removal of the seeds at 2 DAA resulted in slowing of pericarp growth and subsequently abscission (not shown). Treatment of deseeded pericarp with GA3 or 4-Cl-IAA (50 μm) stimulated growth (Fig. 6A), but IAA did not (not shown). Application of GA3 plus 4-Cl-IAA stimulated pericarp growth to a greater extent than either applied alone (Fig. 6A).
Figure 6.
Effect of seeds and hormones on pericarp growth and PsGA3ox1 expression. A, Representative pericarps illustrating the effects of pericarp splitting (SP) and seed removal plus application of GA3 (GA), 4-Cl-IAA (4-Cl), and GA3 plus 4-Cl-IAA (GA + 4-Cl) on pea pericarp growth. Pericarps at 2 DAA were split (SP), or split and deseeded. Tween 80 (0.1% [v/v]) was applied daily to SP, and GA3 and/or 4-Cl-IAA were applied daily to deseeded pericarps (50 μm; 30 μL at 2 and 3 DAA; and 40 μL at 4, 5, and 6 DAA) and harvested for picture at 7 DAA. B, Hormonal regulation of pericarp PsGA3ox1 mRNA levels (fg 200 ng of total RNA−1). Two DAA pericarps were split (SP) or split and deseeded (SPNS) and treated with 50 μm (30 μL) of 4-Cl-IAA, GA3, IAA, 4-Cl-IAA plus GA3, or 0.1% (v/v) aqueous Tween 80 (SPNS control) 12 h after deseeding (indicated by arrow; 0 h on x axis). PsGA3ox1 mRNA levels were monitored 2, 4, 8, and 12 h after hormone application. Data are means ± se, n = 2 to 5.
Two-DAA intact pericarps contained 0.19 ± 0.02 fg 200 ng of total RNA−1 PsGA3ox1 transcript. Pericarp PsGA3ox1 transcript level remained stable 12 h (0 h on Fig. 6B) after splitting of pericarp (SP, 0.15 ± 0.04 fg 200 ng of total RNA−1); however, removal of seeds (SPNS) decreased PsGA3ox1 transcripts to minimally detectable levels 12 h after splitting and deseeding (0.006 ± 0.002 fg 200 ng of total RNA−1, 0 h on Fig. 6B). The effect of seeds and hormones on the expression of PsGA3ox1 in pea pericarp was investigated over a 12-h period (hormones were applied to the pericarps 12 h after deseeding [0 h in Fig. 6B] to allow sufficient time for the pericarps to become depleted of seed-produced factors that might affect pericarp growth). PsGA3ox1 mRNA levels in pericarp with seeds (SP) had increased about 3 times over the 12-h time course (to 0.50 ± 0.16 fg 200 ng of total RNA−1), whereas message from pericarps without seeds (SPNS) remained minimally detectable throughout the time course (Fig. 6B).
When deseeded pericarps were treated with 4-Cl-IAA, a significant increase in PsGA3ox1 mRNA levels was detected within 2 h of hormone application (Fig. 6B). Transcript levels in 4-Cl-IAA-treated deseeded pericarps peaked 4 h after hormone treatment (270-fold increase) and decreased substantially by 12 h after hormone treatment (levels 28-fold above levels at 0 h; Fig. 6B). Application of IAA or GA3 to deseeded pericarps resulted in further reductions in PsGA3ox1 transcript levels compared with the SPNS treatment (Fig. 6B, insert). Simultaneous treatment of deseeded pericarps with 4-Cl-IAA and GA3 (4-Cl-IAA + GA3 treatment) resulted in a much smaller increase in PsGA3ox1 transcript levels 4 h after hormone application than observed in 4-Cl-IAA-induced deseeded pericarps (Fig. 6B).
Comparison of PsGA3ox1 Gene Expression among Pea Tissues
Pericarp tissue at 2 DAA had the lowest expression of PsGA3ox1 among the various tissues assayed (over 1,000 times lower than 3 d after imbibition [DAI] shoots and roots; Table I). By 10 DAA, expression of pericarp PsGA3ox1 expressed as femtogram 200 ng of total RNA−1 was greater than 3-DAI cotyledons and 10-DAA seeds. In 3-DAI seedling tissues, the level of PsGA3ox1 message was: shoot > root tips > cotyledons.
Table I.
