Abstract
Small GTP-binding proteins regulate diverse processes in eukaryotic cells such as signal transduction, cell proliferation, cytoskeletal organization, and intracellular membrane trafficking. These proteins function as molecular switches that cycle between “active” and “inactive” states, and this cycle is linked to the binding and hydrolysis of GTP. The Arabidopsis genome contains 93 genes that encode small GTP-binding protein homologs. Phylogenetic analysis of these genes shows that plants contain Rab, Rho, Arf, and Ran GTPases, but no Ras GTPases. We have assembled complete lists of these small GTPases families, as well as accessory proteins that control their activity, and review what is known of the functions of individual members of these families in Arabidopsis. We also discuss the possible roles of these GTPases in relation to their similarity to orthologs with known functions and localizations in yeast and/or animal systems.
Small GTP-binding proteins are molecular switches that are “activated” by GTP and “inactivated” by the hydrolysis of GTP to GDP. The resulting cycles of binding and hydrolysis of GTP by small GTP-binding proteins represents a ubiquitous regulatory mechanism in eukaryotic cells. Members of this class of proteins are among the largest families of signaling proteins in eukaryotic cells. Their importance in cellular signaling processes is underscored by their conservation throughout evolution of eukaryotic organisms and by the presence of homologs that perform related functions in cells of yeasts, humans, and plants. Although the GTP-hydrolysis “core” of this class of regulatory molecules is highly conserved, the surrounding domains are highly variable and undergo conformational changes as these proteins switch from GTP-associated to GDP-associated states. Eukaryotes have harnessed this diversity of protein conformations, linked to the nucleotide-associated state of the GTP-binding domain, to regulate a myriad of cellular processes (for review, see Takai et al., 2001). Small GTP-binding proteins are involved in regulation of diverse eukaryotic cellular processes, such as cell proliferation, cytoskeletal assembly and organization, and intracellular membrane trafficking (for reviews, see Barbacid, 1987; Boguski and McCormick, 1993; Takai et al., 2001). Physiological control of these GTPase “switches” occurs through association of the GTPase with accessory proteins, termed guanine nucleotide exchange factors (GEFs), that catalyze the conversion of the small GTP-binding protein to their GTP-bound “active” conformation. In their “active” state, small GTPases interact with various downstream “effector” proteins that perform the diverse cellular functions controlled by this class of regulatory molecules. Inactivation occurs through either the intrinsic ability of the small GTP-binding protein to hydrolyze GTP to GDP+Pi, or through association with another set of accessory proteins, GTPase-activating proteins (GAPs), which stimulate this hydrolytic activity. Upon hydrolysis of GTP, the small GTP-binding protein is returned to the “inactive” state and is ready to begin the cycle again (supplemental data can be viewed at www.plantphysiol.org).
Structural and functional similarities between different members of this large superfamily has led to establishment of five distinct families: Ras, Rab, Rho, Arf, and Ran (Kahn et al., 1992). Ras GTPases regulate cell proliferation in yeast and mammalian systems. Members of the Rho GTPase family control actin reorganization and signal transduction pathways associated with MAP kinases. The Rab and Arf GTPase families function in distinct steps of membrane trafficking, whereas Ras-related nuclear protein (Ran) GTPases regulate transport of proteins and RNA across the nuclear envelope. Individual members of these families share higher overall sequence conservation with one another than with any other small GTPase families (Fig. 1). Analysis of the genomes of Saccharomyces cerevisiae, fruitfly (Drosophila melanogaster), Caenorhabditis elegans, Arabidopsis, and human (Homo sapiens) has underscored the conservation of these classes of regulatory molecules and has provided interesting glimpses into the ways in which the small GTPases of these families have evolved and proliferated. Here, we describe the identification and classification of 93 small GTP-binding proteins in Arabidopsis. These GTPases were classified within four of the five small GTPase families: with 57 Rab GTPases; 21 Arf GTPases; 11 Rho GTPases; and 4 Ran GTPases. Interestingly, Arabidopsis does not contain any Ras GTPases that can be identified based on phylogenetic analysis, perhaps reflecting unique mechanisms for control of cell signaling during development in plants (Meyerowitz, 1999, 2002).
Figure 1.
Phylogenetic star diagrams of small GTPases of A. thaliana, S. cerevisiae, and H. sapiens. Unrooted star diagrams were obtained using the program, ClustalW (Thompson et al., 1994). Sequences of small GTPases were obtained from the sequenced genomes of the three indicated organisms using BLAST (Altschul et al., 1997) sequence similarity searches against characterized GTPases. In each organism, distinct families (Ras, Rab, Rho, Arf, and Ran) could be distinguished, except for Arabidopsis in which no apparent Ras GTPase family could be identified.
RESULTS
Identification and Classification of Small GTPase Genes in Arabidopsis
To identify small GTPase genes in the Arabidopsis genome, we first compiled sequences previously documented in S. cerevisiae and humans as members of the small GTP-binding protein superfamily of regulatory proteins. These genes were classified into the five accepted families of small GTPases (Kahn et al., 1992) based on their functions and were then used to generate a template phylogenetic tree for future grouping of Arabidopsis sequences. We then carried out BLASTP (Altschul et al., 1997) searches on the theoretical protein complement of the Arabidopsis Genome Initiative (AGI) database (Arabidopsis Genome Initiative, 2000) with representative sequences of each of the five families of small GTPases (Kahn et al., 1992). After individual protein sequences were identified, we ensured that we had eliminated duplications and contaminating sequences through careful comparison with theoretical cDNA and genomic DNA sequences in the AGI genomic database. In this manner, we identified a total of 93 small GTPase genes in the Arabidopsis genome.
To classify Arabidopsis small GTPase genes into Ras, Rho, Rab, Arf, and Ran families, theoretical protein sequences from these genes were aligned to previously compiled lists of small GTPases of S. cerevisiae and humans using ClustalW (Thompson et al., 1994) and then edited manually to restrict phylogenetic analysis to conserved “core” domains of these protein sequences. Phylogenetic trees were constructed using the neighbor-joining method as implemented in PAUP* (http://paup.csit.fsu.edu/index.html) with 1,000 bootstrap replicates (Swofford et al., 2001). Branches with bootstrap values of less than 80% were collapsed to simplify tree structures. After this phylogenetic analysis, we determined that Arabidopsis contained members of the Rab (57 members), Arf (21 members), Rho (11 members), and Ran (4 members) families, but no small GTPase genes could be determined to cosegregate with yeast or human members of the Ras GTPase family (Tables I–IV; Fig. 1).
Table I.
Arabidopsis RAB GTPase genes
| Genes Name
|
AGI Genea | Accession No.b | Expression/Localizationc | ||
|---|---|---|---|---|---|
| a | b | c | |||
| AtRABA1a | AtRab11E | Ara-2i | At1g06400 | 114086 | |
| AtRABA1b | At1g16920 | 3024526 | |||
| AtRABA1c | At5g45750 | 9758678 | |||
| AtRABA1d | AtRab11B | AthSGBPii | At4g18800 | 7438436 | Highly expressed in roots1 |
| AtRABA1e | At4g18430 | 7438435 | |||
| AtRABA1f | At5g60860 | 10176913 | |||
| AtRABA1g | At3g15060 | 8777489 | |||
| ArRABA1h | At2g33870 | 1707014 | |||
| AtRABA1i | At1g28550 | 6560749 | |||
| AtRABA2a | AtRab11C | At1g09630 | 3024516 | ||
| AtRABA2b | At1g07410 | 8778562 | |||
| AtRABA2c | AtRab11A | At3g46830 | 3915842 | ||
| AtRABA2d | At5g59150 | 9759237 | |||
| AtRABA3 | At1g01200 | 9665141 | |||
| AtRABA4a | At5g65270 | 10178189 | |||
| AtRABA4b | AtRab11G | AtGB3iii | At4g39990 | 7438426 | |
| AtRABA4c | At5g47960 | 9758522 | |||
| AtRABA4d | At3g12160 | 9294115 | |||
| AtRABA4e | At2g22390 | 7438435 | |||
| AtRABA5a | At5g47520 | 9758780 | |||
| AtRABA5b | At3g07410 | 6041856 | |||
| AtRABA5c | AtRab11F | Ara-4i | At2g43130 | 114089 | Ubiquitous expression/Golgi-derived vesicles2 |
| AtRABA5d | At2g31680 | 4582469 | |||
| AtRABA5e | AtRab11D | Ara-1i | At1g05810 | 114085 | |
| AtRABA6a | At1g73640 | 6692728 | |||
| AtRABA6b | At1g18200 | 9719734 | |||
| AtRABB1a | AtRab2B | At4g17160 | 7438380 | ||
| AtRABB1b | AtRab2C | AtGB2iii | At4g35860 | 2129702 | |
| AtRABB1c | AtRab2A | At4g17170 | 1765895 | High expression in cotyledeons, fruits, and pollen3 | |
| AtRABC1 | AtRab18 | At1g43890 | 2231312 | Stems and roots4 | |
| AtRABC2a | AtRab18B | At5g03530 | 11274537 | ||
| AtRABC2b | AtRab18C | At3g09910 | 6681328 | ||
| AtRABD1 | At3g11730 | 4097557 | |||
| AtRABD2a | AtRab1B | Ara-5i | At1g02130 | 5902803 | Ubiquitous expression2 |
| AtRABD2b | AtRab1A | At5g47200 | 2245111 | ||
| AtRABD2c | AtRab1C | At4g17530 | 7268505 | ||
| AtRABE1a | AtRab8B | At3g53610 | 11274528 | ||
| AtRABE1b | AtRab8D | At4g20360 | 10172744 | ||
| AtRABE1c | AtRab8A | Ara-3i | At3g46060 | 114088 | |
| AtRABE1d | AtRab8C | At5g03520 | 11274535 | ||
| AtRABE1e | AtRab8E | At3g09900 | 6681329 | ||
| AtRABF1 | AtRab5C | Ara-6iv | At3g54840 | 13160603 | |
| AtRABF2a | AtRab5A | Rha1v | At5g45130 | 400976 | Stomatal guard cells, stipules, and root tips5 |
| AtRABF2b | AtRab5B | Ara-7iv | At4g19640 | 7438427 | |
| AtRABG1 | At5g39620 | 9758338 | |||
| AtRABG2 | AtRab7A | At2g21880 | 4417298 | ||
| AtRABG3a | At4g09720 | 7438424 | |||
| AtRABG3b | At1g22740 | 3914521 | |||
| AtRABG3c | AtRab7D | At3g16100 | 9294457 | ||
| AtRABG3d | At1g52280 | 18389228 | |||
| AtRABG3e | At1g49300 | 5430767 | |||
| AtRABG3f | AtRab7B | At3g18820 | 9293907 | ||
| AtRABH1a | At5g64990 | 8843763 | |||
| AtRABH1b | AtRab6A | At2g44610 | 7438386 | Highest expression in liquid root culture6 | |
| AtRABH1c | At4g39890 | 7438387 | |||
| AtRABH1d | At2g22290 | 4567198 | |||
| AtRABH1e | At5g10260 | 11274352 | |||
a, Nomenclature used in Periera-leal and Seabra (2001). b, Nomenclature used in Bischoff et al. (1999). c, Nomenclatures used in: i, Anai et al. (1991) and Ueda et al. (1996); ii, Yi and Guerinot (1994); iii, Biermann et al. (1996); iv, Ueda et al. (2001); and v, Terryn et al. (1993).
AGI gene nomenclature.
Genbank protein accession no. (GI:) for a translation of the corresponding gene.
Numbers in superscript refer to articles where expression and localization data can be found: 1, Yi and Guerinot (1994); 2, Anai et al. (1994) and Ueda et al. (1994); 3, Moore et al. (1997); 4, Mikami et al. (1997); 5, Terryn et al. (1993); 6, Bednarek et al. (1994).
Table IV.
