Abstract
Pto confers disease resistance to Pseudomonas syringae pv tomato carrying the cognate avrPto gene. Overexpression of Pto under the cauliflower mosaic virus 35S promoter activates spontaneous lesions and confers disease resistance in tomato (Lycopersicon esculentum) plants in the absence of avrPto. Here, we show that these AvrPto-independent defenses require a functional Prf gene. Several Pto-interacting (Pti) proteins are thought to play a role in Pto-mediated defense pathways. To test if interactions with Pti proteins are required for the AvrPto-independent defense responses by Pto overexpression, we isolated several Pto mutants that were unable to interact with one or more Pti proteins, but retained normal interaction with AvrPto. Overexpression of two mutants, PtoG50S and PtoR150S, failed to activate AvrPto-independent defense responses or confer enhanced resistance to the virulent P. s. pv tomato. When introduced into plants carrying 35S::Pto, 35S::PtoG50S dominantly suppressed the AvrPto-independent resistance caused by former transgene. 35S::PtoG50S also blocked the induction of a number of defense genes by the wild-type 35S::Pto. However, 35S::PtoG50S and 35S::PtoR150S plants were completely resistant to P. s. pv tomato (avrPto), indicating a normal gene-for-gene resistance. Furthermore, 35S::PtoG50S plants exhibited normal induction of defense genes in recognition of avrPto. Thus, the AvrPto-independent defense activation and gene-for-gene resistance mediated by Pto are functionally separable.
R (plant disease resistance) genes encode a large group of surveillance proteins that detect invading pathogens containing cognate avr (avirulence) genes in a highly specific manner (for recent review, see Dangl and Jones, 2001). It is now recognized that the avr gene products normally are involved in pathogen parasitism, and the subsequent evolution in plants enable them to recognize these gene products as signals of pathogen invasion. A simple model for gene-for-gene recognition predicts that R genes encode receptors that bind ligands encoded by pathogen avr genes, and that the recognition triggers downstream signal transduction pathways to activate rapid defense responses. Such a direct interaction between an R gene product and an avr gene product has been demonstrated experimentally for two disease resistance genes, the tomato (Lycopersicon esculentum) Pto gene (Scofield et al., 1996; Tang et al., 1996) and the rice (Oryza sativa) Pi-ta gene (Jia et al., 2000). However, experiments designed to detect R-Avr protein interactions in other gene-for-gene systems have yielded negative results. Although a handful of putative Avr-binding proteins have been identified, they often exist in both susceptible and resistant plants (Kooman-Gersmann et al., 1996; Ji et al., 1998; Ren et al., 2000). As a consequence, it has been proposed that the molecular recognition between many R and Avr proteins is indirect and requires a third protein (van der Niezen and Jones, 1998; Dangl and Jones, 2001; Luderer et al., 2001).
The tomato protein kinase Pto confers gene-for-gene resistance to Pseudomonas syringae pv tomato strains that carry the avrPto gene (Martin et al., 1993). The resistance requires Prf, a nucleotide-binding Leu-rich repeat (LRR) protein (Salmeron et al., 1996). Interestingly, overexpression of either Pto or Prf leads to nonspecific resistance in tomato plants to pathogens in the absence of avrPto (Oldroyd et al., 1998; Tang et al., 1999). Transient expression of PtoY207D alone under the control of the cauliflower mosaic virus (CaMV) 35S promoter induces a hypersensitive response (HR)-like response in plants (Rathjen et al., 1999). The activation of defense by ectopic expression of Pto or Prf, either wild-type or mutant forms, are collectively referred to as “AvrPto-independent defense activation.” It remains unclear whether the AvrPto-independent activation of disease resistance and gene-for-gene resistance share the same molecular basis.
In a previous yeast (Saccharomyces cerevisiae) two-hybrid screen, we identified 10 Pto-interacting (Pti) proteins (Zhou et al., 1998), notably the Pti1 protein kinase and EREBP-like transcription factors Pti4, Pti5, and Pti6 (Zhou et al., 1995, 1997). At least Pti1 and Pti4 appear to be specific substrates of Pto (Zhou et al., 1995; Gu et al., 2000). Although overexpression experiments suggested a role of Pti1 in HR (Zhou et al., 1995) and Pti5 in general resistance (He et al., 2001), expression of antisense RNA of the Pti1 and Pti5 genes did not affect the Pto-mediated gene-for-gene resistance (P. He and J.-M. Zhou, unpublished data).
To test if any of the Pti proteins play a role in the AvrPto-independent resistance mediated by Pto, we utilized a reverse yeast two-hybrid assay to isolate Pto mutants that were unable to interact with Pti proteins, but were completely normal in AvrPto interaction. Two mutants, PtoR150S and PtoG50S, with severely diminished interactions with one or more Pti proteins were examined for their ability to confer gene-for-gene resistance and nonspecific resistance in stable transgenic tomato plants. When expressed under the control of the CaMV 35S promoter, the mutants conferred normal gene-for-gene resistance to P. s. pv syringae (avrPto), but exhibited no detectable resistance to a virulent P. s. pv syringae strain. Moreover, the 35S::PtoG50S mutant, when introduced into a transgenic line carrying the 35S::Pto transgene, dominantly suppressed nonspecific resistance conferred by the latter. The results suggest that the general resistance and gene-for-gene resistance conferred by Pto are functionally separated.
RESULTS
Pto Mutants Defective in Pti Interactions
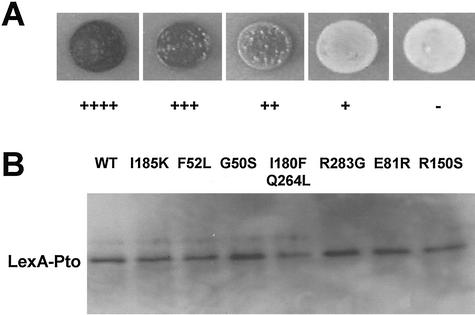
To determine if the interactions with various Pti proteins were required for Pto-mediated resistance, we sought to identify mutations that specifically disrupt interactions of Pto with Pti proteins but not affecting the interaction with AvrPto. We adopted a reverse yeast two-hybrid approach to screen for Pto mutants with diminished interactions with selected Pti proteins but displaying normal interactions with AvrPto (see “Materials and Methods” and Fig. 1A). A Pto mutant library was first screened for mutations that interfere with the interaction with Pti6, a Pti protein exhibiting the strongest interaction with Pto (Zhou et al., 1997, 1998) without affecting the interaction with AvrPto. This led to the identification of six unique mutants. These mutants were tested for interactions with nine other Pti proteins by yeast two-hybrid assays (Zhou et al., 1997, 1998). Four mutants were nonspecifically diminished in interactions with all but one Pti protein, Pti7. Two showed reduced/abolished interactions with eight of the 10 Pti proteins, except for Pti1 and Pti7. Therefore, we decided to isolate additional Pto mutants that are affected specifically in the interaction with Pti7. A single Pto mutant was isolated as a result. The respective mutations and interactions of seven confirmed mutants with AvrPto and each Pti protein are given in Table I. Six mutants carried single amino acid substitutions, and one contained two amino acid substitutions. All seven mutants interacted normally with AvrPto, but their ability to interact with one or more Pti proteins was abolished or reduced. The first class of mutants, such as PtoG50S, was severely affected in the interactions with multiple Pti proteins, but was not affected for Pti7 interaction. PtoR150S is the only mutant showing a lack of interaction with Pti7, whose protein sequence does not suggest a known function (Zhou et al., 1998), but had little effects on interactions with other Pti proteins. All mutant proteins accumulated normally in yeast (Fig. 1B), indicating that the reduced protein-protein interaction was not caused by protein instability.