PsGA3ox1 mRNA levels in vegetative and reproductive tissue of pea cv I3 (Alaska-type)
| Tissue
| ||||||||
|---|---|---|---|---|---|---|---|---|
| PsGA3ox1 mRNA | Cotyledonsa | Rootsa | Root tipsa (4 mm) | Shoota | Pericarp (2 DAA) | Pericarp (10 DAA) | Seeds (2 DAA) | Seeds (10 DAA) |
| fg 200 ng total RNA−1 | 19.20 ± 0b | 206.83 ± 0.89 | 78.50 ± 1.95 | 183.70 ± 0.74 | 0.17 ± 0.01 | 63.48 ± 10.99 | 0.79 ± 0.13 | 13.19 ± 0.82 |
| fg mg fresh weight−1 | 34.07 ± 0 | 495.26 ± 38.59 | 899.01 ± 15.94 | 1,261.4 ± 10.20 | 1.97 ± 0.15 | 58.99 ± 13.09 | 11.07 ± 2.38 | 71.88 ± 3.07 |
Tissue harvested from 3 DAI pea seedlings.
Data are means ± se (cotyledons, n = 1; roots, root tips, and shoots, n = 2; 2-DAA pericarps, n = 5; 10-DAA pericarps and 2- and 10-DAA seeds, n = 3).
DISCUSSION
PsGA3ox1 (Mendel's LE gene) plays an important role in pea plant height (Lester et al., 1997; Martin et al., 1997); however, its role in fruit development has remained unclear. We were able to detect PsGA3ox1 signal in 2-DAA intact pea pericarps on northern blots when 30 μg of total RNA was probed using a [32P]-labeled riboprobe of the full-length PsGA3ox1 gene; however, the signal was very weak (data not shown). Using northern-blot analysis, Martin et al. (1997) were unable to detect PsGA3ox1 signal in unopened pea flowers or 1-cm young fruits. Therefore, a more sensitive method of mRNA detection was required to study regulation of PsGA3ox1 message level in this tissue. Using real-time RT-PCR analysis, the PsGA3ox1 target sequence was amplified clearly and reproducibly, and quantification of plant tissue PsGA3ox1 mRNA in real-time RT-PCR assays was achieved by comparison against serially diluted plasmid-derived PsGA3ox1 mRNA. In addition, a similar trend in Ct values for 2-, 6-, 10-, and 14-DAA pericarp samples assayed using a second set of forward and reverse primers (GA3ox126; compared with the GA3ox87 primers) is further evidence of the specificity of the assay for PsGA3ox1. Using this assay, we found that PsGA3ox1 mRNA levels were regulated during pollination and subsequent development of pericarps and seeds, as well as during hormone-induced pericarp growth.
Pericarps from unpollinated ovaries contained high levels of PsGA20ox1 mRNA (−2 DAA; van Huizen et al., 1997) and GA20 (5 ng g fresh weight−1; emasculated ovaries at the equivalent to 0 and 1 DAA of pea cv Alaska; Garcia-Martinez et al., 1991), but minimally detectable levels of PsGA3ox1 mRNA (Fig. 4) and GA1 (Garcia-Martinez et al., 1991). After pollination (0 DAA), pericarp PsGA3ox1 mRNA levels increased 50-fold, then rapidly decreased to levels 10-fold above prepollinated levels by 1 DAA and remained at this level through 4 DAA (Figs. 4 and 5). Seed PsGA3ox1 mRNA levels increased 19-fold after pollination (0 DAA), then decreased to levels 9-fold above prepollinated levels by 1 DAA (Fig. 4). Although steady-state GA1 levels were reported to be the same in pollinated and non-pollinated ovaries at 0 DAA, GA8 levels were 2 times higher in pollinated than non-pollinated ovaries at this time (Garcia-Martinez et al., 1991). Because GA8 is the immediate biologically inactive product of GA1 (as a result of 2β-hydroxylation), these data suggest that more GA1 was synthesized in pollinated than non-pollinated pericarps and/or ovules at 0 DAA. In addition, the large increase in PsGA3ox1 mRNA levels was not observed in non-pollinated pericarps at the equivalent to 0 DAA; instead, a large increase in PsGA3ox1 message was detected at the equivalent to 3 DAA (Fig. 4). This dramatic increase in PsGA3ox1 message at the equivalent to 3 DAA is likely due to feedback regulation of GA biosynthesis, because at this time levels of GA20, GA1, GA8, and GA3 in non-pollinated ovaries are extremely low or not detectable (Garcia-Martinez et al., 1991). However, even if PsGA3ox1 message was converted to active enzyme and substrate (GA20) was available, the ability of non-pollinated pericarps at the equivalent to 3 DAA to respond in growth to GAs (exogenous) is minimal (Garcia-Martinez and Carbonell, 1980). These data show that pollination triggers the synthesis of pericarp PsGA3ox1 mRNA message and suggest that GA1 is synthesized from the pool of GA20 present in prepollinated pericarps by pericarp GA 3β-hydroxylase, and that this pulse of GA1 stimulates initial fruit set and development.