Arabidopsis RAN GTPase genes
| Gene Name | AGI Genea | Accession No.b | Expression/Localizationc |
|---|---|---|---|
| AtRAN1d | At5g20010 | 2058278 | Ubiquitous, highest in meristematic tissue/cytoplasm and nucleus |
| AtRAN2d | At5g20020 | 1668706 | Same as AtRAN1 |
| AtRAN3d | At5g55190 | 9758116e | Same as AtRAN1 |
| AtRAN4f | At5g55080 | 9758105 | ND |
AGI gene nomenclature.
Genbank protein accession no. (GI:) for a translation of the corresponding gene.
ND, Not determined. Expression and localization data for AtRAN1, -2, and -3 have been published by Haizel et al. (1997).
Nomenclatures used in Haizel et al. (1997).
In the database, several GI numbers refer to the same AtRAN3 sequence (GI: 9758116, GI; 2058280, and GI: 2801433).
AtRAN4 has been described in the database as “salt stress-inducible small GTP-binding protein Ran-1-like protein.”
Rab GTPases
Rab GTPases make up the largest family of the small GTP-binding protein superfamily. Both in vivo and in vitro experiments have demonstrated the roles of this class of proteins in intracellular membrane trafficking. Furthermore, given the specific distributions of Rab GTPases to different cellular membranes, it was hypothesized that Rab GTPases would, in conjunction with SNARE proteins, provide specificity for membrane fusion events (for reviews, see Stenmark and Olkkonen, 2001; Zerial and McBride, 2001).
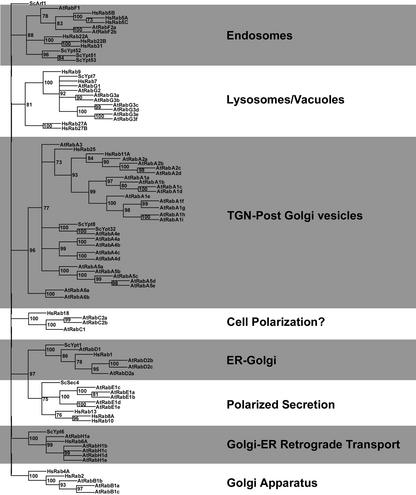
Our analysis of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000) identified 57 Rab GTPase isoforms (Table I; Fig. 2), which we have named AtRAB GTPases. This corresponded with the number recently determined by others (Pereira-Leal and Seabra, 2001). With a few notable exceptions, little is known of the function of AtRAB GTPases. As a result, our discussion of this gene family will focus on organization and classification of members of the Rab GTPase family from Arabidopsis. Examination of the phylogenetic trees generated with protein sequences from human, yeast, and Arabidopsis Rab GTPases indicated that this GTPase family could be further divided into eight subfamilies based on sequence similarity and segregation with yeast and mammalian orthologs (see Fig. 2). A number of plant Rab GTPases have been cloned and named according to various schemes. To simplify and synchronize the nomenclature of the Rab GTPase family in Arabidopsis, we have followed the naming and classification scheme described in recent papers (Pereira-Leal and Seabra, 2000, 2001), and have named them according to their presence in one of the eight subfamilies that arose from our analysis (AtRABA-AtRABH; Fig. 2).
Figure 2.
The Rab GTPase family of Arabidopsis. A neighbor-joining tree of A. thaliana Rab GTPases including representative Rab GTPase sequences of S. cerevisiae and H. sapiens was generated using ClustalW and scoring for amino acid differences (Thompson et al., 1994). The tree was rooted with a S. cerevisiae Arf1 sequence, and branches with percentage bootstrap values of less than 70% (of 1,000) were collapsed to simplify the tree. Labels to the right of the Rab GTPase subfamilies reflect functions determined for distinct Rab GTPases in that subfamily.
Correlation of Sequence Similarity with Localization in Different Species
Rab GTPase functions have been studied extensively in yeast and mammalian systems (for review, see Stenmark and Olkkonen, 2001). Individual members of the Rab GTPase family localize to different intracellular compartments where they regulate vesicle trafficking events (for review, see Simons and Zerial, 1993). S. cerevisiae only contains 11 Rab GTPases. Because of the cellular similarities between eukaryotic cells, we used the known distribution and function of these Rab GTPases as a basis for classification of plant Rab GTPases. S. cerevisiae Rab GTPases, termed yeast protein transport (YPT) proteins, are localized to the endoplasmic reticulum (ER), the Golgi apparatus and trans-Golgi network (TGN), the endosomal/prevacuolar compartments, and vacuoles in S. cerevisiae. They also regulate retrograde trafficking of proteins from Golgi to ER and polarized secretion (for review, see Lazar et al., 1997).
The correlation between sequence similarity and regulation of membrane trafficking through related compartments appears to be a conserved feature in the Rab GTPase family (Fig. 2). When functions of mammalian Rab GTPases sharing significant similarity to yeast counterparts have been studied, they regulate membrane trafficking through compartments with related function. For example, the human Rab8 GTPase is targeted to the protein secretion pathway, and displays high sequence similarity to Sec4p/Ypt2p, which regulates protein secretion in S. cerevisiae. Rab8 complements the ypt2 mutant (Sec4p homolog) in fission yeast (Schizosaccharomyces pombe; Huber et al., 1993a, 1993b). Rab8 and Sec4p cosegregate in the same subfamily in our phylogenetic analysis (Fig. 2). Functions for most AtRAB GTPases have not yet been established. However, when plant Rab GTPase function is known, these isoforms also cosegregate in subfamilies containing their mammalian and yeast counterparts (discussed below). In particular, studies of Rab GTPases that are members of five of the eight subfamilies highlighted by our phylogenetic analysis have been performed and published (AtRABA, Inaba et al., 2002; AtRABB, Cheung et al., 2002; AtRABD, Batoko et al., 2000; AtRABF, Ueda et al., 2001; AtRABH, Bednarek et al., 1994). In every case, their localization and/or function in membrane trafficking correlate with their segregation in subfamilies for yeast and mammalian Rab GTPases whose functions not only are the same, but also are well established. We have therefore indicated the compartments to which mammalian and yeast homologs are localized, and suggest that this may prove a useful initial framework for classification of AtRAB GTPases (Fig. 2).
Rab GTPases of the Endocytic Trafficking Pathway
Endocytosis is the primary means by which proteins and macromolecules too large to pass through the plasma membrane enter the cell. During endocytosis, regions of the plasma membrane with associated cargo molecules are invaginated and pinch off into transport vesicles, which then fuse with endocytic compartments. In addition, many proteins targeted to lysosomal and/or vacuolar compartments are sorted in endosomal compartments.
When compared with other eukaryotic Rab GTPases, three members of the AtRABF subfamily show highest similarity to human Rab5 and yeast Ypt51p family members (Fig. 2). In mammals, Rab5 GTPases localize to early endosomes and regulate fusion of clathrin-coated vesicles to early endosomes and fusion between early endosomes (Bucci et al., 1992). In yeast, Ypt51p family members similarly regulate membrane trafficking through prevacuolar compartments (Horazdovsky et al., 1994; Singer-Krüger et al., 1995). Both AtRABF2a (E. Nielsen, unpublished data) and AtRABF2b (Ueda et al., 2001) localize to compartments observed upon uptake of the fluorescent styryl dye, FM 4–64. Interestingly, the third Arabidopsis member of this subfamily, AtRABF1, appears to be a novel, plant-specific Rab GTPase (Bolte et al., 2000; Ueda et al., 2001). AtRABF1 lacks the traditional carboxy-terminal-CAAX motif for posttranslational isoprenylation but is myristoylated at the amino terminus (Ueda et al., 2001). Despite these differences, AtRABF1 colocalizes with AtRABF2b on the plant endosomal compartment where it may provide novel regulatory functions (Ueda et al., 2001).
The AtRABG subfamily contains eight members with significant similarity to human Rab7 and Ypt7p (Fig. 2). In mammals, Rab7 regulates transport of cargo from early endosomes to late endosomes and lysosomes (Feng et al., 1995; Mukhopadhyay et al., 1997). In yeast, Ypt7p localizes to vacuoles and regulates homotypic fusion between vacuolar compartments (Price et al., 2000). In plants, vacuoles are used as storage organelles in addition to having lytic roles, and single cells can contain multiple vacuole types (Paris et al., 1996; Vitale and Raikhel, 1999). This increased complexity appears to be reflected in the large number of members in this Rab GTPase subfamily in plants.
Rab GTPases of the Biosynthetic Trafficking Pathway
Newly synthesized proteins enter the secretory pathway by translocation through ER membranes. Subsequent transport from the ER to Golgi complexes involves recruitment of cargo proteins into vesicle transport intermediates. The AtRABD subfamily contains five members that display significant similarity with mammalian Rab1 and Ypt1p Rab GTPases (Fig. 2). In mammals, Rab1 GTPase isoforms localize to ER, ER Golgi intermediate compartment, and Golgi compartments and regulate ER-to-Golgi membrane trafficking steps (Tisdale et al., 1992; Nuoffer et al., 1994). In yeast, YPT1 is an essential gene required for ER-to-Golgi trafficking events (Segev et al., 1988). In plants, transient expression of a dominant-negative mutant of AtRABD2a resulted in accumulation of a secreted GFP marker in an intracellular compartment reminiscent of the ER and inhibited movement of Golgi complexes along cytoskeletal elements (Batoko et al., 2000). Arabidopsis contains three AtRABB subfamily members related to the human Rab2 GTPase. Rab2 GTPases are found associated with ER Golgi intermediate compartment membranes and COP-I transport vesicles in mammalian cells (Tisdale et al., 1992). The function and localization of the AtRABB subfamily members as yet remains uncharacterized in plants.
Recycling of ER-resident proteins, such as the KDEL-receptor Erd2p and Sec23p/Sec24p cargo receptor proteins requires the specific enrichment of these proteins in transport vesicles and subsequent delivery back to ER membranes (for review, see Kirchhausen, 2000). In mammals, Rab6 GTPases regulate this process (Martinez et al., 1997; White et al., 1999). Five isoforms of this subfamily of Rab GTPases (AtRABH) are present in the Arabidopsis genome. Of these, the AtRABH1b homolog, previously identified as AtRAB6, was shown to complement a yeast ypt6 mutation, although the function of this Rab GTPase in plants was not determined (Bednarek et al., 1994).
Rab GTPases Involved in Polarized Secretion
Arabidopsis contains five Rab GTPases that cosegregate with yeast and mammalian isoforms that regulate polarized secretion (AtRABE subfamily; Fig. 2). In yeast, Sec4p regulates membrane trafficking to the daughter cell bud site (Salminen and Novick, 1987; Goud et al., 1988). In plants, members of this Rab GTPase subfamily may have roles in cellular responses to bacterial pathogens. In tomato (Lycopersicon esculentum), yeast two-hybrid screening for plant proteins that interact with the Pseudomonas sp. avirulence factor, avrPto, identified a RabE subfamily member with significant similarity to mammalian Rab8 (Bogdanove and Martin, 2000). This interaction only occurred in absence of the tomato resistance protein, Pto (Bogdanove and Martin, 2000), raising the possibility that in susceptible plants, avrPto may interfere with membrane trafficking pathways regulated by this RabE homolog. In an intriguing possibility, the authors suggest this Rab GTPase might regulate polarized secretion of antimicrobial compounds and/or components involved in mounting cellular responses to attack by bacterial pathogens.
A Plethora of Post-Golgi Rab GTPases in Plants
One of the most striking features evident upon examination of the AtRAB GTPases is the large number of AtRABA subfamily members (Fig. 2). This subfamily contains Rab GTPases from mammals and yeast that regulate TGN membrane trafficking pathways. In mammals, human Rab11A and Rab11B isoforms localize to recycling endosomes (Ullrich et al., 1996), and in epithelial cells, mammalian Rab11A is critical for exit of internalized proteins from apical recycling endosomes (Calhoun et al., 1998; Duman et al., 1999). Yeast Ypt31p and Ypt32p Rab GTPases appear to function during exit of membrane traffic from trans-Golgi cisternae (Benli et al., 1996; Jedd et al., 1997).
