Abstract
Phytochromes are photoreceptors that control many plant light responses. Phytochromes have two carboxyl-terminal structural domains called the PAS repeat domain and the histidine kinase-related domain. These domains are each related to bacterial histidine kinase domains, and biochemical studies suggest that phytochromes are light-regulated kinases. The PAS repeat domain is important for proper phytochrome function and can interact with putative signaling partners. We have characterized several new phytochrome B mutants in Arabidopsis that express phyB protein, three of which affect the histidine kinase-related domain. Point mutations in the histidine kinase-related domain cause phenotypes similar to those of null mutants, indicating that this domain is important for phyB signaling. However, a truncation that removes most of the histidine kinase-related domain results in a phyB molecule with partial activity, suggesting that this domain is dispensable. These results suggest that phytochromes evolved in modular fashion. We discuss possible functions of the histidine kinase-related domain in phytochrome signaling.
Phytochromes are a family of red/far-red light photoreceptors in plants that regulate many developmental and cellular responses to light. They are present in all plant species examined and in many algae. The model plant Arabidopsis thaliana has five phytochromes called phyA-phyE that diverge from each other by as much as 50%, and genetic and physiological studies have revealed that they regulate distinct light responses (1–3). Phytochrome proteins interconvert between two stable spectral forms. They are synthesized in the dark in the Pr (red-absorbing) form. Red light converts Pr to the Pfr (far-red-absorbing) form, and far-red light reconverts Pfr to Pr. Pfr (and, for phyA, possibly “cycled” Pr that has passed through the Pfr form) is thought to be the biologically active form of phytochrome (ref. 4 and discussed in ref. 5).
Phytochromes have two major structural domains (6, 7). The amino-terminal domain (≈74 kDa) has a covalently attached linear tetrapyrrole chromophore (phytochromobilin) and is sufficient for light absorption and photoreversibility. The carboxyl-terminal domain (≈55 kDa) is important for dimerization and downstream signaling and consists of two subdomains termed the PAS repeat domain (PRD) and the histidine kinase-related domain (HKRD). These domains are 9%–11% identical to each other within representative phytochromes, and they are each 13%–17% identical to the histidine kinase domain of sensory transducers of bacterial two-component regulatory systems (8).
The cyanobacterium Synechocystis has a phytochrome-like molecule called cph1, which has an amino-terminal chromophore domain very similar to that of higher plant phytochromes and a single carboxyl-terminal histidine kinase domain. Like other two-component sensory transducers, cph1 autophosphorylates on a histidine residue and then transfers this phosphate to an aspartate on a second protein. Both of these activities are higher when cph1 is in the Pr form, and cph1 thus functions as a light-regulated kinase (9). The homology of phytochromes to cph1 suggests that they may function similarly. In fact, phyA can autophosphorylate on serine residues and can function as a serine/threonine kinase in vitro (8). Similarly, phyB-dependent phosphorylation has been detected in vivo (10). It is not known whether the PRD or the HKRD has kinase activity, or whether the kinase activity is essential for phytochrome signaling.
Studies of mutant phytochromes have revealed that the PRD is critical for signaling. Point mutations in the PRD of both phyA and phyB did not affect photoreversibility but eliminated biological activity (11–13). The PRD interacts with the proposed phytochrome signal transducers phytochrome-interacting factor 3 (PIF3) and nucleoside diphosphate kinase 2 (NDPK2) (14–16).
Less is known about the function of the HKRD. A truncated oat phyA lacking the final 36 carboxyl-terminal amino acids conferred no phenotype when overexpressed in tobacco, suggesting that the HKRD is necessary for phyA activity (17). Several other truncated phyA and phyB proteins did not accumulate and were therefore uninformative about HKRD function (12, 17–20). Site-directed point mutations in conserved residues of the G1 and G2 motifs of the HKRD of phyA did not affect function (21). The phyA-105 mutation falls at the start of the HKRD (A893V) and decreases but does not eliminate phyA activity (11). Two other phyA mutations, T928I and A955V, have been reported but have not been described in detail (13). A protein that interacts with the HKRD called PKS1 is a putative negative regulator of phytochrome responses and can be phosphorylated by phyA or phyB (10).
We have collected 25 new phyB mutants, five of which retain wild-type levels of phyB protein, and one that has a substantial amount of protein. Three of these six have mutations in the HKRD. Our analyses of these new mutants reveal that this domain is necessary for phyB activity. However, removal of the HKRD does not eliminate activity. We discuss possible modes of HKRD action and phytochrome evolution suggested by these results.
Materials and Methods
Genetic Material.