After pollination, pericarp PsGA3ox1 levels increased linearly from 6 to 10 DAA (Fig. 5A; from 5–8 DAA when expressed as femtogram per milligram fresh weight−1, Fig. 5B). This increase in PsGA3ox1 mRNA levels coincided with the rapid increase in pericarp diameter to accommodate the developing seeds (Fig. 1, B and C). From 4 to 7 DAA, mesocarp cells are continuing to expand and the only increases in cell number occur in cell layers of the endocarp middle zone layer (pericarp wall thickness; Ozga et al., 2002). GA1 levels in pollinated pericarps (pea cv Alaska) were approximately 1 ng g fresh weight−1 from 4 to 12 DAA, with one exception: 6-DAA pericarps contained about 1.75 ng g fresh weight−1 (Rodrigo et al., 1997). The steady-state GA1 levels do not increase with increasing PsGA3ox1 message level during this period. It is possible that the turnover rate of GA1 is substantially higher during this period, resulting in similar steady-state GA1 levels from 4 to 12 DAA. Alternatively, a substantial amount of the PsGA3ox1 message may be degraded before translation into protein. Monitoring of GA1 turnover rate and GA 3β-hydroxylase protein levels, and sensitive metabolic studies during pericarp development would differentiate between these hypotheses.
A 2-fold increase (femtogram 200 ng of total RNA−1) in seed PsGA3ox1 levels occurs at 4 DAA (16-fold increase when expressed as femtogram milligram fresh weight−1; Fig. 5, A and B), which precedes the peak of GA1 levels in the seeds at 6 DAA (about 90 ng g fresh weight−1; Rodrigo et al., 1997). At 4 DAA, 86% of the GA1 observed in the seeds occurred in the testa, with the remainder in the endosperm (Rodrigo et al., 1997). A subsequent increase in seed PsGA3ox1 mRNA levels was observed from 8 to 12 DAA (Fig. 5A), coincident with rapid embryo (Fig. 2, A and B) and endosperm development (Eeuwens and Schwabe, 1975). When data were expressed as femtogram milligram fresh weight−1 (Fig. 5B), seed PsGA3ox1 mRNA levels were maintained between 50 and 75 fg mg fresh weight−1 from 6 to 14 DAA, whereas seed GA1 levels decreased to 20 ng g fresh weight−1 by 8 DAA and were minimally detectable by 12 DAA (Rodrigo et al., 1997).
Because in vitro metabolism experiments show that [14C]GA12 is converted to [14C]GA19 in 4-DAA seeds (Rodrigo et al., 1997), it is possible that GA1 is synthesized through the early 13-hydroxylation pathway in developing pea seeds; however, using genetic and metabolic studies, an alternative hypothesis for GA1 synthesis via GA4 has been proposed. This model proposes that GA4 is synthesized in the endosperm/embryo (4 DAA) and transported to the testa, where it is 13β-hydroxylated to GA1 (Rodrigo et al., 1997; MacKenzie-Hose et al., 1998). However, conversion of GA4 or other GA substrates to GA1 in any seed tissues has not been demonstrated to date. Although it is possible that expression of a second 3β-hydroxylase with a different substrate specificity occurs in young seeds, the expression pattern of PsGA3ox1 (LE) in seeds (Figs. 4 and 5) suggests that PsGA3ox1 gene is involved in the production of GAs for seed development. Further metabolism experiments and monitoring of GA1 turnover rates and GA 3β-hydroxylase protein levels during seed development are also required to further understand regulation of GA1 biosynthesis in seed tissue.