Figure 1.
A, Criteria used for reverse yeast two-hybrid screen. The number of pluses indicates relative β-galactosidase activity (see Table I). B, Equal expression of LexA fusion proteins in yeast. An anti-LexA polyclonal antibody was used to detect the presence of LexA-Pto fusion proteins in yeast strains carrying the wild-type Pto (WT) or different mutants. Approximately 50 μg of total soluble protein was loaded for western blot.
Table I.
Interactions of Pto mutants with AvrPto and various Pti proteins
| Prey | Bait
|
|||||||
|---|---|---|---|---|---|---|---|---|
| WT Pto | I185K | F52 mol | G50S | I180F, Q264 | R283G | E81R | R150S | |
| AvrPto | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | ++++ |
| Pti1 | ++ | – | ++ | – | + | ++ | – | ++ |
| Pti2 | ++++ | ++ | +++ | +++ | ++ | ++ | + | ++++ |
| Pti3 | +++ | – | – | – | – | – | – | +++ |
| Pti4 | ++ | + | – | – | – | – | – | ++ |
| Pti5 | ++ | + | – | – | – | – | – | ++ |
| Pti6 | ++++ | ++ | + | + | + | ++ | + | ++++ |
| Pti7 | ++++ | ++++ | ++++ | ++++ | ++ | ++ | ++++ | – |
| Pti8 | ++++ | – | – | ++ | – | – | – | ++++ |
| Pti9 | +++ | – | – | – | + | – | – | +++ |
| Pti10 | ++++ | – | – | – | + | – | – | ++++ |
Relative degrees of the two-hybrid interaction were measured by colony color on X-Gal plates (Figure 1).
Overexpression of PtoG50S and PtoR150S Fails to Cause AvrPto-Independent Lesions But Confers HR in Response to avrPto
Next, we asked whether mutations disrupting the interaction with Pti proteins affected the AvrPto-independent resistance of Pto. PtoG50S and PtoR150S were selected as the representatives for the two classes of mutants. We constructed transgenic PtoS plants overexpressing PtoG50S, PtoR150S, and Pto. All three constructs were placed under the control of the CaMV 35S promoter and contained a translational fusion with the FLAG epitope at the carboxyl terminus of Pto (or Pto mutants). For simplicity, these constructs are referred to as 35S::PtoG50S, 35S::PtoR150S, and 35S::Pto. Northern analysis identified five 35S::PtoG50S, 13 35S::PtoR150S, and 11 35S::Pto lines that notably accumulated Pto transcripts.
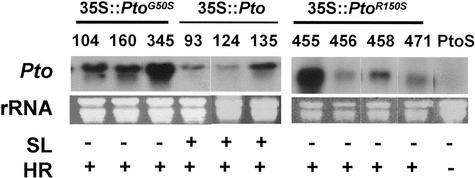
Three lines (104, 160, and 345) of 35S::PtoG50S, four lines (455, 456, 458, and 471) of 35S::PtoR150S, and three lines (93, 124, and 135) of 35S::Pto were examined in the second (T2) generation for spontaneous lesion formation and resistance to P. s. pv tomato (avrPto). As expected, all 35S::Pto lines developed spontaneous lesions in the leaf (Fig. 2). In contrast, none of the 35S::PtoG50S and 35S::PtoR150S lines showed any detectable lesions. However, when inoculated with P. s. pv tomato (avrPto), all 10 transgenic lines reproducibly displayed an HR, suggesting that the mutations had differential effects on the AvrPto-independent and the AvrPto-induced cell death.
Figure 2.
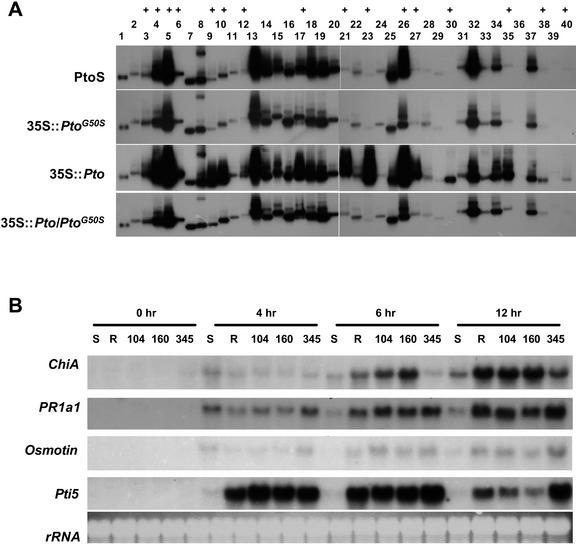
Spontaneous lesions and HR in transgenic plants overexpressing the wild-type and mutant Pto transgenes. Northern blot shows the accumulation of transgene transcripts in primary transgenic plants. Non-transgenic PtoS plants were used as a control. Lines 104, 160, 345, 93, 124, 135, 455, 456, 458, and 471 carrying respective transgenes were examined in the T2 generation for the presence (+) or absence (−) of visible spontaneous lesions (SL) in leaves without infection. The plants were also tested for the presence (+) or absence (−) of HR in response to P. s. pv tomato (avrPto).