The Arabidopsis genome contains 26 distinct AtRABA subfamily members. Whether this multitude of genes provides distinct functions or represents functionally redundant gene families remains to be determined. Antisense inhibition of RABA subfamily GTPases in tomato results in complex developmental abnormalities and delayed fruit ripening (Lu et al., 2001). In pea (Pisum sativum), expression of two RABA GTPases, PRA2 and PRA3, was examined (Nagano et al., 1995). Whereas PRA3 was constitutively expressed, PRA2 was specifically up-regulated in the stem-elongation zone of dark-grown seedlings, a region in which rapid plant cell expansion is observed (Nagano et al., 1995). More recent experiments indicated that Pra2 GTPase localizes predominantly to Golgi and possibly endosomal compartments, whereas Pra3 appears on TGN and/or prevacuolar compartments (Inaba et al., 2002). These RABA isoforms were suggested to play a role in delivery of new cell wall components to the plasma membrane. Interestingly, identification of effector proteins for RABA GTPases may further support roles for this class of GTPases in stem elongation in dark-grown seedlings because one effector protein for this Rab GTPase was identified as a cytochrome p450 involved in biosynthesis of brassinosteroids (Kang et al., 2001). Plant cells display a high degree of polarized deposition of primary and secondary cell wall components, in some cases, adjacent walls of a cell display differential localization of antigenic determinants (Lynch and Staehelin, 1992). The delivery of hemicelluloses, integral cell wall proteins, and probably even delivery of the cellulose synthase complex to the plasma membrane likely occur via post-Golgi membrane trafficking pathways. Could the large numbers of Rab GTPases in the AtRABA subfamily reflect the increased complexity associated with the highly polarized deposition of cell wall material observed in plants?
Rab-Interacting Proteins
Rab GTPases cycle between an inactive GDP-bound form located in the cytosol and an active GTP-bound form that is membrane associated. For most Rab GTPases, membrane association is promoted by posttranslational lipid modification. Stabilization of the lipid-modified Rab GTPase in cytosol occurs through association with Rab GDP-dissociation inhibitor proteins (RabGDI). Three RabGDI homologs (AtRabGDI1-AtRabGDI3; see supplemental data) are present in Arabidopsis. Two of these, AtRabGDI1 and AtRabGDI2 were identified by complementation of yeast sec19 mutants, indicating functional conservation of activity between yeast and plants. Although AtRabGDI1 was ubiquitously expressed, AtRabGDI2 displayed higher expression levels in suspension cultures cells and in roots (Ueda et al., 1996, 1998; Zarsky et al., 1997). AtRabGDI1-AtRabGDI3 homologs all share significant similarity (more than 77% identity). Interestingly, two RabGDI transcripts were determined to be up-regulated during early stages of fungal infection in rice (Oryza sativa; Kim et al., 1999), however the exact roles of these proteins during early plant defense responses to fungal infection remain unknown. A fourth related protein, AtREP1 displays similarity to the AtRabGDI homologs, but appears somewhat diverged (27% identity, 44% similarity), instead displaying higher similarity to mammalian REP-1 (Alexandrov et al., 1994). In mammals, REP-1 associates with newly synthesized Rab GTPases and presents them for posttranslational lipid modification.
GTP/GDP exchange is essential for Rab GTPase function. Upon binding to its target membrane, the Rab GTPase is converted from Rab:GDP to Rab:GTP through the action of RabGEF proteins. RabGEFs have been identified for several Rab GTPases in mammals and yeast (Horiuchi et al., 1997; Wada et al., 1997; Walch-Solimena et al., 1997; Hama et al., 1999; Iwasaki and Toyonaga, 2000; Siniossoglou et al., 2000). In general, significant levels of sequence similarity have not been observed between RabGEFs for different Rab GTPases. This suggests that despite having similar guanine exchange functions, different RabGEF proteins may have diverse evolutionary origins. However, in one notable exception to this trend, Vps9p, the RabGEF for the yeast Rab GTPase, Ypt51p, and Rabex-5, which catalyzes nucleotide exchange on mammalian Rab5, shows significant sequence similarity (Horiuchi et al., 1997; Hama et al., 1999). The Arabidopsis genome contained two sequences with significant similarity to the Vps9p and Rabex-5 RabGEF sequences. We designated these RabGEFs AtVPS9A and AtVPS9B (supplemental data). No Arabidopsis sequences with significant primary amino acid similarities could be detected for non-Vps9p-like RabGEFs.
As opposed to RabGEFs, which activate Rab GTPases, RabGAPs inactivate Rab GTPases by accelerating the slow intrinsic Rab GTPase activity. The first Rab-specific GAP proteins, Gyp6p, Gyp7p, and Gyp1p, were identified in yeast (Strom et al., 1993; Vollmer and Gallwitz, 1995; Vollmer et al., 1999). These proteins all contained six conserved sequence motifs within their catalytic domains (Albert et al., 1999). In other proteins, such as RN-tre and GAPCenA proteins, presence of these motifs successfully predicted their RabGAP activity (Cuif et al., 1999; Lanzetti et al., 2000). Analysis of the Arabidopsis genome revealed 20 proteins with all or most of the RabGAP “catalytic core” motifs, which we have named AtGYP proteins (supplemental data). All AtGYP proteins contained a conserved Arg residue critical for RabGAP activity (Albert et al., 1999) and at least five of the six “catalytic core” motifs. Although additional sequences containing portions of the RabGAP “catalytic core” were detected, these were not included because they (a) were missing two or more conserved motifs, and (b) did not contain essential conserved residues. Apart from the RabGAP motif, AtGYP family members contain no other detectable protein domains and displayed significant sequence diversity. In animals, two RabGAP proteins have been found associated with noncatalytic partner proteins (Nagano et al., 1998; Lanzetti et al., 2000). It seems likely that other RabGAP proteins may also be found in association with partner proteins.
In the last several years, many Rab effector proteins have been characterized. Because Rab GTPases are highly conserved and function similarly in different organisms, one might expect that Rab effector proteins also be conserved. But this is not apparently the case (Zerial and McBride, 2001). The structural heterogeneity of Rab effectors make it unlikely that plant Rab effectors can be identified by sequence similarity alone. However, some established Rab effectors, such as lipid kinases (Christoforidis et al., 1999), are present in Arabidopsis. In addition, in several cases, zinc-finger domains are associated with Rab effector protein functions (Christoforidis et al., 1999; Simonsen et al., 1998), and proteins with these domains are present in Arabidopsis (Jensen et al., 2001; Heras and Drobak, 2002). Identification of plant Rab effector proteins clearly is likely to uncover specialized proteins whose activities have been exclusively tailored for plant-specific transport systems.
Rop GTPases
Members of the Rho GTPase family have emerged as key regulators of the actin cytoskeleton in yeast and animal cells. Transduction of signals from cell surface receptors, which ultimately result in reorganization of the actin cytoskeleton, occurs through interaction of Rho GTPases with regulatory proteins and downstream effector proteins (Hall, 1998). Rho GTPases have been categorized into three major subfamilies: CDC42, RAC, and RHO (capitalized to distinguish them from the collective “Rho” GTPase family), based on their cellular functions and sequence homology (Chant and Stowers, 1995; Hall, 1998). In both vertebrate and invertebrate animals, all subfamilies of the Rho GTPase family (RHO, RAC, and CDC42) are present, but some lower eukaryotes lack certain subfamilies. No RAC ortholog is present in S. cerevisiae or S. pombe, and Dictyostelium discoideum lacks CDC42 and RHO orthologs (Takai et al., 2001). In Arabidopsis, all small GTPases that segregate with the Rho GTPases appear to be members of a unique subfamily (Fig. 3). Because this subfamily has so far only been identified in plants, they have been named Rop GTPases (for Rho-related proteins from plants; Li et al., 1998; Zheng and Yang, 2000a; Yang, 2002). For a detailed discussion on the structure and evolution of the Arabidopsis Rop GTPase subfamily, readers are referred to several recent reviews and research papers (Winge et al., 2000; Zheng and Yang, 2000a; Yang, 2002).
Figure 3.
The Rop GTPase family of Arabidopsis. A neighbor-joining tree of A. thaliana Rop GTPases including representative Rho GTPase sequences of S. cerevisiae and H. sapiens was generated using ClustalW and scoring for amino acid differences (Thompson et al., 1994). The tree was rooted with a S. cerevisiae Ras1 sequence, and branches with percentage bootstrap values of less than 80% (of 1,000) were collapsed to simplify the tree. Arabidopsis Rop GTPases clearly represent a distinct subfamily, most closely related to Brewer's yeast Rho GTPases.
Due to a slightly higher overall similarity with human RAC GTPases, plant Rho-related GTPases have been identified as plant RAC GTPases (Winge et al., 1997; Kost et al., 1999; Winge et al., 2000; Lemichez et al., 2001). However, phylogenetic analysis from three representative species, S. cerevisiae, H. sapiens, and A. thaliana (Fig. 3), and sequence comparisons clearly suggest that plant ROP GTPases are distinct from the three subfamilies found in animals, and in particular, they do not belong to the RAC subfamily as represented by mammalian RAC1. Instead, they represent a novel, plant-specific subfamily of Rho GTPases (Fig. 3).
Analysis of the Arabidopsis genome revealed the existence of 11 ROP GTPases, which we have named AtROP (Fig. 3; Table II). Previous reports have been inconsistent regarding the number of Rho-related GTPases present in Arabidopsis, e.g. 13 in Lemichez et al. (2001) and Valster et al. (2000) and 12 in Bischoff et al. (2000). However, after careful examination of the AGI genomic database, we determined that the extra AtROP GTPases resulted either from duplicated database entries or contaminating sequences (Table II). In particular, the “Rac-like protein” (GI:U88402; Collins and Johnson, 1997) appears to result from contaminating sequences in the cDNA library used for expressed sequence tag sequencing (R. Geng and Z. Yang, unpublished data). Previous classification of AtROP GTPases has led to confusing nomenclature, e.g. Lemichez et al. (2001) used the name AtRAC1 for AtROP6, and AtRAC1/ARAC1 (Winge et al., 1997, 2000) is actually AtROP3. Here, we propose a unifying nomenclature for the 11 AtROP genes, named AtROP1 through AtROP11. To provide continuity to previous naming schemes for these AtROP GTPases, we have also listed nomenclature used in earlier studies (Table II), and, as far as possible, we have maintained the numbering schemes for this class of GTPases.
Table II.
Arabidopsis ROP GTPase genes
| Gene Name
|
AGI Genea | Accession No.b | Expression/Localizationc | ||
|---|---|---|---|---|---|
| a | b | c | |||
| AtROP1 | Arac11/AtRAC11 | At3g51300 | 2558666 | Pollen and flowers only1/specific domains of the PM4 | |
| AtROP2 | Arac4/AtRAC4 | At1g20090 | 1777764 | Vegetative tissues and inflorescence1,2/specific domains of the PM5 | |
| AtROP3d | Arac1/AtRAC1 | At2g17800 | 7211191 | Vegetative tissues, inflorescence, and pollen1,2 | |
| AtROP4e | Arac5/AtRAC5 | At1g75840 | 7211204 | Vegetative tissues and inflorescence1,2/perinuclear organelle6 | |
| AtROP5f | Arac6/AtRAC6 | AtRac2 | At4g35950 | 7211206 | Leaves, stems, flowers, and pollen; low in roots1 |
| AtROP6g | Arac3/AtRAC3 | AtRac1 | At4g35020 | 7211200 | Vegetative tissues and inflorescence (open flowers only)1,2/entire PM6 |
| AtROP7 | Arac2/AtRAC2 | At5g45970 | 7211198 | Roots and stems only7 | |
| AtROP8 | Arac9/AtRAC9 | At2g44690 | 5381420 | ND | |
| AtROP9 | Arac7/AtRAC7 | At4g28950 | 7211208 | Entire PM3 | |
| AtROP10 | Arac8/AtRAC8 | At3g48040 | 7211210h | Entire PM3 | |
| AtROP11 | Arac10/AtRAC10i | At5g62880 | 10177466 | ND | |
a, Nomenclatures used in Li et al. (1998, 1999, 2001), Bischoff et al. (2000), and Molendijk et al. (2000). b, Nomenclature used in Winge et al. (1997, 2000). c, Nomenclature used in Kost et al. (1999) and Lemichez et al. (2001).