Both PsGA20ox1 and PsGA3ox1 mRNA levels substantially decrease after pollination. This decrease in message levels may be important for fine-tuned regulation of GA biosynthesis by a multitude of factors including hormones (auxins and GAs) during this period. The effects of seeds and hormones (4-Cl-IAA, IAA, and GA3) on the expression of PsGA3ox1 in pea pericarp were investigated over a 24-h treatment period from 2 to 3 DAA. The presence of seeds was required for PsGA3ox1 gene expression in the pericarp (Fig. 6B). When 2-DAA deseeded pericarps were treated with 4-Cl-IAA, PsGA3ox1 levels peaked 4 h after hormone treatment (0.006–1.623 fg 200 ng of total RNA−1 or a 270-fold increase). 4-Cl-IAA stimulated the expression of both PsGA20ox1 (van Huizen et al., 1997) and PsGA3ox1 (Fig. 6B) in pea pericarp, suggesting that similar auxin-induced transcription regulatory elements may operate to coordinate regulation of this part of the GA biosynthesis pathway. Also, as with PsGA20ox1, pericarp PsGA3ox1 mRNA levels were stimulated only by biologically active auxin (4-Cl-IAA stimulates pericarp growth and IAA does not; Reinecke et al., 1995; Ngo et al., 2002; Fig. 6B). Similarly, application of IAA to decapitated pea plants restored internode elongation and the level of PsGA3ox1 levels in the elongating internode (Ross et al., 2000). Because PsGA3ox1 mRNA levels were at minimally detectable levels at the time of hormone application (0 h, Fig. 6B), feedback regulation of PsGA3ox1 expression by GA3 was minimal. However, GA3 reduced 4-Cl-IAA stimulation of PsGA3ox1 mRNA levels (Fig. 6B), indicating that GA3 does feedback regulate PsGA3ox1 message levels in the pericarp.
We propose the following working model for hormonal-directed fruit set and seed- and pericarp-coordinated development. Pollination stimulates GA1 synthesis (via an increase in GA3ox mRNA levels) in both the seeds and pericarp, resulting in initial fruit set and growth of both tissues. Subsequently, seeds maintain growth in the pericarp at least in part by transporting auxin (4-Cl-IAA) to the pericarp, where it stimulates both GA20ox and GA3ox message levels maintaining a critical level of GA1 for pericarp growth. Biologically active GA (GA1) also can feedback regulate its synthesis in the pericarp by reducing GA20ox and GA3ox message levels. In addition, 4-Cl-IAA affects fruit growth directly through auxin-mediated responses (van Huizen et al., 1996). Therefore, interaction of both auxin and GAs are required for coordination of fruit and seed development.
PsGA3ox1 mRNA levels were assayed in young pea seedlings to examine relative amounts of PsGA3ox1 transcript in these tissues versus fruit tissues. The levels of PsGA3ox1 mRNA varied considerably among the pea plant organs assayed (Table I), with roots and shoots of 3-DAI seedlings having the greatest levels, followed by 10-DAA pericarps, 10-DAA seeds, 3-DAI cotyledons, 2-DAA seeds, and 2-DAA pericarps in descending order. It is interesting to note that in 3-DAI shoots, PsGA3ox1 message levels were about 1,000 times higher than those of 2-DAA pericarps. It is possible that other GA3ox genes exist in pea (Southern-blot analysis indicated only one gene in the pea genome; Lester et al., 1997; Martin et al., 1997) as in Arabidopsis (four known GA3ox genes; Hedden and Phillips, 2000); however, the low levels of PsGA3ox1 mRNA in 2-DAA pericarp compared with shoots helps explain why conversion of labeled GA20 to GA1 in pericarps has been difficult to obtain using conventional methods of detection.