35S::PtoG50S Dominantly Suppresses 35S::Pto-Induced AvrPto-Independent Defense Responses
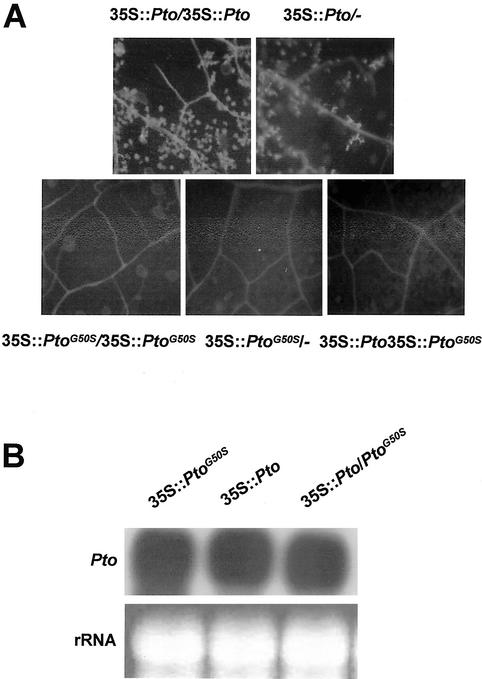
To accurately assess the effect of the PtoG50S mutation, we selected lines 104 (35S::PtoG50S) and 135 (35S::Pto) because they both carried a single copy of transgene expressed at a similar level (Fig. 3). Line 135 ♂ was crossed to line 104 ♀ to generate 35S::PtoG50S/35S::Pto plants (hemizygous for both transgenes). T2 plants of lines 104 and 135 and 35S::PtoG50S/35S::Pto plants were confirmed by Southern analysis for the presence of respective transgenes. These plants were examined for spontaneous cell death, presence of autofluorescent materials, salicylate (SA) accumulation, bacterial resistance, and defense gene expression. Figure 3A shows the presence of a large amount of autofluorescent materials in leaves of 35S::Pto plants, indicative of cell death at the microscopic level. Both hemizygous and homozygous 35S::Pto plants of line 135 displayed similar lesion and accumulated autofluorescent compounds (Fig. 3A). No 35S::PtoG50S plants, either homozygous or hemizygous, accumulated any autofluorescent compounds. Approximately 50 35S::PtoG50S/35S::Pto plants were identified by Southern blot, and none developed lesions in the leaf. These indicate that the presence of 35S::PtoG50S dominantly suppressed lesions caused by the 35S::Pto transgene. Attempts to identify the Pto-Flag protein in transgenic plants by anti-FLAG antibodies were unsuccessful. However, northern analysis indicated that the wild-type Pto transcripts were expressed normally in 35S::PtoG50S/35S::Pto plants. 35S::PtoG50S/35S::Pto plants had similar total Pto transcripts compared with 35S::Pto plants (Fig. 3B). Furthermore, we amplified and cloned cDNAs corresponding to Pto and PtoG50S transcripts from 35S::PtoG50S/35S::Pto plants by reverse transcriptase-PCR. Sequencing analysis indicated that the wild-type and mutant transcripts exist in a 1:1 ratio (27:25). These indicate that the wild-type Pto was expressed normally in 35S::PtoG50S/35S::Pto plants.
Figure 3.
35S::PtoG50S dominantly suppresses the 35S::Pto-dependent spontaneous lesions. A, Accumulation of autofluorescent materials in line 135 (35S::Pto/35S::Pto and 35S::Pto/_) and 104 (35S::PtoG50S/35S::PtoG50S and 35S::PtoG50S/_), and line 104 × line 135 F1 (35S::Pto/35S::PtoG50S) was examined under a fluorescence microscope. The tested T2 plants were self-pollinated and their homozygosity was determined in the T3 generation. B, Total Pto transcripts (wild type and mutant) in homozygous 104 (35S::PtoG50S), homozygous 135 (35S::Pto), and 35S::Pto/35S::PtoG50S plants.
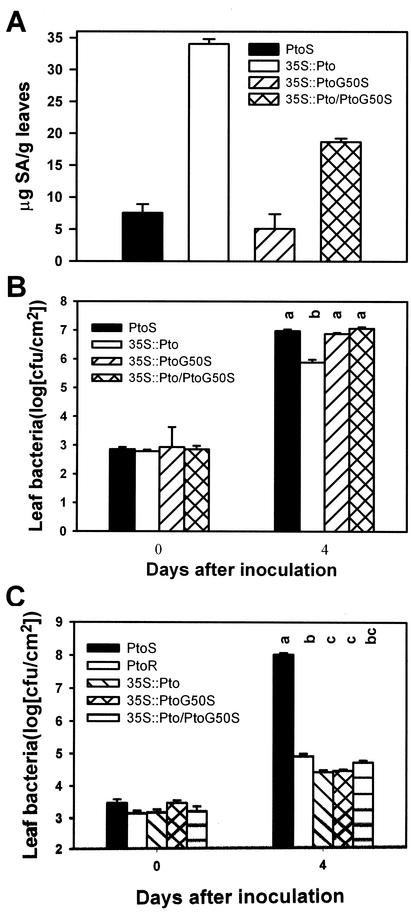
In addition to spontaneous lesions, overexpression of Pto in tomato plants also leads to the accumulation of SA, constitutive expression of defense-related genes, and enhanced resistance to virulent pathogens (Tang et al., 1999). The SA level in 35S::PtoG50S T2 plants was indistinguishable from that in non-transgenic plants (Fig. 4A), indicating that the mutation abolished the SA-inducing ability. 35S::PtoG50S/35S::Pto plants accumulated an intermediate amount of SA compared with 35S::Pto T2 and non-transgenic plants, indicating that the mutant transgene partially suppressed the 35S::Pto-dependent SA accumulation.
Figure 4.
35S::PtoG50S dominantly suppresses the 35S::Pto-mediated AvrPto-independent resistance, but confers gene-for-gene resistance. A, Accumulation of free SA. B, Bacterial growth assay of P. s. pv tomato in plants. C, Bacterial growth assay of P. s. pv tomato (avrPto) in plants. Each data point represents average of three replicates (three plants). Error bars indicate ses. Lines 104 and 135 were in the T2 generation segregating for the transgenes. All plants used in the experiments had been confirmed for the presence of transgene by Southern analysis. Both homozygous and hemizygous plants of lines 104 and 135 were used. Different letters denote significant difference (P = 0.05) as determined by Student's t distribution.
To determine the effect of 35S::PtoG50S on nonspecific disease resistance, we inoculated the plants with P. s. pv tomato, a virulent strain. Figure 4B shows that in 35S::Pto plants, bacterial growth was reduced by approximately 20-fold compared with the non-transgenic plants 4 d after inoculation. In contrast, the bacterial growth in 35S::PtoG50S and 35S::PtoG50S/35S::Pto plants was indistinguishable from that in the non-transgenic plants. Thus, the G50S mutation abolished the AvrPto-independent lesion formation, SA accumulation, and nonspecific resistance to virulent bacteria. It also acted as a dominant suppressor to the 35S::Pto-dependent defense responses and nonspecific resistance.