Details and sources of phyB mutants analyzed in this study are listed in Table 1, which is published as supplementary data on the PNAS web site, www.pnas.org. Mutants characterized in detail were backcrossed once (phyB-15, phyB-18, phyB-19, and phyB-35) or twice (phyB-13, phyB-14, and phyB-28) to their respective wild-type ecotypes.
SDS/PAGE and Immunoblotting.
Between 0.1 and 0.2 g of tissue was ground with a small amount of washed and ignited sea sand (Fisher Scientific) at 100°C in 75–150 μl of boiling 2× SDS loading buffer [100 mM Tris (pH 6.8)/200 mM DTT/4% SDS/20% glycerol/0.2% bromophenol blue/5 μM PMSF] and then briefly centrifuged to pellet debris. Supernatant (15 μl) was immediately loaded onto an 8% polyacrylamide SDS gel with a 4% stack (22). Electrophoresis was carried out as described (23). After electrophoresis, proteins were electroblotted to supported nitrocellulose BA-S 85 (Schleicher & Schuell) for 30 min at 100 V. After transfer, the gel was stained with Coomassie brilliant blue G-250 (Bio-Rad) to assay transfer efficiency. PhyB was detected by using the mAb mBA2 (24), followed by incubation with alkaline phosphatase-linked goat anti-mouse Ig (Sigma) and color development (23). To estimate relative phyB protein levels, total extracted protein was quantitated by Bradford assay (Bio-Rad), and equivalent protein amounts were loaded in a dilution series onto SDS/PAGE gels. Western blots and Coomassie-stained gels were digitized using a scanner, and signal intensity was quantitated by using National Institutes of Health image software (http://rsb.info.nih.gov/nih-image/).
Sequencing of phyB Alleles.
Fragments of the PHYB gene from mutants were amplified by PCR and either sequenced directly or subcloned and then sequenced, by the University of North Carolina-Chapel Hill Automated DNA Sequencing Facility.
Red Light Fluence Rate/Response Experiments.
Seeds were surface sterilized and plated on Murashige and Skoog (MS)/agar plates [1× MS salts (GIBCO), 0.8% phytager (GIBCO), 1× Gamborg's B5 vitamin mix (Sigma)], stored overnight at 4°C, and placed vertically behind various thicknesses of bronze Plexiglass No. 2412 (Golden Rule Plastics, Haw River, NC). Red light-emitting diode (LED) light sources emitting light with a peak at 670 nm and a half bandwidth of 25 nm (Quantum Devices, Barneveld, WI) were placed to project horizontally. After 4 days, hypocotyl lengths were measured by hand against a ruler. Light levels were measured with an LI-189 quantum radiometer (Li-Cor, Lincoln, NE) or extrapolated based on numbers of layers of Plexiglass.
End-of-Day Far-Red Response.
Seeds were surface sterilized, stored overnight at 4°C, and placed on MS/agar/2% sucrose plates. Fluorescent light was provided on a 9h:15h day/night cycle. At the end of each day, a 20-min saturating far-red pulse of light was given from an LED light source emitting with a peak at 730 nm and a half bandwidth of 25 nm (Quantum Devices). After 6 days of treatment, hypocotyl lengths were measured by hand against a ruler.
Flowering Time Experiments.
Seedlings were grown on MS/agar/2% sucrose plates for 10–14 days and then were transplanted to soil. Experiments were performed in a Conviron (Pembina, ND) growth chamber at 21°C. Light was provided on a 9h:15h day/night cycle from 12 fluorescent (F72T12/CW/VHO, 160 W) and 6 incandescent (60 W) bulbs and had an intensity at plant height of 125–225 μmol⋅m−2⋅s−1.
PHYB and phyB-28-Overexpressing Transgenic Plants.
We previously described a transgenic Columbia line that overexpresses PHYB from a 35S∷PHYB construct (25). For construction of the 35S∷phyB-28 transgene, RNA was isolated from the phyB-28 mutant by using TRI Reagent (Sigma). A fragment containing the phyB-28 mutation was then amplified by reverse transcription (RT)-PCR using poly(dT) as a primer for first strand synthesis, and the primers pB7J (5′-TCTGTTTCTTGCAAATCCCGAGC-3′) and pB8 (5′-AAATCTAGAGCTGAACGCAAATAATCTCCC-3′; XbaI site underlined) for PCR amplification. This fragment was digested with PstI and XbaI and cloned into the phyB cDNA clone p41A (26). A SacI fragment from this construct was then inserted into the corresponding location of the BOE overexpression plasmid (27). The resulting plasmid construct was transformed into phyB-9 and Columbia wild-type plants by vacuum infiltration (28). We identified kanamycin-resistant T1 transformants that overexpressed phyB-28 protein in both backgrounds by Western blot (data not shown). After self-fertilization, T2 self-progeny of two of the Columbia 35S∷phyB-28 T1 plants and one of the phyB-9 35S∷phyB-28 T1 plants segregated roughly 3:1 for kanamycin resistance/sensitivity, consistent with the presence of a single transferred DNA (T-DNA) integration locus (data not shown). T3 self-progeny of homozygous T2 plants were identified and used for phenotypic characterization.