AGI gene nomenclature.
Genebank protein accession no. (GI:) for a translation of the corresponding gene.
PM, Plasma membrane; ND, not determined. Numbers in superscript refer to articles where expression and localization data can be found: 1, Li et al. (1998); 2, Winge et al. (1997); 3, Zheng et al. (2002); 4, Li et al. (1999); 5, Jones et al. (2002) and Fu et al. (2002); 6, Bischoff et al. (2000); 7, Z.L. Zheng and Z. Yang, unpublished data.
AtROP3 is the same as ATGP2 (GI: 4097563).
AtROP4 is the same as ATGP3 (GI: 4097565). The accession number GI: 2654009 (“RHO-like GTP binding protein”) refers also to AtROP4.
GI: 2654011 corresponds also to AtROP5, but the sequence is incorrect; the last 100 amino acid residues are missing.
AtROP6 is the same as the clone At43 published by Xia et al. (1996) and defined in the database as “Arabidopsis Rho1Ps homolog” (GI: 1732519). AtROP6 is also found in the database under the accession number GI: 2645643.
The accession no. GI: 4678324 refers also to AtROP10/Arac8, but the splicing suggested in the database is inaccurate; the last exon is missing.
The first intron of AtROP11/Arac10 gene contains a cryptic exon of unknown function (GI: 7211195).
Rop GTPases Are Multifunctional Signaling Molecules
Rho GTPases play central roles in a wide range of cellular processes, many of which are associated with the actin cytoskeleton. In mammalian cells, RHO proteins control assembly of actin stress fibers and focal adhesion complexes, RAC proteins regulate accumulation of actin filaments responsible for lamellipodia formation, and CDC42 is involved in formation of actin-containing microspikes called filopodia (Machesky and Hall, 1997; Kaibuchi et al., 1999). In S. cerevisiae, CDC42p and RHO1p regulate polarization of the actin cytoskeleton and control establishment and maintenance of cell polarity (Adams et al., 1990; Johnson and Pringle, 1990; Yamochi et al., 1994). More recently, Rho GTPases were found to influence microtubule dynamics and organization (Wittmann and Waterman-Storer, 2001). Rho GTPases also play important roles in a multitude of other processes (for reviews, see Erickson and Cerione, 2001; Ridley, 2000; Settleman, 2001).
The expression and localization patterns of the 11 AtROP GTPases (Table II) is consistent with their involvement in complex signaling networks in plants (Yang, 2002). Given the conspicuous absence of Ras-family GTPases and other RHO, RAC, and CDC42 subfamily members, it was hypothesized that the ROPs might replace these GTPases in plant signaling (Li et al., 1998; Winge et al., 2000). Overexpression of wild-type ROP GTPase genes and expression of mutants either deficient for GTPase activity (constitutively active; CA) or that can only bind GDP (dominant negative [DN]) have provided evidence for the above hypothesis (for reviews see, Yang, 2002; Zheng and Yang, 2000a).
Mammalian and fungal Rho GTPases regulate the establishment of cell polarity and influence cell morphogenesis (for review, see Arellano et al., 1999). Plant ROP GTPases appear to have retained this function. Three AtROP genes, AtROP1, AtROP3, and AtROP5, are expressed in pollen and may be functionally redundant during pollen tube growth. AtROP1 and AtROP5 are preferentially localized to the plasma membrane in apical regions of the pollen tube and control tip growth (Kost et al., 1999; Li et al., 1999). AtROP2 and AtROP4 localize to tips of elongating root hairs (Molendijk et al., 2001; Jones, 2002). CA mutants of AtROP2, AtROP4, and AtROP6 either caused isotropic growth or increased length in root hairs of Arabidopsis, whereas DN-AtROP2 mutants inhibited root hair elongation, indicating that AtROP GTPases also control tip growth during root hair development (Molendijk et al., 2001; Jones, 2002). In both root hairs and pollen tubes, AtROP GTPases control tip growth by modulating the formation of both the dynamic fine tip F-actin and a tip-focused cytosolic calcium gradient (Li et al., 1999; Fu et al., 2001; Fu and Yang, 2001; Molendijk et al., 2001; Jones, 2002). AtROP GTPases also regulate formation of cell shape by controlling assembly of dynamic cortical F-actin in cells that do not undergo tip-based growth such as epidermal cells (Fu and Yang, 2002).
In mammals, RAC GTPases control production of reactive oxygen species by directly associating with and regulating the activity of plasma membrane-associated NADPH oxidase complexes (for review, see Ridley, 1995). In Arabidopsis, oxygen deprivation rapidly and transiently activates ROP GTPases, resulting in H2O2 accumulation and alcohol dehydrogenase gene expression (Baxter-Burrell et al., 2002). ROP GTPase-dependent H2O2 production is blocked by treatment with DPI, an inhibitor of the plasma membrane NADPH oxidase (Kawasaki et al., 1999; Baxter-Burrell et al., 2002). These results are consistent with the suggestion that plant ROP GTPases may regulate the plasma membrane NADPH oxidase complex.
AtROP GTPases are also involved in signal transduction pathways mediated by the plant hormone, abscisic acid (ABA). ABA promotion of seed dormancy was enhanced and inhibited respectively by DN-Atrop2 and CA-Atrop2 expression in Arabidopsis (Li et al., 2001). Lemichez et al. (2001) showed that AtROP6 was implicated during the negative regulation of stomatal closure, and studies using loss of function mutants have revealed that AtROP9 and AtROP10, two putative ERA1 targets, act as general negative regulators of ABA responses (Zheng et al., 2002). ERA1 encodes a β-subunit of protein farnesyltransferase involved in the negative regulation of ABA responses both in guard cell movement and seed dormancy.
Plant ROP GTPases may also regulate the accumulation of and/or responses to brassinolide and auxin. CA-AtROP2 plants exhibit diverse morphologies that resemble auxin- or brassinolide-overproduction mutants, whereas DN-AtROP2 plants show opposite phenotypes (Li et al., 2001). Readers are referred to several recent reviews for more detailed discussion of the function of the ROP family GTPases in these processes (Valster et al., 2000; Zheng and Yang, 2000a, 2000b; Fu and Yang, 2001; Yang, 2002).
Rop-Interacting Proteins
Several types of Rho GTPase regulators are known in animals: GAPs and GDIs (guanine nucleotide dissociation factors) that act as negative regulators of Rho GTPase function, and GEFs, which activate these GTPases through exchange of GDP for GTP (see introduction and “RAB GTPases” section).
In most fungal and animal RhoGEFs, GDP-GTP exchange activity typically is localized in Dbl-homology domains (Cerione and Zheng, 1996). Surprisingly, plants lack proteins containing obvious Dbl-homology domains. This suggests that plants may have evolved different mechanisms to activate the GDP-bound ROP GTPases. One possible mechanism of activation may be through direct association of ROP GTPases with plant receptor-like kinases (RLKs). A plant ROP GTPase was co-immunoprecipitated with the plant RLK, CLAVATA1 (Trotochaud et al., 1999). Therefore, plant ROP GTPases could be activated by RLKs directly, although the functional significance of ROP-RLK association remains to be determined.
Three RhoGDI homologs are present in Arabidopsis. AtRhoGDI1 is expressed in all tissues examined and has been shown to interact specifically with AtROP GTPases (Bischoff et al., 2000), but so far, no functional data is available. Using the yeast two-hybrid method, Wu et al. (2000) identified a family of Rop-specific, Rho GAPs called RopGAPs. Arabidopsis contains six RopGAPs, all with an N-terminal Cdc42/Rac-interactive binding (CRIB) motif located adjacent to a conserved RhoGAP catalytic domain. In yeast and animals, CRIB domains mediate GTP-dependent interaction between Rho GTPases and their effector proteins, but are not found in conjunction with RhoGAP domains. In plants, presence of the CRIB domain is critical for the Rop-specific regulation of GAP activity (Wu et al., 2000). RopGAP1 localizes in the apical PM region in pollen tube and acts as a negative regulator of AtROP1 function during pollen tube growth (G. Wu and Z. Yang, unpublished data). A role for RopGAP4 in Arabidopsis responses to oxygen deprivation has been revealed using a ropgap4 knockout mutant (Baxter-Burrell et al., 2002).
No obvious homologs of animal and yeast Rho effector proteins, except for phosphatidyl-inositol-phosphate kinases, are present in the Arabidopsis genome. Because the CRIB motif is a hallmark structure of many CDC42/RAC effectors, plant proteins containing these domains make attractive Rop GTPase effector protein candidates. Arabidopsis contains a novel plant-specific family of 11 putative ROP GTPase effector proteins, Rop-interactive CRIB motif-containing proteins (RICs; Wu et al., 2001). RIC1 associates with AtROP1 in a nucleotide-dependent manner, and this association is dependent upon the CRIB domain. Because RIC family members display little sequence similarity with other proteins or even with one another (apart from the CRIB domain), it seems unlikely that the variable regions of RICs function as enzymes to regulate downstream events. RICs could act as adaptor proteins linking Rop GTPases to effector proteins, which in turn would activate specific downstream events. Such a linker function would allow the generation of a greater functional diversity for the Rop GTPase switch and would present a novel mechanism for the activation of G protein effectors (Wu et al., 2001; Yang, 2002).
Arf GTPases
ADP-ribosylation factors (Arfs) were initially identified due to their ability to stimulate the ADP-ribosyltransferase activity of cholera toxin A (Moss and Vaughan, 1998). GTPases that shared significant similarity (40%–60% identity), but that could not activate cholera toxin A or could not rescue S. cerevisiae mutants were termed Arf-like GTPases (Arl GTPases; Clark et al., 1993). Today these two subfamilies are classified together as Arf GTPases (Moss and Vaughan, 1998). The Arabidopsis genome (Arabidopsis Genome Initiative, 2000) contains 21 Arf GTPase family members, with isoforms present in both Arf and Arl GTPase subfamilies (Table III; Fig. 4).
Table III.
Arabidopsis ARF GTPase genes
| Genes Name
|
AGI Genea | Accession No.b | ||
|---|---|---|---|---|
| a | b | c | ||
| AtARFA1a | AtArf1 | AtArf | At1g23490 | 8778579 |
| AtARFA1b | At5g14670 | 7573316 | ||
| AtARFA1c | At2g47170 | 166586 | ||
| AtARFA1d | At1g70490 | 6175141 | ||
| AtARFA1e | At3g62290 | 6899939 | ||
| AtARFA1f | At1g10630 | 6573751 | ||
| AtARFB1a | At2g15310 | 4662630 | ||
| AtARFB1b | At5g17060 | 9755707 | ||
| AtARFB1c | At3g03120 | 6714424 | ||
| AtARFC1 | At3g22950 | 11994726 | ||
| AtARFD1a | At1g02440 | 9972392 | ||
| AtARFD1b | At1g02430 | 9857537 | ||
| AtARLA1a | At5g37680 | 9757978 | ||
| AtARLA1b | At3g49860 | 6723431 | ||
| AtARLA1c | At3g49870 | 6723432 | ||
| AtARLA1d | At5g67560 | 9757877 | ||
| AtARLB1 | At5g52210 | 1184981 | ||
| AtARLC1 | At2g18390 | 4309728 | ||
| AtSARA1a | AtSar1 | At1g09180 | 3249104 | |
| AtSARA1b | At1g56330 | 12321751 | ||
| AtSARA1c | AtSar2 | At4g02080 | 1314860 | |
a, Nomenclature used in this manuscript. b, Nomenclature used in Bischoff et al. (1999). c, Nomenclatures used in Regad et al. (1993).
AGI gene nomenclature.
Genbank protein accession no. (GI:) for a translation of the corresponding gene.
Figure 4.