35S::PtoG50S Confers Gene-for-Gene Resistance
To determine quantitatively if the G50S mutation affected the gene-for-gene resistance mediated by Pto-avrPto interaction, we measured bacterial growth of P. s. pv tomato (avrPto) in lines 104 and 135 (T2 plants), 35S::PtoG50S/35S::Pto F1 plants, and non-transgenic PtoS and PtoR plants (Fig. 4C). All transgenic plants showed strong resistance to the bacterium. Lines 104 and 135, 35S::PtoG50S/35S::Pto, and PtoR plants were nearly indistinguishable. The plants were also inoculated with a high concentration of P. s. pv tomato (avrPto) to induce HR. However, no detectable difference in the timing and appearance of HR development was observed among lines 104, 135, and 35S::PtoG50S/35S::Pto plants (data not shown). Thus, the G50S mutation, although completely abolishing the AvrPto-independent resistance, had no detectable effect on the gene-for-gene resistance.
PtoG50S Differentially Affects Defense Gene Inductions by 35S::Pto and Pto-AvrPto Interaction
To further understand the effects of PtoG50S on plant defense at the molecular level, we examined the expression of defense-related genes in plants carrying the wild-type and/or mutant transgenes. We previously identified a large number of tomato genes whose transcripts accumulated to a higher level in plants overexpressing Pto (Tang et al., 1999; Xiao et al., 2001). These genes likely represent a variety of downstream signal transduction and metabolic pathways activated by the overexpression of Pto. We compared the expression of 39 genes among non-transgenic PtoS, 35S::Pto, 35S::PtoG50S, and 35S::PtoG50S/35S::Pto plants by “reverse northern” (Fig. 5A; Xiao et al., 2001). In brief, DNA blots containing the 39 cDNA clones were hybridized with total cDNA probes reverse transcribed from mRNA samples. We detected 16 genes whose transcripts accumulated to a markedly higher level in the 35S::Pto plants compared with the non-transgenic plants. The remaining 23 genes were not induced in line 135, although some of these genes were slightly induced (2-fold) in another transgenic line 48, from which these cDNA clones were isolated (Xiao et al., 2001). The discrepancy between lines 135 and 48 may be explained by differences of cultivars and lesion severity. Line 48, which was constructed in the Money Maker background, had more severe lesions than line 135, which was constructed in Rio Grande PtoS background. Nevertheless, none of the 16 genes showed increased expression in 35S::PtoG50S and 35S::PtoG50S/35S::Pto plants compared with non-transgenic plants, indicating that PtoG50S globally blocked gene expression that was normally activated by 35S::Pto.
Figure 5.
PtoG50S suppresses the 35S::Pto-induced defense gene expression, but mediates normal defense gene activation by AvrPto. A, Reverse northern analysis of gene expression in untreated non-transgenic PtoS and transgenic lines 104 (35S::PtoG50S), 135 (35S::Pto), and line 104 × line 135 F1 (35S::Pto/35S::PtoG50S) plants. cDNA probes were hybridized to duplicated DNA blots containing PCR products of cDNA clones (Xiao et al., 2001). Lane 1 contains an actin cDNA as a constitutive control. Lanes 2 thorough 40 are 39 cDNA clones isolated previously (Xiao et al., 2001). The GenBank accession numbers of corresponding cDNA clones that showed a differential hybridization (+) between 35S::Pto and non-transgenic PtoS plants are: BG352044 (3), BG351997 (4), BG352022 (5), BG351998 (6), BG352013 (9), BG352014 (10), BG352025 (12), BG351999 (17), BG352007 (21), BG352005 (23), BG352012 (26), BG352008 (27), BG352006 (30), BG352009 (35), BG352020 (38), and BG352046 (40). B, Northern analysis of gene expression in PtoS (S), PtoR (R), and mutant 35S::PtoG50S transgenic lines 104, 160, and 345 after inoculation with P. s. pv tomato (avrPto). Samples were collected at the indicated hours postinoculation, and RNA blots were hybridized with the indicated probes. Lines 104, 160, 345, and 135 were in the T2 generation segregating for the transgenes. All plants used in the experiments had been confirmed for the presence of transgene by Southern analysis.
To assess the effect of the G50S mutation on gene-for-gene resistance, we examined the expression of several PR genes that are induced very strongly by the Pto-avrPto interaction. Figure 5B shows that three PR genes, ChiA, PR1a1, and Osmotin, were similarly induced by P. s. pv tomato (avrPto) in PtoR and three 35S::PtoG50S lines compared with PtoS plants. Pto-avrPto recognition also induces Pti5 transcripts (Thara et al., 1999). Overall, this induction occurred very early (4 h after bacterial infection) in both PtoR and the three 35S::PtoG50S lines. The only exception is the ChiA expression in line 345 at the 6-h time point, but the significance of this is not clear. Thus, PtoG50S does not apparently affect defense-related gene expression during the gene-for-gene interaction.
35S::PtoR150S Confers Normal Gene-for-Gene Resistance But Not Nonspecific Resistance
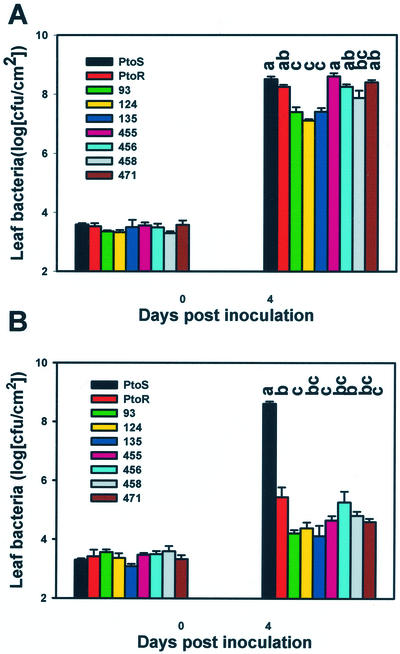
We also quantitatively determined resistance of 35S::PtoR150S plants to P. s. pv tomato and P. s. pv tomato (avrPto) by bacterial growth assay. Figure 6A shows that the three lines carrying the wild-type 35S::Pto transgene were significantly more resistant to P. s. pv tomato compared with non-transgenic control plants. In contrast, the four 35S::PtoR150S lines were nearly indistinguishable from the non-transgenic control plants, indicating that the 35S::PtoR150S transgene confers no measurable resistance to the compatible bacterium. However, the 35S::PtoR150S transgenic lines showed virtually the same level of resistance to P. s. pv tomato (avrPto) compared with the 35S::Pto transgenic lines (Fig. 6B). The results indicate that, similar to PtoG50S, PtoR150S also bears a specific defect in AvrPto-independent resistance.