It has been proposed that the sensitivity of the fruit to GA may be substantially greater than that of the stem internode. Santes et al. (1993) investigated the effect of the le mutation on the growth and GA content of developing fruits in near-isogenic lines of pea 205+ (LELE, PsGA3ox1) and 205− (lele, 1-bp point mutation of LE that greatly increases the Km of GA 3β-hydroxylase; Martin et al., 1997). Stem elongation is reduced in 205− plants (about 65%), but the growth of pods and seeds are less affected by the le mutation. GA20 was less active in stimulating ovary growth on 205− than on 205+, whereas GA1 had similar high activity on both lines. The content of GA1 in 6-DAA seeds and pericarps of 205− was 2.7 and 7 times (nanograms gram fresh weight−1) or 5 and 10 times lower (nanograms ovary−1) than 205+, respectively. The levels of GA20 and GA29 were substantially elevated in 205− compared with 205+ in both tissues (Santes et al., 1993). These results suggest that 3β-hydroxylation of GA20 to GA1 is reduced in pericarps and seeds by the le mutation, even though it does not affect final fruit phenotype.
In wild-type (LE) Alaska plants, when the size of the pericarp was plotted against the GA1 concentration in non-pollinated fruit growing after application of GA1 or GA3 to the leaf subtending the fruit, there was a linear relationship from about 0.1 (minimum amount necessary for fruit set and growth) to 2 ng g fresh weight−1 (Rodrigo et al., 1997). Higher concentrations of GA1 in the pericarp (20 ng g fresh weight−1, obtained by applying 2 μg to the leaf) did not produce substantial further growth. Therefore, the content of GA1 found in le pods at 6 DAA (0.1 ng g fresh weight−1; Santes et al., 1993) may be sufficient, if not optimal, to stimulate fruit set and development.
In addition, application of a GA biosynthesis inhibitor (LAB 198 999; blocks the later steps in the GA biosynthesis pathway including 3β-hydroxylation) to elongating pollinated pericarps inhibited pericarp growth (fresh weight decreased by 26%), and reduced pericarp GA1 levels 3-fold; and the inhibition was reversed by treatment with GA1 or GA3 (Santes and Garcia-Martinez, 1995). Because LAB also produced an enhancement of GA19 and GA20 levels, despite a reduction of fruit growth, these GAs must not be active per se but become so after conversion to an active GA (GA1). LAB also decreased seed GA1 content 4-fold and seed growth about 16% in 6-DAA fruits (Santes and Garcia-Martinez, 1995). Our work along with the work of others suggests a role for GA 3β-hydroxylase and GA1 in growth and development of pea pericarps and likely in seeds.
In summary, our work shows the effectiveness of real-time RT-PCR in quantitating mRNA transcripts from genes expressed at very low levels. This work would not have been possible using standard northern-blot analysis. Real-time RT-PCR enabled sensitive, reproducible, and specific detection and quantification of PsGA3ox1 mRNA in pea tissues. We found that PsGA3ox1 mRNA levels were regulated during pollination and subsequent development of the pericarp and seed, as well as during hormone-induced pericarp growth. The presence and developmental pattern of PsGA3ox1 message in pea pericarp suggest that GA1 is synthesized in the pericarp for fruit growth. 4-Cl-IAA stimulation of pea pericarp PsGA20ox1 and PsGA3ox1 expression (and the lack of stimulation by IAA) suggests biologically active auxin acts in a concerted fashion on the GA biosynthesis pathway to stimulate production of active GAs in the fruit. Further research at the gene, protein, and enzyme level is required to understand fully the role PsGA3ox1 plays in GA biosynthesis during pea fruit development.
MATERIALS AND METHODS
Plant Material and Treatments
Plants of pea (Pisum sativum L. line I3 [Alaska-type, LE]) were grown under 16-h-light/8-h-dark photoperiod (19°C/17°C) with an average photon flux density of 402 μE m−2 s−1 (van Huizen et al., 1995). Pericarps (−2, 0, 1, 2, 3, 4, 5, 6, 7, 8, 10, 12, 14, and 20 DAA) and ovules (−2 DAA) or seeds (0, 1, 2, 3, 4, 5, 6, 7, 8, 10, 12, 14, and 20 DAA) were collected from the first to the fifth flowering node for experiments monitoring PsGA3ox1 expression during fruit development. Non-pollinated pericarps (first to fifth flowering node) were emasculated −2 DAA and harvested from the plant at equivalent to −1, 0, 1, 2, and 3 DAA (ovules removed at harvest). For hormone-treated pericarps, one fruit between the third and fifth flowering nodes was used per plant; subsequent flowers and lateral buds were removed as they developed. Terminal apical meristems of plants were intact and pericarps remained attached to the plant during the entire experiment.