The Arf GTPase family of Arabidopsis. A neighbor-joining tree of A. thaliana Arf and Arl GTPases including representative Arf and Arl GTPase sequences of S. cerevisiae and H. sapiens was generated using ClustalW and scoring for amino acid differences (Thompson et al., 1994). The tree was rooted with the S. cerevisiae Rab GTPase, Ypt51, and branches with percentage bootstrap values of less than 80% (of 1,000) were collapsed to simplify the tree.
Arf GTPases Recruit Coat Proteins to Transport Vesicles
Arf GTPases play important roles during membrane trafficking steps in eukaryotic cells (for review, see Chavrier and Goud, 1999). In all cases, Arf GTPases act to recruit cytosolic coat proteins to sites of vesicle budding. Three types of protein coats have been described for transport vesicles, COP-I, COP-II, and clathrin coats (Kirchhausen, 2000), and formation of these coats are attributed to two distinct subsets of the Arf GTPase family. Arf GTPases recruit COPI and clathrin protein coats to transport vesicles, whereas a specific subset of the Arf GTPase family, the Sar1p GTPases, recruit COP-II coats.
The best understood mechanism by which Arf GTPases recruit coat proteins to transport vesicles is in S. cerevisiae, where recruitment of COP-II protein coat by Sar1p has been extensively studied (for review, see Kirchhausen, 2000). Sar1p GTPases regulate formation of COP-II coated vesicles carrying cargo from the ER to the cis-Golgi compartment (Wieland and Harter, 1999). In Arabidopsis, three Sar1p homologs have been identified (Fig. 4). Because these GTPases cosegregated with animal and yeast Sar1 GTPases, we have named them AtSARA1a, AtSARA1b, and AtSARA1c, respectively (Fig. 4; Table III). AtSARA1a, previously identified as AtSAR1, was shown to functionally complement mutants in S. cerevisiae (d'Enfert et al., 1992), and AtSARA1a expression was correlated to levels of secretion activity from ER membranes in plant cells, because blockage of transport from the ER resulted in up-regulation of mRNA levels for AtSARA1a (Bar-Peled et al., 1995).
In S. cerevisiae, two Arf GTPases, Arf1p and Arf2p, are functionally interchangeable and act to recruit COP-I coat proteins to transport vesicles during transport of cargo from cis-Golgi compartments back to the ER (Cosson and Letourneur, 1997; Gaynor and Emr, 1997). The yeast Arf1p and Arf2p GTPase functions are distinct from that of Sar1p, which acts in ER-to-Golgi trafficking (Barlowe et al., 1994). In Arabidopsis, 12 Arf GTPase isoforms that cosegregated with members of the Arf GTPase subfamily were identified (Table III; Fig. 4). It should be noted, however, that we also observed mammalian and yeast Arl GTPase sequences within this grouping. Six AtARF GTPases cosegregated with human Arf1 and Arf3 sequences (AtARFA subfamily). This included AtARFA1a, previously identified as AtArf1 (Regad et al., 1993), which has been localized to peripheral Golgi stacks along with Atγ-COP, an Arabidopsis homolog of the COP-I coat protein complex (Ritzenthaler et al., 2002). Six additional Arabidopsis sequences cosegregated with the Arf GTPase subfamily (AtARFB-AtARFD), however, no significance could be ascribed to these groupings outside that of the AtARFA subfamily, which cosegregated with a distinct subset of mammalian Arf GTPases.
Arl GTPases: Regulatory Proteins with Mystery Functions
Six of the 21 Arabidopsis Arf GTPases segregate in groups that contain neither Sar1p nor Arf GTPase subfamily members (Fig. 4). As such, they have been classified as Arl GTPases. Little is known of the function of these GTPases, although at least one member provides essential functions as demonstrated in fruitfly (Tamkun et al., 1991). In plants, mutation of the TITAN5 gene, which corresponds to AtARLC1, results in dramatic alterations of mitosis and cell cycle control during seed development (McElver et al., 2000). The authors speculated that this gene might play a role in membrane trafficking steps necessary for proper cell plate deposition during cytokinesis in developing embryos (McElver et al., 2000). However, much remains to be determined regarding the role(s) of these proteins in any eukaryotic organism.
Arf GTPase-Interacting Proteins
Like other small GTPases, Arf GTPases cycle between an active, membrane-bound form when associated with GTP, and an inactive, predominantly cytosolic form when bound to GDP. Similarly, this GTPase cycle is regulated by GEFs (ArfGEFs) and GAPs (ArfGAPs; Donaldson and Jackson, 2000). However, unlike Rho and Rab GTPases, Arf GTPases do not require GDIs to catalyze their delivery to membranes. Whereas Rab and Rho GTPases are lipid modified at their carboxy terminus, Arf GTPases are myristoylated at their amino terminus. Due to this myristoylation, GDP-bound Arf GTPases display a weak interaction with membranes (Franco et al., 1995). Activation of the membrane-associated Arf GTPase through GEF-catalyzed exchange of GDP for GTP causes conformational changes resulting in stabilization of the Arf GTPase-membrane interaction (Antonny et al., 1997).
Much of the research into Arf GTPase function has focused upon their roles in coat protein recruitment during vesicle formation (see above), and several proteins that interact with Arf GTPases during these processes such as Sec12p and Sec23p, display Sar1p-specific GEF and GAP activities, respectively (Springer et al., 1999). However, Arf GTPases also have other roles such as alteration of membrane lipid compositions, and actin remodeling upon various membrane compartments of the cell (Donaldson, 2000). Distinct ArfGEF or ArfGAP proteins regulate these additional activities.
To date, all ArfGEFs possess a Sec7 domain (Donaldson and Jackson, 2000). This 200-amino acid domain is sufficient to catalyze GDP for GTP exchange on Arf GTPases in vitro (Chardin et al., 1996). Arabidopsis encodes eight proteins containing Sec7 domains. One of these ArfGEFs, GNOM, was identified in mutant screen for Arabidopsis with defective body organization in embryos (Mayer et al., 1991, 1993). Polarized distribution of the auxin-efflux carrier, PIN1, was disrupted in gnom mutants, indicating that this ArfGEF is involved in trafficking of PIN1 proteins during development (Steinmann et al., 1999; Geldner et al., 2001). Little is known of the functions of the other seven Arabidopsis ArfGEF proteins. However, given the central role of the ArfGEF GNOM during organization of the early embryo, this class of proteins is likely to fulfill important roles in growth and development in plants.
In animals, ArfGAPs are a diverse family of multidomain proteins (Donaldson, 2000) that contain a zinc-finger motif and a conserved Arg residue within the ArfGAP catalytic domain (Cukierman et al., 1995; Randazzo et al., 2000). Arabidopsis contains 15 proteins with ArfGAP domains, and we have termed these Arf GAP domain (AGD) proteins. We grouped the AtAGD proteins into four distinct classes based on phylogenetic analysis (data not shown) and overall domain organization (Fig. 5). The first class of AtAGD proteins, consisting of AtAGD1-AtAGD4, is a novel, plant-specific family of putative Arf-GAP proteins. These proteins all contain pleckstrin homology (PH) domains, and two or three ankyrin repeat domains in addition to the AGD (Fig. 5). Presence of PH domains in these AtAGD proteins raises the possibility that they may coordinate their ArfGAP activity with phospholipid signaling events as PH domains bind to phosphoinositides (for review, see Lemmon et al., 2002). In addition, AtAGD1, AtAGD2, and AtAGD3 contain amino-terminal Bin1-amphiphysin-Rvs167p/Rvs161p (BAR) domains. BAR domains are found in adaptor proteins implicated in numerous biological functions including actin regulation and synaptic vesicle endocytosis (Wigge and McMahon, 1998; Balguerie et al., 1999). However, presence of BAR domains in ArfGAP proteins appears to be plant specific. Class 2 AtAGD proteins (AtAGD5–AtAFGD10; Fig. 5) contain AGDs at the amino terminus and no other discernible protein domains. These proteins show similarity both at sequence level and in overall domain organization to Golgi-localized ArfGAPs in mammals and in yeast (Cukierman et al., 1995; Poon et al., 1999). Class 3 AtAGD proteins (AtAGD11–AtAGD13; Fig. 5) are distinguished by the presence of a centrally located C2 domain. C2 domains bind a range of ligands, such as phospholipids, phosphoinositides, and other proteins, in calcium-dependent fashion (Sutton et al., 1995; Essen et al., 1996; Shao et al., 1997). In Class 4 AtAGD proteins (AtAGD14 and AtAGD15; Fig. 5), the AGD comprises almost the entire open reading frames of these proteins. In AtAGD14, the AGD is followed by an apparent membrane-spanning α-helix, suggesting this ArfGAP could be an integral membrane protein. As in mammals, plant ArfGAPs have diversified into a large family of proteins. Several classes of the AtAGD proteins contain additional domains, and these may act to coordinate the timing of ArfGAP activity within the cell or may serve functions beyond simply acting as negative regulators of Arf GTPases. Understanding the mechanisms by which temporal and spatial activities of plant ArfGAPs are regulated will be an interesting area for future investigation.
Figure 5.
ArfGAP proteins of Arabidopsis. AtAGD proteins were identified using BLASTP (Altschul et al., 1997) with the AGD of ASAP1 (GenBank accession no. NP_060952). Domains within the AtAGD sequences were detected using SMART (Letunic et al., 2002). AGI numbers of AtAGD1 to AtAGD15 are as follows: At5g61980 (AtAGD1), At1g60680 (AtAGD2), At4g13300 (AtAGD3), At1g10870 (AtAGD4), At5g54310 (AtAGD5), At3g53710 (AtAGD6), At2g37550 (AtAGD7), At4g17890 (AtAGD8), At5g46750 (AtAGD9), At2g35210 (AtAGD10), At3g07490 (AtAGD11), At4g21160 (AtAGD12), At4g05330 (AtAGD13), At1g08680 (AtAGD14), and At3g17660 (AtAGD15).
In yeast and animals, increasing attention is being paid to the roles of Arf GTPases in lipid modification. Mammalian and yeast Arf GTPases activate phosphatidylinositol 4-phosphate, 5-kinases (Honda et al., 1999; Walch-Solimena and Novick, 1999). Arabidopsis also contains phosphatidylinositol 4-phosphate, 5-kinase proteins, but whether AtARF GTPases interact with these remains unknown. A new class of Arf GTPase effector proteins that facilitate membrane trafficking at the TGN has recently been described. These Golgi-localizing, γ-adaptin ear homology, Arf-binding (GGA) proteins contain multiple domains: an amino-terminal VHS (Vps27, Hrs, and STAM) domain, followed by the GAT (GGA1 and TOM proteins) region and the γ-adaptin homology domain at the carboxy terminus. The VHS domain is thought to mediate interactions with cargo molecules, whereas the GAT domain is responsible for association with Arf GTPases, and the γ-adaptin homology domain interacts with clathrin (for review, see Boman, 2001). Five GGA-like proteins are present in the Arabidopsis genome. Intriguingly, although these proteins contain the amino-terminal VHS and GAT domains, they appear to lack the carboxy-terminal γ-adaptin homology domain. Given the apparent central role of GGA proteins during sorting of cargo proteins and recruitment of vesicle coat proteins, it will be interesting to determine the role of this class of proteins in plant Arf GTPase functions.
Ran GTPases
In animals, Ran GTPases together with their regulatory factors, the Ran-binding proteins (RanBPs), RCC1 (a GEF, RanGEF) and RanGAP (a GAP), play key roles in controlling nuclear processes throughout the cell mitotic cycle (for review, see Clarke and Zhang, 2001). Ran GTPases, like other small GTPases, cycle between GDP- and GTP-bound states. However, for Ran GTPases, GTP binding and hydrolysis is linked to transport into or out of the nucleus (Moore, 1998). Also, unlike other small GTPases, Ran GTPases are not posttranslationally lipid modified and do not associate with cellular membranes (Rush et al., 1996). Four Ran GTPases are present in Arabidopsis (Table IV). AtRAN1, AtRAN2, and AtRAN3 were identified by sequence similarity (Haizel et al., 1997; see below). The fourth gene, AtRAN4, is annotated as “salt stress-inducible small GTP-binding protein Ran1-like protein,” but so far no data has been published.