Figure 6.
35S::PtoR150S does not confer nonspecific resistance but confers normal gene-for-gene resistance to P. s. pv tomato (avrPto). Tomato lines PtoS, PtoR, wild-type 35S::Pto transgenic lines (93, 124, and 135), and 35S::PtoR150S lines (455, 456, 458, and 471) were inoculated with the P. s. pv tomato (A) or P. s. pv tomato (avrPto) strain (B), and leaf bacterial populations were determined. Each data point is averaged from three replicates. Error bars indicate ses. Different letters denote significant difference (P = 0.05) as determined by Student's t distribution. All transgenic lines were in the T2 generation segregating for the transgenes. All plants used in the experiments had been confirmed for the presence of transgene by Southern analysis.
prf-3 Dominantly Suppresses Spontaneous Lesions Caused by 35S::Pto
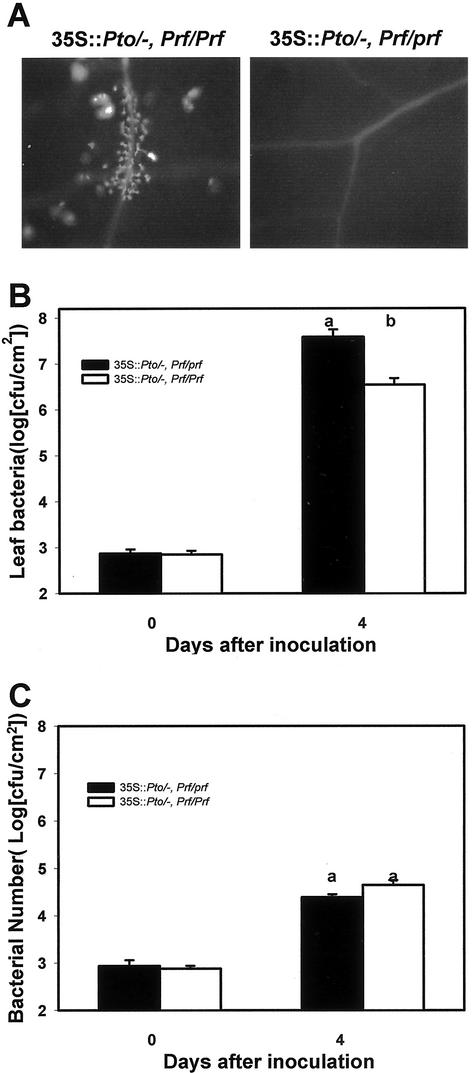
Our results described above suggest that the gene-for-gene resistance mediated by Pto-avrPto interaction and the AvrPto-independent resistance caused by the Pto overexpression are functionally separated. We tested if the AvrPto-independent defense induced by Pto overexpression required a functional Prf. Prf is necessary for resistance mediated by the Pto-avrPto interaction (Salmeron et al., 1996). prf-3, bearing a deletion that truncates the Prf protein before the LRR domain, is a recessive mutation that abolishes the avrPto-induced resistance. We crossed the prf-3 mutant (in the Rio Grande 76R background; Salmeron et al., 1993) with the transgenic line 48 (Loh et al., 1998; Tang et al., 1999). This line carries an HA-tagged Pto under the control of the CaMV 35S promoter. Figure 7A shows that the F1 plants carrying a heterozygous prf-3 mutation and a hemizygous 35S::Pto transgene no longer develop spontaneous lesions as indicated by the lack of autofluorescent materials in the leaf. In contrast, the control (PtoR × line 48) F1 plants developed lesions as expected. The lesion formation was also examined in 80 prf-3 × line 48 F2 plants (Table II). Forty-one plants were found to carry both the 35S::Pto transgene and the prf-3 mutation (heterozygous or homozygous), and none displayed any visible lesions. Twelve plants carried the 35S::Pto but not the prf-3 mutation, and all developed spontaneous lesions.
Figure 7.
prf-3 dominantly suppresses the 35S::Pto-dependent lesions and disease resistance. A, Accumulation of autofluorescent materials. B, Bacterial growth assay with P. s. pv tomato. C, Bacterial growth assay with P. s. pv tomato (avrPto). Plants used were PtoR × line 48 F1 (35S::Pto/_ and Prf/Prf) and prf-3 × line 48 F1 (35S::Pto/_ and Prf/prf). Each data point is averaged from three replicates. Different letters denote significant difference (P = 0.05) as determined by Student's t distribution.
Table II.
prf-3 suppresses 35S::Pto-induced lesions
| Crosses | Total Plants Examined | Plants with Lesions |
|---|---|---|
| MM × line 48 F1 (35S::Pto /− ; Prf/Prf) | 12 | 12 |
| PtoR x MM F1 (− /− ; Prf/Prf) | 12 | 0 |
| PtoR × line 48 F1 (35S::Pto /− ; Prf/Prf) | 9 | 9 |
| prf-3 × line 48 F1 (35S::Pto/− ; prf-3/Prf) | 10 | 0 |
| prf-3 × 48 F2 | 41 (35S::Pto/± ;prf-3/−) | 0 |
| 12 (35S::Pto/±; Prf/Prf) | 12 | |
| 27 (no 35S::Pto) | 0 |
Line 48 was used as pollen donor in all crosses. The Pto locus in prf-3 (from L. pimpinellifolium) is polymorphic to the pto locus in MM and line 48 plants (from tomato), enabling the identification of F2 plants carrying the prf-3 mutation and the 35S::Pto transgene by RFLP analysis (data not shown). prf-3/− indicates plants homozygous or heterozygous for the prf-3 mutation. Lesions were scored 5 weeks after germination. MM, Money Maker.
We also determined the effect of the prf-3 mutation on disease resistance to P. s. pv tomato and P. s. pv tomato (avrPto). Figure 7B shows that plants carrying a heterozygous prf-3 mutation were significantly less resistant to P. s. pv tomato than homozygous Prf plants. Thus, the prf-3 mutation dominantly suppressed the 35S::Pto-dependent spontaneous lesions and disease resistance. In contrast, the heterozygous prf had no effect on resistance to P. s. pv tomato (avrPto), confirming the earlier report that prf-3 is a recessive mutation for Pto-avrPto-mediated resistance (Fig. 7C).
DISCUSSION
In this report, we describe the characterization of stable transgenic plants expressing two Pto mutants, PtoG50S and PtoR150S, that are defective in interactions with one or more Pti proteins. The mutations had differential effects on AvrPto-independent and gene-for-gene resistance. Moreover, 35S::PtoG50S dominantly suppressed/diminished all 35S::Pto-dependent defense responses, including cell death, SA accumulation, defense gene expression, and resistance to P. s. pv tomato, but it conferred normal gene-for-gene resistance to P. s. pv tomato (avrPto). The contrasting effects of the mutants suggest that the AvrPto-independent resistance and gene-for-gene resistance conferred by Pto are functionally separable.