Pericarps were treated with hormones using a split-pod technique (Ozga et al., 1992). Fruits at 2 DAA measuring 15 to 20 mm in length were split down the dorsal suture 1 h before the 8-h dark period, and seeds were either left intact (SP) or removed (SPNS). Surgical manipulation of the pea fruit was completed 12 h before all hormone applications. Pericarps were treated with IAA, 4-Cl-IAA, and/or GA3 (50 μm in 0.1% [v/v] aqueous Tween 80; 30 μL total). All solutions were applied directly to the inside surface of the pericarp wall (endocarp). The SP and SPNS controls were treated with 30 μL of 0.1% (v/v) aqueous Tween 80. Treated pericarps were covered with plastic bags to maintain high humidity. Pericarps were harvested at 2, 4, 8, and 12 h after the hormone treatment. Seeds, if present, were removed from the pericarp at harvest.
For seedling tissues, seeds of pea line I3 (Alaska-type, LE) were surface sterilized in 80% (v/v) ethanol for 1 min, then gently shaken in 1.5% (v/v) sodium hypochlorite with 0.15% (v/v) Tween 80 at 22°C ± 0.5°C for 20 min. The seeds were rinsed five times with sterile deionized water, and those with intact seed coats were immediately placed 2.5 cm deep into moist sterilized sand in 1-L plastic containers. The containers were placed at 22°C/20°C (16/8 h) in a growth chamber for 3 d. No shoot emergence from the sand had occurred at the time of harvest. All tissues were harvested into liquid N2 and subsequently stored at −80°C until extraction.
RNA Isolation
Two whole or half pericarps, seeds, and 3-DAI seedling tissues (root tips [first 4 mm], remainder of roots, shoots, and cotyledons) were ground to a fine powder in liquid N2; and 100 to 500 mg fresh weight pericarp, 10 to 500 mg fresh weight seed, or 60 to 500 mg fresh weight seedling tissue subsamples were used for RNA extraction. Total RNA was extracted using a modified TRIzol (Invitrogen, Carlsbad, CA) procedure. In brief, after initial extraction with the TRIzol reagent and centrifugation, the supernatant was cleaned by chloroform partitioning (0.2 mL per 1 mL of TRIzol reagent). Subsequently, for further purification, the following steps were carried out in order: high salt precipitation (1.2 m NaCitrate and 0.8 m NaCl) to remove polysaccharides and proteoglycans, 4 m LiCl precipitation, followed by precipitation with 1:20 (v/v) solution of 3 m NaAcetate (pH 5):100% ethanol. The total RNA samples were then treated with DNase (DNA-free kit, Ambion, Austin, TX) and stored at −80°C before real-time RT-PCR analysis.
Oligonucleotide Primers and Probe
Oligonucleotide primers for the GA3ox87 amplicon were designed from the GenBank accession number AF001219 sequence to bind to separate exons of PsGA3ox1 to avoid false positive results arising from amplification of contaminating genomic DNA using Primer Express software (PE-Applied Biosystems). For PsGA3ox1 quantitation, the following primers and probe were used to produce an 87-bp amplicon that spans nucleotides 498 to 584 of AF001219: forward primer, 5′-TTCGAGAACTCTGGCCTCAAG-3′; reverse primer, 5′-ATGTTCCTGCTAACTTTTTCATGGTT-3′; and TaqMan MGB fluorescent dye-labeled complement strand probe (PE-Applied Biosystems), 3′-6FAM-ACAATATCACAGAATCTGGT-MGBNFQ-5′. The GA3ox87 amplicon is outside the region proposed as Fe+- and 2-oxoglutarate-binding regions for 2-oxoglutarate-dependent dioxygenases of which GA3ox is a member (Martin et al., 1997).
For pea 18S small subunit nuclear ribosomal RNA quantitation, primers were designed from the GenBank accession number U43011 sequence to produce a 62-bp amplicon that spans nucleotides 1,575 to 1,636 of this sequence as follows: forward primer, 5′-ACGTCCCTGCCCTTTGTACA-3′; reverse primer, 5′-CACTTCACCGGACCATTCAAT-3′; and TaqMan fluorescent dye-labeled probe (PE-Applied Biosystems), 5′-VIC-ACCGCCCGTCGCTCCTACCG-TAMRA-3′.