Results
Isolation of New phyB Mutations That Affect Protein Function.
We isolated eighteen new Arabidopsis phyB mutants in several screens for plants with long hypocotyls and obtained seven new mutants from colleagues. All new phyB mutations failed to complement the long hypocotyl phenotype caused by a known null phyB mutation and were recessive to the wild-type allele in strong white light (data not shown). We have given them the allele designations phyB-11 to phyB-35. Seventeen of the new phyB mutants are in the Columbia background (phyB-11 to phyB-27), and eight are in the Landsberg erecta background (phyB-28 to phyB-35). A summary of these new mutants can be found as supplementary data in Table 1 published on the PNAS web site at www.pnas.org.
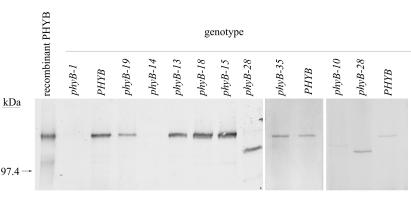
To determine whether the new mutants had phyB protein, we probed Western blots of total protein from mutant plants with an anti-phyB mAb (Fig. 1). Eight mutants had detectable phyB protein. Five of these (phyB-13, phyB-15, phyB-18, phyB-28, and phyB-35) had roughly wild-type levels of phyB. The phyB protein in phyB-28 was smaller than in wild-type and the other mutants (Fig. 1). phyB-19 plants had approximately 40% less phyB than wild-type (Fig. 1; data not shown). phyB-14 and phyB-34 plants each had less than 5% of the level of phyB in wild-type plants, as did the previously described phyB-10 T-DNA insertion mutant (18). The other 17 mutants, including phyB-26, had no detectable phyB (data not shown). By this criterion, these 17 mutants carry null alleles.
Figure 1.
Immunoblot analysis of various phyB mutants. Total protein from adult leaves was detected with mAb mBA2. Recombinant PHYB was from a yeast strain engineered to overexpress PHYB (26). phyB-13, phyB-15, phyB-18, phyB-28, and phyB-35 mutants had approximately wild-type levels of phyB when normalized to loaded total protein. phyB-19 had approximately 40% less protein than wild-type. Although not visible on this blot, phyB-14 had a very low level of phyB protein in other experiments (data not shown). phyB-34 also had a very low level of phyB protein (data not shown). Seventeen other mutants listed as supplementary data in Table 1 had no detectable phyB protein (data not shown). The position of a 97.4-kDa molecular mass standard is indicated.
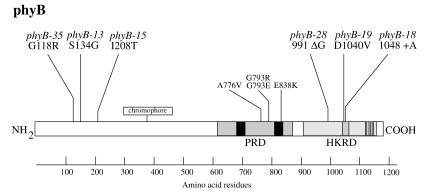
As the phyB-13, phyB-15, phyB-18, phyB-19, phyB-28, and phyB-35 mutations had either no effect, or a moderate effect, on the level of phyB protein, they may define portions of phyB important for photochemistry or signaling rather than folding or stability. We sequenced the PHYB gene from these six mutants and found mutations in each of them (Fig. 2). The phyB-13, phyB-15, and phyB-35 alleles are each missense mutations in the amino-terminal portion of the molecule at amino acid residues 134, 208, and 118, respectively. As this domain is important for chromophore coordination and photoreversibility, these three mutations may affect light perception or photoreversibility.
Figure 2.
Locations of sequenced phyB mutations in the PHYB protein. The PRD and the HKRD are labeled. Black boxes represent the two PAS repeats in the PRD, and dark stippled boxes represent the highly conserved N, G1, F, and G2 motifs of the HKRD. Previously described phyB mutations in the PRD are also shown (12).
The remaining three mutations affect the carboxyl-terminal HKRD. phyB-18 changes a highly conserved aspartate at position 1040 into a valine, and phyB-19 inserts an extra alanine residue next to the alanine at position 1049. Histidine kinase domains share five conserved motifs called H, N, G1, F, and G2, of which the last three are important for ATP binding and hydrolysis (29). The phyB-18 and phyB-19 mutations each fall in the N domain.