The 35S::Pto-dependent, AvrPto-independent defense activation is distinct from gene-for-gene resistance in defense gene induction. The AvrPto-independent defense in 35S::Pto plants and the Pto-avrPto interaction appear to induce overlapping but distinct sets of defense genes. The majority of the genes induced by the 35S::Pto transgene are induced equally in compatible and incompatible interactions (data not shown), indicating a lack of involvement in the gene-for-gene interaction. The only known exceptions are Pti5 and several PR genes whose expression is activated by both the AvrPto-independent defense and gene-for-gene interaction (Tang et al., 1999; Thara et al., 1999). Together, these support that Pto activates the AvrPto-independent resistance and gene-for-gene resistance by two distinct pathways.
A less likely explanation for the differential effects of the Pto mutations may be that the AvrPto-independent resistance require a higher signaling threshold that is achieved by the overexpression of Pto, whereas the gene-for-gene resistance is activated at a lower signaling threshold. The mutations might have reduced the Pto activity below the threshold for AvrPto-independent resistance, but the residual activity was sufficient for gene-for-gene resistance. However, this hypothesis has difficulty explaining why PtoG50S acted as a dominant negative mutant to suppress 35S::Pto-induced spontaneous lesions, whereas it functioned positively to activate HR when induced by AvrPto.
Pto and Prf appear to act in the same pathway to activate the AvrPto-independent resistance. First, both Pto and Prf, when overexpressed, confer general resistance to pathogens independent of avrPto (Oldroyd and Staskawicz, 1998; Tang et al., 1999). Transiently overexpressing certain Pto mutants also induces a Prf-dependent necrosis reminiscent of HR (Rathjen et al., 1999). This is consistent with the observation that prf-3 dominantly suppressed lesions in plants carrying 35S::Pto (Fig. 7). Prf may be a haplo-insufficient gene if it functions downstream of or coincides with Pto to activate the AvrPto-independent lesion formation. Alternatively, this could be due to a dominant negative effect of the prf-3 mutant lacking the LRR domain. We recently isolated a null prf mutant in the 35S::Pto background. This prf mutant showed no spontaneous lesions in the heterozygous state (X.Y. Tang, unpublished data). Furthermore, overexpression of Prf results in nonspecific resistance in the prf-3 mutant background (Oldroyd and Staskawicz, 1998). These indicate that Prf is a haplo-insufficient gene required for the AvrPto-independent defense activation. Taken together, we conclude that Prf functions downstream of or coincident with Pto for the AvrPto-independent resistance. It remains to be determined if Prf overexpression enhances resistance in the absence of Pto.
AvrPto-independent induction of HR-like lesions by the transient overexpression of certain Pto mutants were thought to be equivalent to defense activation in the Pto-avrPto interaction, because a functional Prf is required for lesion induction (Rathjen et al., 1999). However, the 35S::Pto-dependent lesion formation and gene-for-gene resistance, although both depended on a functional Prf, were differentially affected by mutations in Pto and Prf. These caution the use of AvrPto-independent defense responses in gene-for-gene resistance studies.
Two models have been proposed for the Pto-AvrPto-mediated disease resistance pathway. In one, the binding of AvrPto stimulates the kinase activity of Pto that subsequently activates downstream defense responses (Scofield et al., 1996; Tang et al., 1996). Gly-50 is a conserved residue in the Pto kinase family. We did not test if the G50S mutation affected kinase activity. However, two reports demonstrate that the kinase activity is required for the elicitation of HR (Rathjen et al., 1999; Sessa et al., 2000). The Pti proteins were isolated as candidate components downstream of Pto. The lack of correlation between Pti-Pto mutant interaction and gene-for-gene resistance suggests that these Pti proteins are not essential for disease resistance triggered by AvrPto. Furthermore, we have constructed and characterized numerous transgenic tomato plants (in the PtoR background) expressing antisense RNA of eight of the 10 Pti genes (except for Pti2 and Pti7). All antisense plants showed HR and disease resistance when challenged with P. s. pv tomato carrying avrPto (P. He and J.-M. Zhou, unpublished data). Although our results do not exclude the possibility that redundant genes in addition to the tested Pti genes may account for the lack of an effect from antisense RNA expression and Pto mutations, we are inclined to suggest that the interaction of Pto with these Pti proteins is required for AvrPto-independent resistance but not essential for gene-for-gene resistance. This is inconsistent with the previous finding that overexpression of Pti1 in tobacco (Nicotiana tabacum) enhances HR in response to P. s. pv tabaci carrying avrPto (Zhou et al., 1995). It is possible that an effect of Pti1 on HR only occurs when overexpressed. Alternatively, AvrPto may activate distinct signaling pathways in tobacco and tomato. Two AvrPto motives are differentially required for gene-for-gene resistance in tobacco and tomato (Shan et al., 2000b).
The second model, referred to as the guard model, suggests that the Pto kinase and Pti proteins normally function in basal resistance (van der Biezen and Jones, 1998; Dangl and Jones, 2001), and that Pto-AvrPto interaction by itself is intended by the bacterium to suppress host defense. The Prf protein guards Pto by detecting the Pto-AvrPto interaction as a signal of bacterial invasion. Upon Pto-AvrPto interaction, Prf induces gene-for-gene resistance through a different pathway.
Several lines of evidence are consistent with the guard model. First, 35S::Pto confers AvrPto-independent resistance in plants. Weak resistance to virulent P. s. pv tomato was observed in tomato plants carrying the native Pto gene (Tang et al., 1999). Pti5 appears to regulate defense gene expression and confers resistance to P. s. pv tomato bacteria in the absence of avrPto when overexpressed (He et al., 2001). The facts that AvrPto contributes to bacterial virulence (Chang et al., 2000; Shan et al., 2000a) and that 35S::PtoG50S dominantly suppressed AvrPto-independent resistance raise the possibility that a bacterial effector protein can modulate Pto in such a way that the latter suppresses basal defense. Finally, our findings that the AvrPto-independent resistance and gene-for-gene resistance are functionally separable are also consistent with this model.