To test the specificity of the GA3ox87 quantitation primers above, a second set of primers (GA3ox126) were constructed to a region that spanned 126 bp from nucleotides 451 to 576 of AF001219 as follows: forward primer, 5′-CATGTGGTATGAGGGATTTACTATCGT-3′; reverse primer, 5′-GCTAACTTTTTCATGGTTTCATCATATT-3′; and TaqMan MGB fluorescent dye-labeled complement strand probe (PE-Applied Biosystems), 3′-6FAM-ACAATATCACAGAATCTGGT-MGBNFQ-5′.
Preparation of RNA Standard Curve
RNA transcripts were generated from a recombinant plasmid carrying the complete cDNA of PsGA3ox1 (GenBank accession no. AF001219). The plasmid was linearized with EcoRI, treated with proteinase K at 37°C for 1 h, extracted with phenol-chloroform, then chloroform, and precipitated with 1:20 (v/v) solution of 3 m NaAcetate (pH 5):100% ethanol at −20°C. RNA transcripts were generated at 37°C for 2 to 4 h using T3 RNA polymerase with the RiboMax in vitro transcription kit (Promega, Madison, WI). Plasmid DNA was subsequently digested with RNase-free DNase I at 37°C for 15 min, and the transcripts purified by one phenol-chloroform extraction and RNeasy Mini Kit (Qiagen, Mississauga, ON). RNA concentration was determined spectrophotometrically at A260.
Cts for a dilution series of RNA transcripts (0.05, 0.5, 5, 50, and 500 fg) were plotted to yield a standard curve for each 96-well reaction plate. Ct was initially calculated by the TaqMan software to indicate significant fluorescence signals above noise during the early cycles of amplification. When necessary, Ct was adjusted manually to cross an exponential portion of the amplification curves of all samples being compared on the 96-well plate.
TaqMan RT-PCR Assay
The TaqMan One-Step RT-PCR reagent kit (PE-Applied Biosystems) was used throughout as 50-μL reactions in a model 7700 Sequence Detector (PE-Applied Biosystems). The RT-PCR mixture contained 25 μL of 2× Master Mix (containing AmpliTaq Gold DNA polymerase), 1.25 μL of 40× MultiScribe (reverse transcriptase) and RNase Inhibitor Mix, 300 nm forward primer, 300 nm reverse primer, 100 nm probe, and 200 ng of total RNA for PsGA3ox1 or 10 pg of total RNA for 18S rRNA quantitation. Thermal cycling conditions were 48°C for 30 min for RT, 95°C for 10 min for Taq activation, and 40 cycles of 95°C for 15 s and 60°C for 1 min for PCR. Each sample was assayed twice, and the average of the two assays was used to calculate the PsGA3ox1 concentration.
ACKNOWLEDGMENT
We thank Dr. Steve Moore for use of the real-time RT-PCR Sequence Detector.
Footnotes
This research was supported by the National Sciences and Engineering Research Council of Canada (award no. OGP0138166 to J.A.O.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.015974.
LITERATURE CITED
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Eeuwens CJ, Schwabe WW. Seed and pod wall development in Pisum sativum L. in relation to extracted and applied hormones. J Exp Bot. 1975;26:1–14. [Google Scholar]
- Garcia-Martinez JL, Carbonell J. Fruit-set of unpollinated ovaries of Pisum sativum L. Planta. 1980;147:451–456. doi: 10.1007/BF00380187. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Santes C, Croker SJ, Hedden P. Identification, quantitation and distribution of gibberellins in fruits of Pisum sativum L. cv. Alaska during pod development. Planta. 1991;184:53–60. doi: 10.1007/BF00208236. [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB. Mendel's stem length gene (Le) encodes a gibberellin 3β-hydroxylase. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie-Hose AK, Ross JJ, Davies NW, Swain SM. Expression of gibberellin mutations in fruits of Pisum sativum L. Planta. 1998;204:397–403. [Google Scholar]
- Magnus V, Ozga JA, Reinecke DM, Pierson GL, Larue TA, Cohen JD, Brenner ML. 4-chloroindole-3-acetic acid and indole-3-acetic acid in Pisum sativum. Phytochemistry. 1997;46:675–681. [Google Scholar]