The phyB-28 mutation is a deletion of a single guanosine nucleotide at codon 991. This mutation introduces a frameshift that adds four missense residues followed by a stop codon. As this mutation occurs in a stretch of three guanosines at the splice donor site of the second intron, we amplified the PHYB mRNA from the phyB-28 mutant by reverse transcription-PCR and sequenced the product. We found that splicing occurs normally (data not shown). The phyB-28 mutation therefore causes a truncation of 182 aa, consistent with the size of the phyB protein in plants carrying this mutation (Fig. 1). The truncation removes just over two-thirds of the HKRD, including the N, G1, F, and G2 motifs.
Mutations in the HKRD Disturb phyB Function.
Arabidopsis plants that lack phyB have long hypocotyls in red light and flower early (18, 30–32). To determine how our new phyB mutations affect phyB function, we tested the hypocotyl elongation responses of the mutants to constant red light and to end-of-day far-red (EOD-FR) light, and we measured flowering times of some of them in short days.
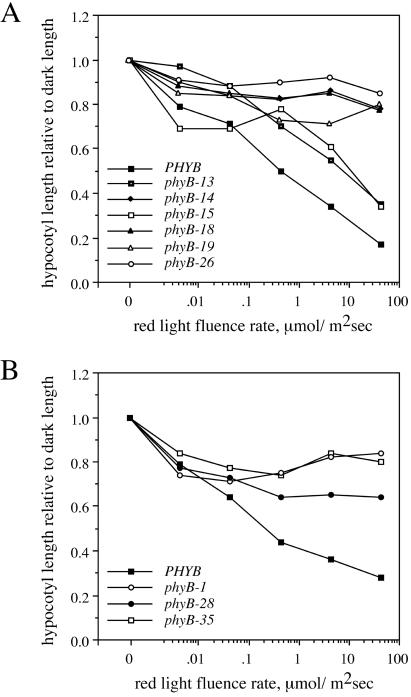
A wide range of red light fluence rates tested inhibited hypocotyl elongation of wild-type plants of both Columbia and Landsberg erecta ecotypes (Fig. 3). In contrast, representative null mutants that lack phyB protein, phyB-1 and phyB-26, had almost no response to any fluence rate. The phyB-14 mutant, which has very little phyB protein, responded as weakly as the null mutants. Two of the amino-terminal domain mutants, phyB-13 and phyB-15, responded almost as much as the wild-type, indicating that these mutants retain significant phyB activity. The third amino-terminal domain mutant, phyB-35, and the carboxyl-terminal domain mutants phyB-18, phyB-19, and phyB-28, had responses more similar to those of the null mutants. Of these, phyB-19 and phyB-28 had shorter hypocotyls than the corresponding null mutants at several fluence rates, suggesting that they may retain some phyB activity.
Figure 3.
Hypocotyl lengths of phyB mutants grown under different fluence rates of red light. (A) Columbia mutants; (B) Landsberg erecta mutants. Each genotype was tested between two and four times, and a single representative experiment is shown. Each point represents the mean hypocotyl length from 10–15 plants of each genotype, normalized to hypocotyl length in the dark. Standard deviations are omitted for clarity and were generally 10%–20% of the mean. Data in A are from two experiments. The apparent increased response of the phyB-15 mutant at low fluence rates was not consistent between experiments (data not shown).
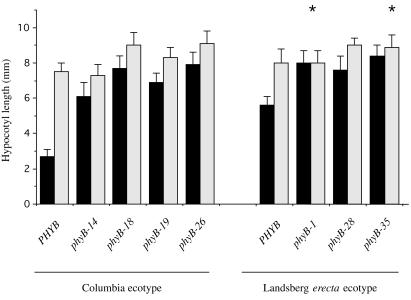
EOD-FR treatments of wild-type plants of both ecotypes caused a significant lengthening of the hypocotyl (Fig. 4). This response is caused by persistence of phyB in the active Pfr form during the night (33). EOD-FR treatments convert phyB to Pr, thereby inactivating it and relieving inhibition of hypocotyl elongation. phyD also makes a minor contribution to this response (34). The ecotype Columbia phyB mutants phyB-14, phyB-18, and phyB-19 and the null mutant phyB-26 showed only a slight increase in hypocotyl length on EOD-FR treatment, consistent with a lack of phyB activity. The ecotype Landsberg erecta null mutant phyB-1 and the amino-terminal missense mutant phyB-35 had no EOD-FR elongation response. The absent EOD-FR response of Landsberg erecta phyB null mutants may be due to a lesser importance of phyD in this ecotype than in Columbia (26, 33). In contrast to the null mutant, the phyB-28 mutant retained a significant response to EOD-FR (Fig. 4).
Figure 4.