However, the guard model does not readily explain the following facts. AvrPto enhances virulence of P. s. pv tomato in plants lacking Pto. At least three AvrPto mutants that do not interact with Pto confer normal virulence activity in tomato plants (Shan et al., 2000a). One argument may be that AvrPto also targets other Pto-like kinases involved in defense. However, no Pto homologs have been shown to interact with AvrPto. Prf does not confer resistance in the absence of Pto and, thus, is unlikely to guard a Pto homolog. The guard model provides a plausible explanation for the presence of Avr-binding sites in both susceptible and resistant plants (Kooman-Gersmann et al., 1996; Ji et al., 1998; Ren et al., 2000). The guardee is a common host target, whereas the LRR protein is specific to the resistant plant. However, the opposite appears to be true for the Pto/Prf-mediated resistance. The Prf alleles in tomato and Lycopersicon pimpinellifolium are nearly identical (99% amino acid identity), both conferring the same function. In contrast, much greater functional and sequence divergence exists among the Pto family members both within and between species. The members share approximately 87% amino acid identity (Jia et al., 1997), and only the L. pimpinellifolium Pto gene is known to confer resistance. Furthermore, as discussed above, Prf is required for AvrPto-independent resistance in 35S::Pto plants. Therefore, the function of Prf is not limited to “guard.”
In summary, we show that Pto and Prf function in the same pathway to activate the AvrPto-independent resistance. However, this resistance appears to be distinct from gene-for-gene resistance in terms of the requirement of downstream components, sensitivity to mutations in Pto, and defense gene activation. Current models are not sufficient to explain the data collected. Knowledge about Pto and Prf at the protein-protein interaction level is required to understand how these two proteins function in gene-for-gene resistance and AvrPto-independent resistance.
MATERIALS AND METHODS
Plants and Bacterial Inoculation
Rio Grande PtoR and PtoS are tomato (Lycopersicon esculentum) isogenic lines carrying either a native Pto gene or no Pto, respectively. The transgenic line 48 carries a 35S::Pto transgene in the Money Maker background (Tang et al., 1999). Money Maker does not carry the native Pto gene. Six-week-old plants grown in the greenhouse at 28°C (day) and 20°C (night) were used for all experiments. The virulent strain Pseudomonas syringae pv tomato and avirulent strain P. s. pv tomato (avrPto; T1 and T1 [pPTE6], respectively; Ronald et al., 1992) were used for inoculation as described (Xiao et al., 2001). Bacteria (2 × 105 colony forming units [cfu] mL−1) was vacuum infiltrated into tomato plants for bacterial growth assays. All bacterial growth assays were repeated with similar results. HR was assayed by syringe infiltration of 108 cfu mL−1 of P. s. pv tomato (avrPto) and scoring for leaf necrosis 12 h after inoculation.
Isolation of Pto mutants
The yeast (Saccharomyces cerevisiae) two-hybrid assay was carried out as described previously (Zhou et al., 1995).
A PCR-based random mutagenesis was used to create mutations in the Pto cDNA (Shan et al., 2000a). Approximately 7,000 mutated Pto clones were first screened for mutations that interfere with the interaction with Pti6 or Pti7 (Zhou et al., 1997, 1998) without affecting the interaction with AvrPto (M. Lu and J.-M. Zhou, unpublished data). Approximately 2% of clones were white or light blue on X-Gal plates, indicating potential mutations in Pto. We verified the authenticity of putative Pto mutants by isolating plasmid DNA individually from these colonies, shuttling through Escherichia coli, and reintroduced plasmid into yeast strains carrying avrPto, Pti6, or Pti7 as prey. The confirmed mutants were sequenced to determine the mutations. The mutants were then tested for interactions with other Pti proteins (Zhou et al., 1998).
The expression of mutant proteins was determined by western blot using an anti-LexA antibody (CLONTECH, Palo Alto, CA) following the manufacturer's instructions.
Generation of Transgenic Plants Expressing Wild-Type and Mutant Pto
For the 35S::Pto construct, the EcoRI fragment of Pto cDNA in pTC3 (Martin et al., 1993) was reversed by EcoRI digestion and religation. The resulting plasmid was used as a template for PCR amplification of the Pto cDNA with a forward primer (T7) and a reverse primer (5′-AGAATTCACTTGTCATCGTCGTCCTTGTAATCGATAACAGACTCTTGGAG-3′). The reverse primer introduced a FLAG epitope-coding sequence fused in-frame to the C terminus of the Pto protein. It also introduced an EcoRI site after FLAG and a ClaI site at the junction of Pto and FLAG. The PCR product was digested with EcoRI and inserted into the pBluescript SK− plasmid (Stratagene, La Jolla, CA) to create pBS::Pto::FLAG. Sequencing analysis confirmed that the plasmid did not carry any mutations during PCR amplification. The Pto::FLAG fragment was excised with EcoRI and reinserted into the EcoRI site of pGEM7Z(+) (Promega, Madison, WI). The resulting plasmid clone was digested with XbaI (5′ to Pto::FLAG) and SacI (3′ to Pto::FLAG), and the insert was ligated into the corresponding sites of pBI121 (CLONTECH). This gave rise to the 35S::Pto::FLAG construct (called 35S::Pto throughout the text). To construct 35S::Pto mutants, internal XhoI-BglII or XhoI-Bsu36I fragments carrying the mutations were excised from the respective mutant clones (from pEG202) and used to replace the corresponding wild-type Pto fragment in a modified pBS::Pto::FLAG plasmid. The modified pBS::Pto::FLAG plasmid was made by removing the Acc656I-SalI fragment from the linker, which carried an undesirable XhoI site, blunt ended by klenow fill-in, and religated. The mutant Pto::FLAG fragments were excised with EcoRV and SacI, and inserted between the SmaI and SacI sites of pBI121 to create 35S::Pto mutants.
The resulting wild-type and mutant 35S::Pto constructs were introduced into Agrobacterium tumefaciens strain LBA4404 to transform tomato (PtoS) following standard protocols (Joao and Brown, 1993). Unless indicated otherwise, all results were collected from the second or third generation of transgenic plants that had been individually verified for the presence of transgenes by Southern-blot analysis.
SA Measurement and Microscopy
Accumulation of free SA in plants was determined as described by Li et al. (2002). Fluorescence microscopy was used to determine the accumulation of fluorescent materials in tomato leaves (Tang et al., 1999).
Northern and Reverse Northern Analyses
Six-week-old plants were either untreated or vacuum infiltrated with P. s. pv tomato strain T1 (avrPto) at 2 × 106 cfu mL−1 in the presence of 0.004% (w/v) Silwet l-77 (Osi, Danbury, CT). Expanded leaves were harvested at the indicated times for total RNA isolation. Northern-blot analysis was carried out as described by Tang et al. (1999). Reverse northern analysis was done as described using an actin cDNA clone as a constitutive control (Xiao et al., 2001).
ACKNOWLEDGMENTS
We thank Drs. Frank White, Barbara Valent, Xueming Wang, and Randall Warren for critical reading of the manuscript.