At the protein level, AtRAN1, AtRAN2, and AtRAN3 are nearly identical (95%–96% of identity) differing only in their C-terminal regions, whereas AtRAN4 is more divergent with only 65% identity to the other AtRAN sequences. All AtRan GTPases contain sequence motifs involved in GTP binding/hydrolysis and an effector-binding domain for interaction with RanGAPs. This effector-binding motif is 100% identical in AtRAN1 to AtRAN3 and in tomato and tobacco Ran GTPases (KKYEPTIGVEV) but diverges strikingly in AtRAN4, with only five residues of 11 conserved. Moreover, although other plant Ran GTPases possess conserved C-terminal acidic domains (DDDDD/E), this sequence is absent in AtRAN4. In animals, this acidic domain is necessary for interaction with Ran-binding proteins (Haizel et al., 1997; see below). These observations suggest that AtRAN1 to AtRAN3 are likely Ran orthologs involved in nucleocytoplasmic transport (see below), whereas AtRAN4 may have distinct functions in Arabidopsis.
Ran GTPases Establish Nuclear Compartment Identity
In interphase cells, Ran GTPases direct nucleocytoplasmic transport. RanGAP and RanBP2 (which stimulate the intrinsic GTPase activity of the Ran GTPase) are cytoplasmic proteins, whereas RCC1 (which generates Ran:GTP) is confined to the nucleus. These distributions ensure that Ran GTPases are bound with GTP in the nucleus and GDP in the cytoplasm. The restriction of Ran:GTP to the nuclear interior has been proposed to be crucial for establishing directional transport of proteins and RNA through the nuclear pore (Moore and Blobel, 1993; Izaurralde et al., 1997). Ran GTPases also play important roles during cellular mitosis (Kalab et al., 1999; Wilde and Zheng, 1999). Association of RCC1 with chromatin results in localized generation of Ran:GTP in the vicinity of chromosomes, which in turn promotes microtubule spindle assembly (Carazo-Salas et al., 1999; Ohba et al., 1999; Wiese et al., 2001). At the end of mitosis, the cycling of Ran:GDP and Ran:GTP induces nuclear envelope reassembly, probably by controlling vesicle binding and fusion (Hetzer et al., 2000; Zhang and Clarke, 2000).
Functions of Arabidopsis Ran GTPases and Associated Proteins
AtRAN1 to AtRAN3 are ubiquitously expressed during development, with higher levels in meristematic tissues and developing embryos (Haizel et al., 1997). Overexpression of plant Ran GTPases suppressed cell cycle defects in mutant fission yeast, indicating that they functioned similarly to their yeast homologs (Ach and Gruissem, 1994; Merkle et al., 1994; Haizel et al., 1997). In animals, Exportin-1 is the nuclear export receptor for proteins carrying nuclear export signals (NES). Exportin-1 binds NES-containing proteins and Ran GTPases cooperatively, and this triple complex undergoes nuclear export (Fornerod et al., 1997). In Arabidopsis, AtRAN1 interacts with AtXPO1, an Arabidopsis Exportin-1 homolog, and a NES-containing protein, AtRanBP1a (Haizel et al., 1997; Haasen et al., 1999). This suggests the nuclear export machinery may be functionally conserved in plants (Haasen et al., 1999). Antisense expression of an additional AtRanBP isoform in Arabidopsis, AtRanBP1c, enhanced primary root growth, suppressed lateral root growth and rendered transgenic roots hypersensitive to auxin (Kim et al., 2001). The authors proposed a model where AtRanBP1c plays key roles in delivery of nuclear proteins responsible for suppression of auxin action and regulation mitosis in root tips. However, direct evidence is still required to clarify the role(s) of Ran GTPases and RanBPs in plant growth and development.
The mammalian RanGEF, RCC1, contains seven tandem repeats of a 50- to 60-amino acid domain, and these repeats constitute the majority of the protein. Arabidopsis contains 18 proteins containing one to five RCC1 domains (see supplemental data). One of these, uvr8, is a mutant hypersensitive to UV-B (Kliebenstein et al., 2002). The UVR8 protein contains five RCC1 repeats but interestingly does not posses nuclear localization sequences conserved in animal and yeast RCC1 homologs (Kliebenstein et al., 2002). Further work is necessary to determine whether UVR8, or other Arabidopsis proteins with RCC1 domains act as RanGEFs.
Two RanGAP sequences have been identified in Arabidopsis: AtRanGAP1 (accession no. AF2214559) and AtRanGAP2 (accession no. AF214560). Both AtRanGAPs complemented yeast RanGAP mutants (Pay et al., 2002), suggesting that these proteins are functional orthologs of the yeast RanGAP. AtRanGAP-GFP fusions associate with the nuclear envelope, and this localization is dependent upon a unique N-terminal domain (Rose and Meier, 2001). AtRanGAP1 localization is consistent with a role for AtRAN GTPases in nucleocytoplasmic transport. However, whether plant Ran GTPases function to regulate these transport steps or different plant Ran GTPases, especially the divergent AtRAN4, have distinct functions remains undetermined.
CONCLUSIONS
From these studies and observations, it is clear that small GTPases represent an important and diverse set of regulatory molecules in plants, with Arabidopsis containing members of the Rab, Rho, Arf, and Ran classes of small GTPases. In many cases, plant small GTPases appear to maintain similar functions as their animal and yeast counterparts. The identification of additional families of putative GAP and GEF proteins for most of these families further highlights the highly conserved nature of these regulatory molecules throughout evolution of eukaryotes. However, plants contain some unique variations on this conserved regulatory mechanism.
Although small GTPases related to Rab, Rho, Arf and Ran GTPases were identified in Arabidopsis, Ras GTPases were not detected. This, in principal, concurs with the evolution of eukaryotic regulatory systems. The notable absence of plant Ras GTPases correlates with lack of Tyr kinase receptors, which act upstream of Ras signaling in animals. Plant Rop GTPases segregate as a distinct subfamily within the Rho GTPase family. While maintaining conserved functions in regulation of actin cytoskeleton and cellular signaling pathways, newly established roles in ABA-signaling events and interactions with plant RLKs highlight potential plant-specific variations of the functions of these Rop GTPases. Plant Rab GTPases segregate into distinct subfamilies that appear to be organized around the types of compartments upon which they are localized. Increased numbers of Arabidopsis Rab GTPases in subfamilies with predicted functions in vacuolar and post-Golgi trafficking may indicate plant-specific elaborations of these pathways. Plant Arf GTPases and Ran GTPases functionally complement mutations of their respective counterparts in yeast. However, plants contain multiple, novel ArfGEFs and ArfGAPs, probably with novel functions. Also, AtRAN4 is significantly different in sequences conserved throughout Ran GTPases from plants, yeast, and animals, perhaps indicating novel roles during plant growth and development. A wealth of knowledge of the mechanisms of action of small GTPases and of some of their important activator proteins (GEFs) and inhibitor proteins (GAPs) will clearly serve as a strong basis for understanding how these conserved regulatory mechanisms have been modified to carry out plant-specific processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Warren Lathe III for initial help with phylogenetic analysis of the small GTPases and Jannie Santos-Serna for help with the tables and figures. We also thank Tony Sanderfoot, Daniel Schachtman, and Christiane Wobus for critical reading of the manuscript, and Ian Moore for helpful comments regarding Rab GTPase nomenclature.
Footnotes
This work was supported by the National Aeronautics and Space Administration (grant no. NAG2–1525 to E.N.), by the U.S. Department of Agriculture (grant no. 2000–01586), and by the Department of Energy (grant no. DE–FG03–00ER15060 to Z.Y.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.013052.
LITERATURE CITED
- Ach RA, Gruissem W. A small nuclear GTP-binding protein from tomato suppresses a Schizosaccharomyces pombe cell-cycle mutant. Proc Natl Acad Sci USA. 1994;91:5863–5867. doi: 10.1073/pnas.91.13.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AE, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert S, Will E, Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999;18:5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov K, Horiuchi H, Steele-Mortimer O, Seabra MC, Zerial M. Rab escort protein-1 is a multifunctional protein that accompanies newly prenylated rab proteins to their target membranes. EMBO J. 1994;13:5262–5273. doi: 10.1002/j.1460-2075.1994.tb06860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B, Beraud-Dufour S, Chardin P, Chabre M. N-Terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Arellano M, Coll PM, Perez P. RHO GTPases in the control of cell morphology, cell polarity, and actin localization in fission yeast. Microsc Res Tech. 1999;47:51–60. doi: 10.1002/(SICI)1097-0029(19991001)47:1<51::AID-JEMT5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Balguerie A, Sivadon P, Bonneu M, Aigle M. Rvs167p, the budding yeast homolog of amphiphysin, colocalizes with actin patches. J Cell Sci. 1999;112:2529–2537. doi: 10.1242/jcs.112.15.2529. [DOI] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Bar-Peled U, Maltz E, Bruckental I, Folman Y, Kali Y, Gacitua H, Lehrer AR, Knight CH, Robinzon B, Voet H et al. Relationship between frequent milking or suckling in early lactation and milk production of high producing dairy cows. J Dairy Sci. 1995;78:2726–2736. doi: 10.3168/jds.s0022-0302(95)76903-x. [DOI] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I. A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell. 2000;12:2201–2218. doi: 10.1105/tpc.12.11.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter-Burrell A, Yang Z, Springer PS, Bailey-Serres J. RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science. 2002;296:2026–2028. doi: 10.1126/science.1071505. [DOI] [PubMed] [Google Scholar]
- Bednarek SY, Reynolds TL, Schroeder M, Grabowski R, Hengst L, Gallwitz D, Raikhel NV. A small GTP-binding protein from Arabidopsis thaliana functionally complements the yeast YPT6 null mutant. Plant Physiol. 1994;104:591–596. doi: 10.1104/pp.104.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benli M, Doring F, Robinson DG, Yang X, Gallwitz D. Two GTPase isoforms, Ypt31p and Ypt32p, are essential for Golgi function in yeast. EMBO J. 1996;15:6460–6475. [PMC free article] [PubMed] [Google Scholar]
- Bischoff F, Vahlkamp L, Molendijk A, Palme K. Localization of AtROP4 and AtROP6 and interaction with the guanine nucleotide dissociation inhibitor AtRhoGDI1 from Arabidopsis. Plant Mol Biol. 2000;42:515–530. doi: 10.1023/a:1006341210147. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Martin GB. AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato. Proc Natl Acad Sci USA. 2000;97:8836–8840. doi: 10.1073/pnas.97.16.8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Bolte S, Schiene K, Dietz KJ. Characterization of a small GTP-binding protein of the rab 5 family in Mesembryanthemum crystallinum with increased level of expression during early salt stress. Plant Mol Biol. 2000;42:923–936. doi: 10.1023/a:1006449715236. [DOI] [PubMed] [Google Scholar]
- Boman AL. GGA proteins: new players in the sorting game. J Cell Sci. 2001;114:3413–3418. doi: 10.1242/jcs.114.19.3413. [DOI] [PubMed] [Google Scholar]
- Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- Calhoun BC, Lapierre LA, Chew CS, Goldenring JR. Rab11a redistributes to apical secretory canaliculus during stimulation of gastric parietal cells. Am J Physiol. 1998;275:C163–C170. doi: 10.1152/ajpcell.1998.275.1.C163. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Cerione RA, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- Chant J, Stowers L. GTPase cascades choreographing cellular behavior: movement, morphogenesis, and more. Cell. 1995;81:1–4. doi: 10.1016/0092-8674(95)90363-1. [DOI] [PubMed] [Google Scholar]
- Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson CL, Chabre M. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature. 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- Chavrier P, Goud B. The role of ARF and Rab GTPases in membrane transport. Curr Opin Cell Biol. 1999;11:466–475. doi: 10.1016/S0955-0674(99)80067-2. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Chen CY, Glaven RH, de Graaf BH, Vidali L, Hepler PK, Wu HM. Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell. 2002;14:945–962. doi: 10.1105/tpc.000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- Clark J, Moore L, Krasinskas A, Way J, Battey J, Tamkun J, Kahn RA. Selective amplification of additional members of the ADP-ribosylation factor (ARF) family: cloning of additional human and Drosophila ARF-like genes. Proc Natl Acad Sci USA. 1993;90:8952–8956. doi: 10.1073/pnas.90.19.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke PR, Zhang C. Ran GTPase: a master regulator of nuclear structure and function during the eukaryotic cell division cycle? Trends Cell Biol. 2001;11:366–371. doi: 10.1016/s0962-8924(01)02071-2. [DOI] [PubMed] [Google Scholar]
- Collins CC, Johnson DI. An Arabidopsis thaliana expressed sequence tag cDNA that encodes a Rac-like protein (accession no. U88402) Plant Physiol. 1997;113:1463. [Google Scholar]
- Cosson P, Letourneur F. Coatomer (COPI)-coated vesicles: role in intracellular transport and protein sorting. Curr Opin Cell Biol. 1997;9:484–487. doi: 10.1016/s0955-0674(97)80023-3. [DOI] [PubMed] [Google Scholar]
- Cuif MH, Possmayer F, Zander H, Bordes N, Jollivet F, Couedel-Courteille A, Janoueix-Lerosey I, Langsley G, Bornens M, Goud B. Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J. 1999;18:1772–1782. doi: 10.1093/emboj/18.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E, Huber I, Rotman M, Cassel D. The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- d'Enfert C, Gensse M, Gaillardin C. Fission yeast and a plant have functional homologues of the Sar1 and Sec12 proteins involved in ER to Golgi traffic in budding yeast. EMBO J. 1992;11:4205–4211. doi: 10.1002/j.1460-2075.1992.tb05514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG. Filling in the GAPs in the ADP-ribosylation factor story. Proc Natl Acad Sci USA. 2000;97:3792–3794. doi: 10.1073/pnas.97.8.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. Regulators and effectors of the ARF GTPases. Curr Opin Cell Biol. 2000;12:475–482. doi: 10.1016/s0955-0674(00)00119-8. [DOI] [PubMed] [Google Scholar]
- Duman JG, Tyagarajan K, Kolsi MS, Moore HP, Forte JG. Expression of rab11a N124I in gastric parietal cells inhibits stimulatory recruitment of the H+-K+-ATPase. Am J Physiol. 1999;277:C361–C372. doi: 10.1152/ajpcell.1999.277.3.C361. [DOI] [PubMed] [Google Scholar]
- Erickson JW, Cerione RA. Multiple roles for Cdc42 in cell regulation. Curr Opin Cell Biol. 2001;13:153–157. doi: 10.1016/s0955-0674(00)00192-7. [DOI] [PubMed] [Google Scholar]
- Essen LO, Perisic O, Cheung R, Katan M, Williams RL. Crystal structure of a mammalian phosphoinositide-specific phospholipase C delta. Nature. 1996;380:595–602. doi: 10.1038/380595a0. [DOI] [PubMed] [Google Scholar]
- Feng Y, Press B, Wandinger-Ness A. Rab 7: an important regulator of late endocytic membrane traffic. J Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Franco M, Chardin P, Chabre M, Paris S. Myristoylation of ADP-ribosylation factor 1 facilitates nucleotide exchange at physiological Mg2+ levels. J Biol Chem. 1995;270:1337–1341. doi: 10.1074/jbc.270.3.1337. [DOI] [PubMed] [Google Scholar]
- Fu Y, Wu G, Yang Z. Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J Cell Biol. 2001;152:1019–1032. doi: 10.1083/jcb.152.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Yang Z. Rop GTPase: a master switch of cell polarity development in plants. Trends Plant Sci. 2001;6:545–547. doi: 10.1016/s1360-1385(01)02130-6. [DOI] [PubMed] [Google Scholar]
- Fu Y, Yang Z. Rop GTPase controls plant cell morphogenesis via a novel form of cortical F-actin. Plant Cell. 2002;14:763–776. [Google Scholar]
- Gaynor EC, Emr SD. COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol. 1997;136:789–802. doi: 10.1083/jcb.136.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jurgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- Goud B, Salminen A, Walworth NC, Novick PJ. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- Haasen D, Kohler C, Neuhaus G, Merkle T. Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 1999;20:695–705. doi: 10.1046/j.1365-313x.1999.00644.x. [DOI] [PubMed] [Google Scholar]
- Haizel T, Merkle T, Pay A, Fejes E, Nagy F. Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J. 1997;11:93–103. doi: 10.1046/j.1365-313x.1997.11010093.x. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hama H, Tall GG, Horazdovsky BF. Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J Biol Chem. 1999;274:15284–15291. doi: 10.1074/jbc.274.21.15284. [DOI] [PubMed] [Google Scholar]
- Heras B, Drobak BK. PARF-1: an Arabidopsis thaliana FYVE-domain protein displaying a novel eukaryotic domain structure and phosphoinositide affinity. J Exp Bot. 2002;53:565–567. doi: 10.1093/jexbot/53.368.565. [DOI] [PubMed] [Google Scholar]
- Hetzer M, Bilbao-Cortes D, Walther TC, Gruss OJ, Mattaj IW. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol Cell. 2000;5:1013–1024. doi: 10.1016/s1097-2765(00)80266-x. [DOI] [PubMed] [Google Scholar]
- Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman MA et al. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- Horazdovsky BF, Busch GR, Emr SD. VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J. 1994;13:1297–1309. doi: 10.1002/j.1460-2075.1994.tb06382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi H, Lippé R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- Huber LA, de Hoop MJ, Dupree P, Zerial M, Simons K, Dotti C. Protein transport to the dendritic plasma membrane of cultured neurons is regulated by rab8p. J Cell Biol. 1993a;123:47–55. doi: 10.1083/jcb.123.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber LA, Pimplikar S, Parton RG, Virta H, Zerial M, Simons K. Rab8, a small GTPase involved in vesicular traffic between the TGN and the basolateral plasma membrane. J Cell Biol. 1993b;123:35–45. doi: 10.1083/jcb.123.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T, Nagano Y, Nagasaki T, Sasaki Y. Distinct localization of two closely related Ypt3/Rab11 proteins on the trafficking pathway in higher plants. J Biol Chem. 2002;277:9183–9188. doi: 10.1074/jbc.M111491200. [DOI] [PubMed] [Google Scholar]
- Iwasaki K, Toyonaga R. The Rab3 GDP/GTP exchange factor homolog AEX-3 has a dual function in synaptic transmission. EMBO J. 2000;19:4806–4816. doi: 10.1093/emboj/19.17.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Jarmolowski A, Beisel C, Mattaj IW, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G, Mulholland J, Segev N. Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. J Cell Biol. 1997;137:563–580. doi: 10.1083/jcb.137.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, La Cour T, Albrethsen J, Nielsen M, Skriver K. FYVE zinc-finger proteins in the plant model Arabidopsis thaliana: identification of PtdIns3P-binding residues by comparison of classic and variant FYVE domains. Biochem J. 2001;1:165–173. doi: 10.1042/0264-6021:3590165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI, Pringle JR. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA. The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell. 2002;14:777–794. doi: 10.1105/tpc.010359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Der CJ, Bokoch GM. The ras superfamily of GTP-binding proteins: guidelines on nomenclature. FASEB J. 1992;6:2512–2513. doi: 10.1096/fasebj.6.8.1592203. [DOI] [PubMed] [Google Scholar]
- Kaibuchi K, Kuroda S, Amano M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu Rev Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pu RT, Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kang JG, Yun J, Kim DH, Chung KS, Fujioka S, Kim JI, Dae HW, Yoshida S, Takatsuto S, Song PS et al. Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell. 2001;105:625–636. doi: 10.1016/s0092-8674(01)00370-1. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K. The small GTP-binding protein rac is a regulator of cell death in plants. Proc Natl Acad Sci USA. 1999;96:10922–10926. doi: 10.1073/pnas.96.19.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Arnold D, Lloyd A, Roux SJ. Antisense expression of an Arabidopsis ran binding protein renders transgenic roots hypersensitive to auxin and alters auxin-induced root growth and development by arresting mitotic progress. Plant Cell. 2001;13:2619–2630. doi: 10.1105/tpc.010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Kim CY, Cheong NE, Choi YO, Lee KO, Lee SH, Park JB, Nakano A, Bahk JD, Cho MJ et al. Characterization of two fungal-elicitor-induced rice cDNAs encoding functional homologues of the rab-specific GDP-dissociation inhibitor. Planta. 1999;210:143–149. doi: 10.1007/s004250050663. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Three ways to make a vesicle. Nat Rev Mol Cell Biol. 2000;1:187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 2002;130:234–243. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, Chua NH. Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J Cell Biol. 1999;145:317–330. doi: 10.1083/jcb.145.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetti L, Rybin V, Malabarba MG, Christoforidis S, Scita G, Zerial M, Di Fiore PP. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature. 2000;408:374–377. doi: 10.1038/35042605. [DOI] [PubMed] [Google Scholar]
- Lazar T, Gotte M, Gallwitz D. Vesicular transport: How many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem Sci. 1997;22:468–472. doi: 10.1016/s0968-0004(97)01150-x. [DOI] [PubMed] [Google Scholar]
- Lemichez E, Wu Y, Sanchez JP, Mettouchi A, Mathur J, Chua NH. Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 2001;15:1808–1816. doi: 10.1101/gad.900401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Abrams CS. Pleckstrin homology domains and the cytoskeleton. FEBS Lett. 2002;513:71–76. doi: 10.1016/s0014-5793(01)03243-4. [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lin Y, Heath RM, Zhu MX, Yang Z. Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell. 1999;11:1731–1742. doi: 10.1105/tpc.11.9.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Shen JJ, Zheng ZL, Lin Y, Yang Z. The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol. 2001;126:670–684. doi: 10.1104/pp.126.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wu G, Ware D, Davis KR, Yang Z. Arabidopsis Rho-related GTPases: differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 1998;118:407–417. doi: 10.1104/pp.118.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Zainal Z, Tucker GA, Lycett GW. Developmental abnormalities and reduced fruit softening in tomato plants expressing an antisense Rab11 GTPase gene. Plant Cell. 2001;13:1819–1833. doi: 10.1105/TPC.010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MA, Staehelin LA. Domain-specific and cell type-specific localization of two types of cell wall matrix polysaccharides in the clover root tip. J Cell Biol. 1992;118:467–479. doi: 10.1083/jcb.118.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Hall A. Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. J Cell Biol. 1997;138:913–926. doi: 10.1083/jcb.138.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez O, Antony C, Pehau-Arnaudet G, Berger EG, Salamero J, Goud B. GTP-bound forms of rab6 induce the redistribution of Golgi proteins into the endoplasmic reticulum. Proc Natl Acad Sci USA. 1997;94:1828–1833. doi: 10.1073/pnas.94.5.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Büttner G, Jürgens G. Apical-basal formation in the Arabidopsis embyro: studies on the role of the gnom gene. Development. 1993;117:149–162. [Google Scholar]
- Mayer U, Torrez-Ruiz RA, Berleth T, Misera S, Jürgens G. Mutations affecting body organization in the Arabidopsis embryo. Nature. 1991;353:402–407. [Google Scholar]
- McElver J, Patton D, Rumbaugh M, Liu C, Yang LJ, Meinke D. The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell. 2000;12:1379–1392. doi: 10.1105/tpc.12.8.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle T, Haizel T, Matsumoto T, Harter K, Dallmann G, Nagy F. Phenotype of the fission yeast cell cycle regulatory mutant pim1–46 is suppressed by a tobacco cDNA encoding a small, Ran-like GTP-binding protein. Plant J. 1994;6:555–565. doi: 10.1046/j.1365-313x.1994.6040555.x. [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM. Plants, animals and the logic of development. Trends Cell Biol. 1999;9:M65–M68. [PubMed] [Google Scholar]