Effect of EOD-FR light on the hypocotyl length of phyB mutants. Solid bars, no EOD-FR treatments; gray bars, EOD-FR treatments. Each genotype was tested at least twice, and a single representative experiment is shown. Each bar represents the mean hypocotyl length from 12–15 plants of each genotype ± SD. Asterisks indicate no significant difference from wild type by t test (P > 0.05).
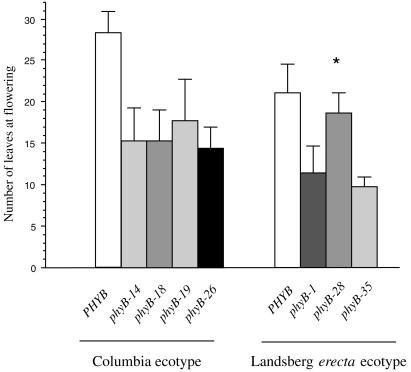
The time it took plants of different PHYB genotypes to flower correlated with the number of leaves at the time of flowering (data not shown). phyB-14, phyB-18, phyB-19, and phyB-35 plants flowered substantially earlier and with fewer leaves than wild-type plants, similarly to the null mutants phyB-1 and phyB-26 (Fig. 5; data not shown). In contrast, phyB-28 mutant plants flowered only slightly earlier than wild-type plants and substantially later than null mutant plants (Fig. 5).
Figure 5.
Flowering of selected phyB mutants in short days. The mean number of leaves from 10 plants at the time of flowering is shown ± SD. Asterisks indicate no significant difference from wild type by t test (P > 0.05).
The phyB-28 Truncated Protein Retains Activity.
The phyB-28 mutant responded slightly to red light and to EOD-FR treatment and flowered almost as late as wild-type plants. These phenotypes suggested that the truncated protein made in the phyB-28 mutant retained substantial activity for inhibition of flowering and slight activity for inhibition of hypocotyl elongation. To assess whether a second mutation in the phyB-28 mutant background might cause the late flowering phenotype of this mutant, we crossed the phyB-28 mutant with a phyB-1 null mutant and compared the flowering time of the resulting F1 plants to that of the homozygous null mutant. We found that phyB-1/phyB-28 F1 plants flowered significantly later than the homozygous null mutant (data not shown), indicating that, if a second mutation delays flowering in the phyB-28 mutant, such a mutation is likely to be dominant.
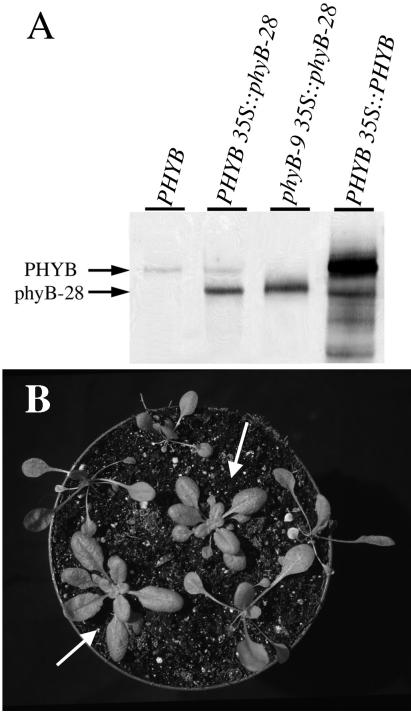
To determine more directly whether the phyB-28 protein retains activity, and to exclude the possibility that a second mutation might modify the phyB-28 flowering time phenotype, we overexpressed a phyB-28 mutant cDNA behind the strong 35S promoter in transgenic phyB-9 null mutant and Columbia (PHYB) wild-type plants (see Materials and Methods). When overexpressed, the wild-type PHYB cDNA confers exaggerated red light responses, including a short hypocotyl and rounded leaves and also (paradoxically) causes early flowering (32, 35). We characterized phenotypes of two PHYB 35S∷phyB-28 lines and one phyB-9 35S∷phyB-28 line. In all cases, both Columbia 35S∷phyB-28 lines gave similar results (data not shown). The phyB-9 35S∷phyB-28 line had approximately 5-fold more phyB-28 protein than did wild type and the PHYB 35S∷phyB-28 lines had approximately 3-fold more protein than did wild type. These levels are much less than the approximately 20-fold overexpression seen in a PHYB 35S∷PHYB control line (Fig. 6A). When grown in white light, the phyB-9 35S∷phyB-28 T1 plants had a compact rosette of round leaves with short petioles, as wild type or adult phyB-28 plants do (Fig. 6B). Homozygous T3 phyB-9 35S∷phyB-28 plants flowered slightly earlier than the wild type but significantly later than the phyB-9 null mutant (Fig. 7B). These phenotypes show that the 35S∷phyB-28 transgene rescued the adult morphological and flowering time phenotypes of the phyB-9 mutant.