- Maki SL, Brenner ML. [14C]GA12-aldehyde, [14C]GA12, and [2H]- and [14C]GA53 metabolism by elongating pea pericarp. Plant Physiol. 1991;97:1359–1366. doi: 10.1104/pp.97.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P. Mendel's dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc Natl Acad Sci USA. 1997;94:8907–8911. doi: 10.1073/pnas.94.16.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumo S, Hattori H, Abe H, Munakata K. Isolation of 4-chloroindolyl-3-acetic acid from immature seeds of Pisum sativum. Nature. 1968;219:959–960. doi: 10.1038/219959b0. [DOI] [PubMed] [Google Scholar]
- Ngo P, Ozga JA, Reinecke DM. Specificity of auxin regulation of gibberellin 20-oxidase gene expression in pea pericarp. Plant Mol Biol. 2002;49:439–448. doi: 10.1023/a:1015522404586. [DOI] [PubMed] [Google Scholar]
- Ozga JA, Brenner ML, Reinecke DM. Seed effects on gibberellin metabolism in pea pericarp. Plant Physiol. 1992;100:88–94. doi: 10.1104/pp.100.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Reinecke DM. Interaction of 4-chloroindole-3-acetic acid and gibberellins in early pea fruit development. Plant Growth Regul. 1999;27:33–38. [Google Scholar]
- Ozga JA, van Huizen R, Reinecke DM. Hormone and seed-specific regulation of pea fruit growth. Plant Physiol. 2002;128:1379–1389. doi: 10.1104/pp.010800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke DM, Ozga JA, Magnus V. Effect of halogen substitution of indole-3-acetic acid on biological activity in pea fruit. Phytochemistry. 1995;40:1361–1366. [Google Scholar]
- Reinecke DM. 4-Chloroindale-3-acetic acid and plant growth. Plant Growth Regul. 1999;27:3–13. [Google Scholar]
- Reinecke DM, Ozga JA, Ilic N, Magnus V, Kojic-Prodic B. Molecular properties of 4-substituted indole-3-acetic acids affecting pea pericarp elongation. Plant Growth Regul. 1999;27:39–48. [Google Scholar]
- Rodrigo MJ, Garcia-Martinez JL, Santes CM, Gaskin P, Hedden P. The role of gibberellins A1 and A3 in fruit growth of Pisum sativum L. and the identification of gibberellins A4 and A7 in young seeds. Planta. 1997;201:446–455. [Google Scholar]
- Ross JJ, O'Neill DP, Smith JJ, Huub L, Kerckhoffs J, Elliott RC. Evidence that auxin promotes gibberellin A1 biosynthesis in pea. Plant J. 2000;21:547–552. doi: 10.1046/j.1365-313x.2000.00702.x. [DOI] [PubMed] [Google Scholar]
- Santes CM, Garcia-Martinez JL. Effect of the growth retardant 3,5-dioxo-4-butyryl-cyclohexane carboxylic acid ethyl ester, an acylcyclohexanedione compound, on fruit growth and gibberellin content of pollinated and unpollinated ovaries in pea. Plant Physiol. 1995;108:517–523. doi: 10.1104/pp.108.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santes CM, Hedden P, Sponsel VM, Reid JB, Garcia-Martinez JL. Expression of the le mutation in young ovaries of Pisum sativum and its effect on fruit development. Plant Physiol. 1993;101:759–764. doi: 10.1104/pp.101.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponsel VM. Effects of applied gibberellins and naphthylacetic acid on pod development in fruits of Pisum sativum L. cv. Progress No. 9. J Plant Growth Regul. 1982;1:147–152. [Google Scholar]
- Sponsel VM. The biosynthesis and metabolism of gibberellins in higher plants. In: Davies PJ, editor. Plant Hormones: Physiology, Biochemistry and Molecular Biology. Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 66–97. [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM. Influence of auxin and gibberellin on in vivo protein synthesis during early pea fruit growth. Plant Physiol. 1996;112:53–59. doi: 10.1104/pp.112.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM. Seed and hormonal regulation of gibberellin 20-oxidase expression in pea pericarp. Plant Physiol. 1997;115:123–128. doi: 10.1104/pp.115.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM, Twitchin B, Mander LN. Seed and 4-chloroindole-3-acetic acid regulation of gibberellin metabolism in pea pericarp. Plant Physiol. 1995;109:1213–1217. doi: 10.1104/pp.109.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]