- Meyerowitz EM. Plants compared to animals: the broadest comparative study of development. Science. 2002;295:1482–1485. doi: 10.1126/science.1066609. [DOI] [PubMed] [Google Scholar]
- Molendijk AJ, Bischoff F, Rajendrakumar CS, Friml J, Braun M, Gilroy S, Palme K. Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 2001;20:2779–2788. doi: 10.1093/emboj/20.11.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moss J, Vaughan M. Molecules in the ARF orbit. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Funato K, Stahl PD. Rab7 regulates transport from early to late endocytic compartments in Xenopus oocytes. J Biol Chem. 1997;272:13055–13059. doi: 10.1074/jbc.272.20.13055. [DOI] [PubMed] [Google Scholar]
- Nagano F, Sasaki T, Fukui K, Asakura T, Imazumi K, Takai Y. Molecular cloning and characterization of the noncatalytic subunit of the Rab3 subfamily-specific GTPase-activating protein. J Biol Chem. 1998;273:24781–24785. doi: 10.1074/jbc.273.38.24781. [DOI] [PubMed] [Google Scholar]
- Nagano Y, Okada Y, Narita H, Asaka Y, Sasaki Y. Location of light-repressible, small GTP-binding protein of the YPT/rab family in the growing zone of etiolated pea stems. Proc Natl Acad Sci USA. 1995;92:6314–6318. doi: 10.1073/pnas.92.14.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C, Davidson HW, Matteson J, Meinkoth J, Balch WE. A GDP-bound of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J Cell Biol. 1994;125:225–237. doi: 10.1083/jcb.125.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Paris N, Stanley CM, Jones RL, Rogers JC. Plant cells contain two functionally distinct vacuolar compartments. Cell. 1996;85:563–572. doi: 10.1016/s0092-8674(00)81256-8. [DOI] [PubMed] [Google Scholar]
- Pay A, Resch K, Frohnmeyer H, Fejes E, Nagy F, Nick P. Plant RanGAPs are localized at the nuclear envelope in interphase and associated with microtubules in mitotic cells. Plant J. 2002;30:699–709. doi: 10.1046/j.1365-313x.2002.01324.x. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol. 2000;301:1077–1087. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- Poon PP, Cassel D, Spang A, Rotman M, Pick E, Singer RA, Johnston GC. Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 1999;18:555–564. doi: 10.1093/emboj/18.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A, Seals D, Wickner W, Ungermann C. The docking stage of yeast vacuole fusion requires the transfer of proteins from a cis-SNARE complex to a Rab/Ypt protein. J Cell Biol. 2000;148:1231–1238. doi: 10.1083/jcb.148.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo PA, Andrade J, Miura K, Brown MT, Long YQ, Stauffer S, Roller P, Cooper JA. The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc Natl Acad Sci USA. 2000;97:4011–4016. doi: 10.1073/pnas.070552297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad F, Bardet C, Tremousaygue D, Moisan A, Lescure B, Axelos M. cDNA cloning and expression of an Arabidopsis GTP-binding protein of the ARF family. FEBS Lett. 1993;316:133–136. doi: 10.1016/0014-5793(93)81201-a. [DOI] [PubMed] [Google Scholar]
- Ridley A. Rho. In: Hall A, editor. GTPases. Oxford: Oxford University Press; 2000. pp. 89–136. [Google Scholar]
- Ridley AJ. Intracellular regulation: Rac and Bcr regulate phagocytic phoxes. Curr Biol. 1995;5:710–712. doi: 10.1016/s0960-9822(95)00140-0. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler C, Nebenfuhr A, Movafeghi A, Stussi-Garaud C, Behnia L, Pimpl P, Staehelin LA, Robinson DG. Reevaluation of the effects of brefeldin A on plant cells using tobacco bright yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell. 2002;14:237–261. doi: 10.1105/tpc.010237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A, Meier I. A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci USA. 2001;98:15377–15382. doi: 10.1073/pnas.261459698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush MG, Drivas G, D'Eustachio P. The small nuclear GTPase Ran: How much does it run? Bioessays. 1996;18:103–112. doi: 10.1002/bies.950180206. [DOI] [PubMed] [Google Scholar]
- Salminen A, Novick PJ. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- Segev N, Mulholland J, Botstein D. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell. 1988;52:915–924. doi: 10.1016/0092-8674(88)90433-3. [DOI] [PubMed] [Google Scholar]
- Settleman J. Rac 'n Rho: the music that shapes a developing embryo. Dev Cell. 2001;1:321–331. doi: 10.1016/s1534-5807(01)00053-3. [DOI] [PubMed] [Google Scholar]
- Shao X, Li C, Fernandez I, Zhang X, Sudhof TC, Rizo J. Synaptotagmin-syntaxin interaction: the C2 domain as a Ca2+-dependent electrostatic switch. Neuron. 1997;18:133–142. doi: 10.1016/s0896-6273(01)80052-0. [DOI] [PubMed] [Google Scholar]
- Simons K, Zerial M. Rab proteins and the road maps for intracellular transport. Neuron. 1993;11:789–799. doi: 10.1016/0896-6273(93)90109-5. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3) K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Singer-Krüger B, Stenmark H, Zerial M. Yeast Ypt51p and mammalian Rab5: counterparts with similar function in the early endocytic pathway. J Cell Sci. 1995;108:3509–3521. doi: 10.1242/jcs.108.11.3509. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Peak-Chew SY, Pelham HR. Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J. 2000;19:4885–4894. doi: 10.1093/emboj/19.18.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer S, Spang A, Schekman R. A primer on vesicle budding. Cell. 1999;97:145–148. doi: 10.1016/s0092-8674(00)80722-9. [DOI] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Galweiler L, Palme K, Jurgens G. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;286:316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2:Reviews 3007.1–3007.7. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M, Vollmer P, Tan TJ, Gallwitz D. A yeast GTPase-activating protein that interacts specifically with a member of the Ypt/Rab family. Nature. 1993;361:736–739. doi: 10.1038/361736a0. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Davletov BA, Berghuis AM, Sudhof TC, Sprang SR. Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- Swofford DL, Waddell PJ, Huelsenbeck JP, Foster PG, Lewis PO, Rogers JS. Bias in phylogenetic estimation and its relevance to the choice between parsimony and likelihood methods. Syst Biol. 2001;50:525–539. [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Kahn RA, Kissinger M, Brizuela BJ, Rulka C, Scott MP, Kennison JA. The arflike gene encodes an essential GTP-binding protein in Drosophila. Proc Natl Acad Sci USA. 1991;88:3120–3124. doi: 10.1073/pnas.88.8.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, Balch WE. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol. 1992;119:749–761. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell. 1999;11:393–406. doi: 10.1105/tpc.11.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Matsuda N, Anai T, Tsukaya H, Uchimiya H, Nakano A. An Arabidopsis gene isolated by a novel method for detecting genetic interaction in yeast encodes the GDP dissociation inhibitor of Ara4 GTPase. Plant Cell. 1996;8:2079–2091. doi: 10.1105/tpc.8.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A. Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 2001;20:4730–4741. doi: 10.1093/emboj/20.17.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Yoshizumi T, Anai T, Matsui M, Uchimiya H, Nakano A. AtGDI2, a novel Arabidopsis gene encoding a Rab GDP dissociation inhibitor. Gene. 1998;206:137–143. doi: 10.1016/s0378-1119(97)00584-2. [DOI] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valster AH, Hepler PK, Chernoff J. Plant GTPases: the Rhos in bloom. Trends Cell Biol. 2000;10:141–146. doi: 10.1016/s0962-8924(00)01728-1. [DOI] [PubMed] [Google Scholar]
- Vitale A, Raikhel NV. What do proteins need to reach different vacuoles? Trends Plant Sci. 1999;4:149–155. doi: 10.1016/s1360-1385(99)01389-8. [DOI] [PubMed] [Google Scholar]
- Vollmer P, Gallwitz D. High expression cloning, purification, and assay of Ypt-GTPase-activating proteins. Methods Enzymol. 1995;257:118–128. doi: 10.1016/s0076-6879(95)57017-9. [DOI] [PubMed] [Google Scholar]
- Vollmer P, Will E, Scheglmann D, Strom M, Gallwitz D. Primary structure and biochemical characterization of yeast GTPase-activating proteins with substrate preference for the transport GTPase Ypt7p. Eur J Biochem. 1999;260:284–290. doi: 10.1046/j.1432-1327.1999.00192.x. [DOI] [PubMed] [Google Scholar]
- Wada M, Nakanishi H, Satoh A, Hirano H, Obaishi H, Matsuura Y, Takai Y. Isolation and characterization of a GDP/GTP exchange protein specific for the Rab3 subfamily small G proteins. J Biol Chem. 1997;272:3875–3878. doi: 10.1074/jbc.272.7.3875. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C, Collins RN, Novick PJ. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- White J, Johannes L, Mallard F, Girod A, Grill S, Reinsch S, Keller P, Tzschaschel B, Echard A, Goud B et al. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J Cell Biol. 1999;147:743–760. doi: 10.1083/jcb.147.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland F, Harter C. Mechanisms of vesicle formation: insights from the COP system. Curr Opin Cell Biol. 1999;11:440–446. doi: 10.1016/s0955-0674(99)80063-5. [DOI] [PubMed] [Google Scholar]
- Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Wigge P, McMahon HT. The amphiphysin family of proteins and their role in endocytosis at the synapse. Trends Neurosci. 1998;21:339–344. doi: 10.1016/s0166-2236(98)01264-8. [DOI] [PubMed] [Google Scholar]
- Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Winge P, Brembu T, Bones AM. Cloning and characterization of rac-like cDNAs from Arabidopsis thaliana. Plant Mol Biol. 1997;35:483–495. doi: 10.1023/a:1005804508902. [DOI] [PubMed] [Google Scholar]
- Winge P, Brembu T, Kristensen R, Bones AM. Genetic structure and evolution of RAC-GTPases in Arabidopsis thaliana. Genetics. 2000;156:1959–1971. doi: 10.1093/genetics/156.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM. Cell motility: Can Rho GTPases and microtubules point the way? J Cell Sci. 2001;114:3795–3803. doi: 10.1242/jcs.114.21.3795. [DOI] [PubMed] [Google Scholar]
- Wu G, Gu Y, Li S, Yang Z. A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell. 2001;13:2841–2856. doi: 10.1105/tpc.010218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Li H, Yang Z. Arabidopsis RopGAPs are a novel family of rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for rop-specific GTPase stimulation. Plant Physiol. 2000;124:1625–1636. doi: 10.1104/pp.124.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamochi W, Tanaka K, Nonaka H, Maeda A, Musha T, Takai Y. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J Cell Biol. 1994;125:1077–1093. doi: 10.1083/jcb.125.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Small GTPases: versatile signaling switches in plants. Plant Cell. 2002;14:S375–S388. doi: 10.1105/tpc.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarsky V, Cvrckova F, Bischoff F, Palme K. At-GDI1 from Arabidopsis thaliana encodes a rab-specific GDP dissociation inhibitor that complements the sec19 mutation of Saccharomyces cerevisiae. FEBS Lett. 1997;403:303–308. doi: 10.1016/s0014-5793(97)00072-0. [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhang C, Clarke PR. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science. 2000;288:1429–1432. doi: 10.1126/science.288.5470.1429. [DOI] [PubMed] [Google Scholar]
- Zheng ZL, Nafisi M, Tam A, Li H, Crowell DN, Chary SN, Schroeder JI, Shen J, Yang Z. The plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell. 2002;14:2787–2797. doi: 10.1105/tpc.005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZL, Yang Z. The Rop GTPase: an emerging signaling switch in plants. Plant Mol Biol. 2000a;44:1–9. doi: 10.1023/a:1006402628948. [DOI] [PubMed] [Google Scholar]
- Zheng ZL, Yang Z. The Rop GTPase switch turns on polar growth in pollen. Trends Plant Sci. 2000b;5:298–303. doi: 10.1016/s1360-1385(00)01654-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.