Footnotes
This work was supported by the National Science Foundation (grant no. MCB9808701 to J.-M.Z.) and by the U.S. Department of Agriculture (grant no. 9802511 to X.Y.T.). This is Kansas Agricultural Experimental Station contribution no. 02–170–A.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.016113.
LITERATURE CITED
- Chang JH, Rathjen JP, Bernal AJ, Staskawicz BJ, Michelmore RW. avrPto Enhances Growth and Necrosis Caused by Pseudomonas syringae pv. tomato in Tomato Lines Lacking Either Pto or Prf. Mol Plant-Microbe Interact. 2000;13:568–571. doi: 10.1094/MPMI.2000.13.5.568. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Gu YQ, Yang C, Thara V, Zhou J-M, Martin GB. Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell. 2000;12:771–785. doi: 10.1105/tpc.12.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Warren RF, Shan L, Zhao T, Tang X, Zhou J-M. Overexpression of Pti5 potentiates pathogen-induced defense gene expression in tomato. Mol Plant-Microbe Interact. 2001;14:1453–1457. doi: 10.1094/MPMI.2001.14.12.1453. [DOI] [PubMed] [Google Scholar]
- Ji C, Boyd C, Slaymaker D, Okinaka Y, Takeuchi Y, Midland SL, Sims JJ, Herman E, Keen N. Characterization of a 34-kDa soybean binding protein for the syringolide elicitors. Proc Natl Acad Sci USA. 1998;95:3306–3311. doi: 10.1073/pnas.95.6.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Loh Y-T, Zhou J, Martin GB. Alleles of Pto and Fen occur in bacterial speck-susceptible and fenthion-insensitive tomato lines and encode functional protein kinases. Plant Cell. 1997;9:61–73. doi: 10.1105/tpc.9.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joao KHL, Brown TA. Enhanced transformation of tomato-co-cultivated with Agrobacterium tumefaciens C58C1Rifr::pGS-FR1161 in the presence of acetosyringone. Plant Cell Rep. 1993;12:422–425. doi: 10.1007/BF00234705. [DOI] [PubMed] [Google Scholar]
- Kooman-Gersmann M, Honée G, Bonnema G, deWit PJGM. A high-affinity binding site for theAVR9 peptide elicitor of Cladosporium fulvum is present on plasma membranes of tomato and other solanaceous plants. Plant Cell. 1996;8:929–938. doi: 10.1105/tpc.8.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Shan L, Zhou J-M, Tang X. Overexpression of Pto induces a salicylate-independent cell death but inhibits necrotic lesions caused by salicylate deficiency in tomato plants. Mol Plant-Microbe Interact. 2002;15:654–661. doi: 10.1094/MPMI.2002.15.7.654. [DOI] [PubMed] [Google Scholar]
- Loh Y-T, Zhou J-M, Martin GB. The myristylation motif of Pto is not required for disease resistance. Mol Plant-Microbe Interact. 1998;11:572–576. doi: 10.1094/MPMI.1998.11.6.572. [DOI] [PubMed] [Google Scholar]
- Luderer R, Rivas S, Nurnberger T, Mattei B, Van den Hooven HW, Van der Hoorn RA, Romeis T, Wehrfritz JM, Blume B, Nennstiel D et al. No evidence for binding between resistance gene product Cf-9 of tomato and avirulence gene product AVR9 of Cladosporium fulvum. Mol Plant-Microbe Interact. 2001;14:867–876. doi: 10.1094/MPMI.2001.14.7.867. [DOI] [PubMed] [Google Scholar]
- Martin GB, Brommonschenkel S, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Staskawicz BJ. Genetically engineered broad-spectrum disease resistance in tomato. Proc Natl Acad Sci USA. 1998;95:10300–10305. doi: 10.1073/pnas.95.17.10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen JP, Chang JH, Staskawicz BJ, Michelmore RW. Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 1999;18:3232–3240. doi: 10.1093/emboj/18.12.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Qu F, Morris TJ. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell. 2000;12:1917–1926. doi: 10.1105/tpc.12.10.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald PC, Salmeron JM, Carland FM, Staskawicz BJ. The cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J Bacteriol. 1992;174:1604–1611. doi: 10.1128/jb.174.5.1604-1611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Barker SJ, Carland FM, Mehta AY, Staskawicz BJ. Tomato mutants altered in bacterial disease resistance provide evidence for a new locus controlling pathogen recognition. Plant Cell. 1993;6:511–520. doi: 10.1105/tpc.6.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Oldroyd GED, Rommens CMT, Scofield SR, Kim H-S, Lavelle DT, Dahlbeck D, Staskawicz BJ. Tomato Prf is a member of the leucine-rich-repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, Staskawicz BJ. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- Sessa G, D'Ascenzo M, Martin GB. Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto-Pto-mediated hypersensitive response. EMBO J. 2000;19:2257–2269. doi: 10.1093/emboj/19.10.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Zhou J-M, Tang X. A cluster of mutations disrupt the avirulence but not the virulence function of AvrPto. Mol Plant-Microbe Interact. 2000a;13:592–598. doi: 10.1094/MPMI.2000.13.6.592. [DOI] [PubMed] [Google Scholar]
- Shan L, Thara V, Martin G, Zhou J-M, Tang X. The PseudomonasAvrPto protein is differentially recognized by tomato and tobacco and is localized to the plant plasma membrane. Plant Cell. 2000b;12:2323–2338. doi: 10.1105/tpc.12.12.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- Tang X, Xie M, Kim YJ, Zhou J, Klessig DF, Martin GB. Overexpression of Pto activates Defense responses and confers broad resistance. Plant Cell. 1999;11:15–29. doi: 10.1105/tpc.11.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thara VK, Gu YQ, Tang X, Martin GB, Zhou J-M. Pseudomonas syringae pv. tomato induces the expression of tomato EREBP-like genes Pti4 and Pti5 independent of ethylene, salicylate and jasmonate. Plant J. 1999;20:475–483. doi: 10.1046/j.1365-313x.1999.00619.x. [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Jones JDG. Plant-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- Xiao FM, Tang X, Zhou J-M. Expression of 35S::Pto globally activates defense gene expression in tomato plants. Plant Physiol. 2001;126:1637–1645. doi: 10.1104/pp.126.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Loh Y-T, Bressan RA, Martin GB. The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell. 1995;83:925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- Zhou J, Tang T, Frederick R, Martin GB. Pathogen recognition and signal transduction by the Pto kinase. J Plant Res. 1998;111:353–356. [Google Scholar]
- Zhou J, Tang T, Martin GB. The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 1997;16:3207–3218. doi: 10.1093/emboj/16.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]