Figure 6.
(A) Immunoblot analysis of phyB protein in homozygous 35S∷PHYB and 35S∷phyB-28 plants. An equivalent amount of protein was loaded in each lane. PhyB protein was detected with mAb mBA2. (B) Phenotype of 20-day-old T1 phyB-9 35S∷phyB-28 plants in short days. Arrows indicate the plants overexpressing phyB-28 protein. The other plants are T1 transformants not expressing the transgene and equivalent to phyB-9 null mutant plants.
Figure 7.
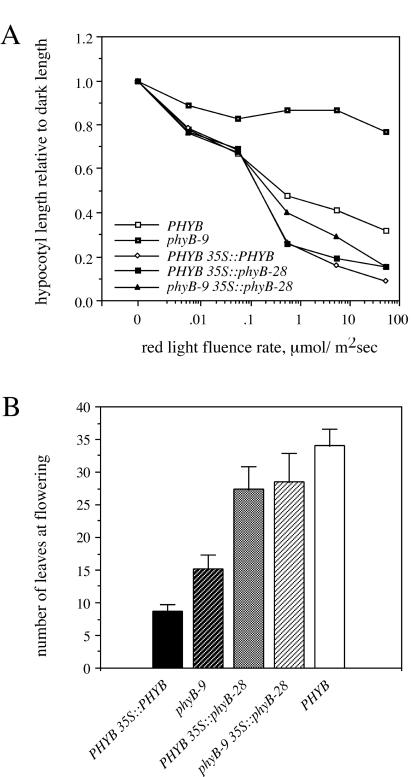
(A) Hypocotyl lengths of homozygous 35S∷PHYB and 35S∷phyB-28 seedlings grown under different fluence rates of red light. Each point represents the mean hypocotyl length from 20 plants of each genotype, normalized to hypocotyl length in the dark. SD are omitted for clarity and were generally 10%–20% of the mean. (B) Flowering of homozygous 35S∷PHYB and 35S∷phyB-28 plants in short days. The mean number of leaves from 18 plants at the time of flowering is shown ± SD.
The 35S∷phyB-28 transgene also rescued the long hypocotyl phenotype of the phyB-9 mutant. Thus, phyB-9 35S∷phyB-28 seedlings responded to the full fluence rate range of red light tested and, in fact, were more sensitive to red light inhibition of hypocotyl elongation than were wild-type plants (Fig. 7A). PHYB 35S∷phyB-28 plants were also hypersensitive to red light, to a degree similar to that of PHYB 35S∷PHYB plants (Fig. 7A). When grown in the dark, all seedlings of each of these transgenic lines had hypocotyls as long as those of wild-type seedlings (data not shown). These data show that, when overexpressed, the phyB-28 protein can mediate red light inhibition of hypocotyl elongation.
Whereas phyB-28 retained activity to inhibit hypocotyl elongation and flowering in these transgenic plants, we did not observe the contrasting promotion of flowering when phyB-28 was overexpressed in a wild-type background. As seen previously (32), PHYB 35S∷PHYB plants flowered much earlier than wild-type plants (Fig. 7B). In contrast, PHYB 35S∷phyB-28 plants flowered only slightly earlier than wild-type plants, and much later than PHYB 35S∷PHYB plants (Fig. 7B). This result may be due to the lower level of phyB-28 protein in the 35S∷phyB-28 lines compared with the level of phyB in PHYB 35S∷PHYB plants (Fig. 6A). Another possibility is that the HKRD is necessary to promote flowering when phyB is overexpressed and that the truncated phyB-28 protein lacks this activity.
Discussion
Our results provide insight into the function of the HKRD of phyB. phyB-28 mutant plants had weaker hypocotyl elongation and early-flowering phenotypes than phyB-1 null mutant plants. Moreover, overexpression of phyB-28 mutant protein enhanced red light inhibition of hypocotyl elongation in both wild type and phyB null backgrounds and delayed flowering in a phyB null background. These results show that the truncated phyB-28 protein retains activity to regulate both hypocotyl elongation and flowering and that the HKRD is therefore dispensable for at least these two phyB functions.
These results agree with previously published models that the PRD is the primary signaling domain of phytochromes (13). However, the HKRD mutations phyB-18 and phyB-19 decreased inhibition of both hypocotyl elongation and flowering, indicating that the HKRD is also important for phyB signaling. One hypothesis to explain how truncation of the HKRD causes less severe phenotypes than mutations that change amino acids in this domain is that both the PRD and HKRD domains may have to be “activated” for full Pfr function. If a point mutation prevents HKRD activation, then the inactive domain may prevent signaling by the PRD. Similar interdomain negative regulation has been postulated in the transmitter module of the Escherichia coli histidine kinase NtrB. Nitrogen starvation both activates the kinase activity of NtrB and suppresses its phosphatase activity. NtrB derivatives mutated in or lacking the G domain constitutively dephosphorylate a substrate and therefore repress signaling (36)
Such putative inhibition by the phyB HKRD may act between subunits of the phyB homodimer, or indirectly through other proteins such as PKS1. In either case, the HKRD may affect the conformation or phosphorylation state of the PRD or another portion of the phytochrome molecule. PKS1 protein interacts with both the phyA and phyB HKRD domains, can be phosphorylated by phyA in vitro or phyB in vivo, and is thought to inhibit phyB signaling (10). Perhaps, on conversion to Pfr, the HKRD normally abrogates repression by bound PKS1. Absence of the HKRD might bypass the need for such regulation by preventing PKS1 binding in the first place.
The phyB-28 mutation affects flowering time less than hypocotyl elongation. As overexpression of phyB-28 affects both of these phenotypes, it seems most likely that the threshold phyB activity needed to inhibit hypocotyl elongation is higher than that needed to inhibit flowering. The wild-type level of phyB-28 mutant protein may confer a level of activity between these thresholds, whereas overexpressed phyB-28 exceeds both thresholds. That phyB-18 and phyB-19 plants flower early also suggests that the HKRD normally regulates both hypocotyl elongation and flowering.
The phyB-18 and phyB-19 mutations fall in the N subdomain of the HKRD. Based on the crystal structure of the Thermatoga maritima CheA histidine kinase, this subdomain forms an α−helix close to the ATP-binding pocket formed by the G1, F, and G2 subdomains (37). Therefore, these mutations may disrupt ATP or Mg2+ binding, or they may have a more general effect on the conformation of the HKRD. It will be interesting to test whether the phyB-18, phyB-19, and phyB-28 mutations affect kinase activity or interaction with putative signaling partners. Moreover, the phyB-28 protein may serve as a useful tool for studies of phyB signaling, potentially free of complicating activities of the HKRD.
Although optimal phyB function apparently requires both the PRD and HKRD subdomains, the functionality of a truncated phyB lacking most of the HKRD suggests that these domains can work apart from each other, and may have done so in an ancestor of phytochromes. Consistent with this idea, known bacterial phytochrome-like photoreceptors have just one histidine kinase domain rather than two related domains, as higher plant phytochromes do. Other bacterial and plant photoreceptors have phytochrome chromophore domains and/or histidine kinase domains in a variety of different contexts. For example, a predicted protein of the moss Ceratodon purpureus has a phytochrome-like amino-terminal chromophore domain and a carboxyl-terminal domain with high homology to serine/threonine kinases (38). A photoreceptor from the fern Adiantum, PHY3, has a phytochrome-like amino-terminal chromophore domain and a carboxyl-terminal domain similar to NPH1, a blue light receptor kinase that mediates phototropism (39). Finally, the Ppr protein of the purple photosynthetic bacterium Rhodospirillum centenum has an amino-terminal domain similar to that of photoactive yellow protein, a blue light photoreceptor; a central domain similar to the amino terminus of phytochrome (but lacking the conserved cysteine used for the covalent attachment of phytochromobilin); and a single carboxyl-terminal histidine kinase domain (40). Further biochemical and structural comparisons may reveal how such light sensing and signal transduction modules may be combined in these different ways to create new light sensors.
Supplementary Material
Acknowledgments
We thank Tedd Elich for supplying the recombinant phyB extract used in Western blots, Akira Nagatani for the mBA2 Ab, Margaret Ahmad, Eva Huala, Enrique López-Juez, and Aron Silverstone for providing new phyB mutants, Alan Jones for the boiling SDS protein extraction method, Susan Whitfield for help with figures, and members of the Reed lab for helpful discussion. This work was supported by Grant R29-GM52456 from the National Institutes of Health to J.W.R.
Abbreviations
- phyA
phytochrome A holoprotein
- phyB
phytochrome B holoprotein
- EOD-FR
end-of-day far-red
- HKRD
histidine kinase-related domain
- PRD
PAS repeat domain
- Pr
red-absorbing form of phytochrome
- Pfr
far-red-absorbing form of phytochrome
- MS
Murashige and Skoog
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140520097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140520097
References
- 1.Deng X-W, Quail P H. Sem Cell Dev Biol. 1999;10:121–129. doi: 10.1006/scdb.1999.0287. [DOI] [PubMed] [Google Scholar]
- 2.Fankhauser C, Chory J. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- 3.Neff M M, Fankhauser C, Chory J. Genes Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- 4.Shinomura T, Uchida K, Furuya M. Plant Physiol. 2000;122:147–156. doi: 10.1104/pp.122.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reed J W. Curr Opin Plant Biol. 1999;2:393–397. doi: 10.1016/s1369-5266(99)00011-4. [DOI] [PubMed] [Google Scholar]
- 6.Quail P H. Plant Cell Environ. 1997;20:657–665. [Google Scholar]
- 7.Furuya M. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:617–645. [Google Scholar]
- 8.Yeh K-C, Lagarias J C. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh K-C, Wu S-H, Murphy J T, Lagarias J C. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- 10.Fankhauser C, Yeh K-C, Lagarias J C, Zhang H, Elich T D, Chory J. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Parks B M, Short T W, Quail P H. Plant Cell. 1995;7:1433–1443. doi: 10.1105/tpc.7.9.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner D, Quail P H. Proc Natl Acad Sci USA. 1995;92:8596–8600. doi: 10.1073/pnas.92.19.8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quail P H, Boylan M T, Parks B M, Short T W, Xu Y, Wagner D. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 14.Choi G, Yi H, Lee J, Kwon Y K, Soh M S, Shin B, Luka Z, Hahn T R, Song P S. Nature (London) 1999;401:610–613. doi: 10.1038/44176. [DOI] [PubMed] [Google Scholar]
- 15.Ni M, Tepperman J M, Quail P H. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 16.Ni M, Tepperman J M, Quail P H. Nature (London) 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- 17.Cherry J R, Hondred D, Walker J M, Keller J M, Hershey H P, Vierstra R D. Plant Cell. 1993;5:565–575. doi: 10.1105/tpc.5.5.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reed J W, Nagpal P, Poole D S, Furuya M, Chory J. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dehesh K, Franci C, Parks B M, Seeley K A, Short T W, Tepperman J M, Quail P H. Plant Cell. 1993;5:1081–1088. doi: 10.1105/tpc.5.9.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clough R C, Jordan-Beebe E T, Lohman K N, Marita J M, Walker J M, Gatz C, Vierstra R D. Plant J. 1999;17:155–167. doi: 10.1046/j.1365-313x.1999.00360.x. [DOI] [PubMed] [Google Scholar]
- 21.Boylan M T, Quail P H. Protoplasma. 1996;195:12–17. [Google Scholar]
- 22.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M. Proc Natl Acad Sci USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reed J W, Nagpal P, Bastow R, Solomon K S, Dowson-Day M J, Elumalai R P, Millar A J. Plant Physiol. 2000;122:1149–1160. doi: 10.1104/pp.122.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elich T D, Chory J. Plant Cell. 1998;9:2271–2280. doi: 10.1105/tpc.9.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirschfeld M, Tepperman J M, Clack T, Quail P H, Sharrock R A. Genetics. 1998;149:523–535. doi: 10.1093/genetics/149.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bechtold N, Ellis J, Pelletier G. C R Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- 29.Parkinson J S, Kofoid E C. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 30.Koornneef M, Rolff E, Spruit C J P. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- 31.Goto N, Kumagai T, Koornneef M. Physiol Plant. 1991;83:209–215. [Google Scholar]
- 32.Bagnall D J, King R W, Whitelam G C, Boylan M T, Wagner D, Quail P H. Plant Physiol. 1995;108:1495–1503. doi: 10.1104/pp.108.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robson P R H, Whitelam G C, Smith H. Plant Physiol. 1993;102:1179–1184. doi: 10.1104/pp.102.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aukerman M J, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino R M, Sharrock R A. Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner D, Tepperman J M, Quail P H. Plant Cell. 1991;3:1275–1288. doi: 10.1105/tpc.3.12.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer G, Weiss V. Proc Natl Acad Sci USA. 1999;96:604–609. doi: 10.1073/pnas.96.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bilwes A M, Alex L A, Crane B R, Simon M I. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 38.Thümmler F, Dufner M, Kreisl P, Dittrich P. Plant Mol Biol. 1992;20:1003–1017. doi: 10.1007/BF00028888. [DOI] [PubMed] [Google Scholar]
- 39.Nozue K, Kanegae T, Imaizumi T, Fukuda S, Okamoto H, Yeh K C, Lagarias J C, Wada M. Proc Natl Acad Sci USA. 1998;95:15826–15830. doi: 10.1073/pnas.95.26.15826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang Z Y, Swem L R, Rushing B G, Devanathan S, Tollin G, Bauer C E. Science. 1999;285:406–409. doi: 10.1126/science.285.5426.406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.