Abstract
Eurycoma longifolia Jack. is a treelet that grows in the forests of Southeast Asia and is widely used throughout the region because of its reported medicinal properties. Widespread harvesting of wild-grown trees has led to rapid thinning of natural populations, causing a potential decrease in genetic diversity among E. longifolia. Suitable genetic markers would be very useful for propagation and breeding programs to support conservation of this species, although no such markers currently exist. To meet this need, we have applied a genome complexity reduction strategy to identify a series of single nucleotide polymorphisms (SNPs) within the genomes of several E. longifolia accessions. We have found that the occurrence of these SNPs reflects the geographic origins of individual plants and can distinguish different natural populations. This work demonstrates the rapid development of molecular genetic markers in species for which little or no genomic sequence information is available. The SNP markers that we have developed in this study will also be useful for identifying genetic fingerprints that correlate with other properties of E. longifolia, such as high regenerability or the appearance of bioactive metabolites.
Eurycoma longifolia Jack., from the family of Simaroubaceae is commonly distributed in South East Asia including Myanmar, Thailand, Laos, Cambodia, Indo-China, and Malaysia. This tree has achieved considerable attention from the public for its medicinal properties and is used traditionally as a blood coagulant for complications during childbirth, as a treatment for dysentery, and as an aphrodisiac, among other applications. Extracts from E. longifolia also contain biologically active compounds with antiplasmodial activity (Chan et al., 1986). Increased harvesting of wild-grown trees for their medicinal use has led to rapid thinning of natural populations and a potential loss of genetic diversity in this species. Genetic diversity studies are essential for providing information for propagation, domestication, and breeding programs as well as conservation of genetic resources for this species.
Molecular markers have proven to be powerful tools for assessing genetic variation within and between populations of plants. Several criteria should be considered in choosing molecular techniques for genetic diversity studies including the following: whether the techniques are highly reproducible between laboratories and whether the data that is generated can be reliably transferred; whether markers are dominant or codominant, allowing homozygotes and heterozygotes to be distinguished; the amount of genomic sequence information required; and whether the markers detect highly polymorphic loci. At present, various molecular techniques are available for assessing genetic diversity in plants including identification of isozymes (Gomory et al., 2001; Nassar et al., 2001); amplified fragment length polymorphisms (Creswell et al., 2001; Quagliaro et al., 2001), random amplified polymorphic DNA, RFLP, and microsatellites (Maguire et al., 2000; Walter and Epperson, 2001). However, the limitations of these techniques include low numbers of polymorphic loci, their requirements for large amounts of DNA, or their poor reproducibility and labor intensity.
Because little or no information on genetic diversity in E. longifolia has been generated, we have tested a highly polymorphic marker to investigate the level of genetic diversity between and within populations of this species using single nucleotide polymorphism (SNPs). SNPs have become popular tools for identifying genetic loci that contribute to phenotypic variation based on linkage disequilibrium. Compared with other genetic markers, SNPs are more abundant in the genome and are much more stably inherited. Another advantage of SNP-based genotyping is that SNP detection does not involve gel electrophoresis, which is relatively slow and labor intensive. Many different strategies have been developed for high throughput detection of SNPs including high-density oligonucleotide hybridization arrays (Wang et al., 1998), dynamic allele-specific hybridization (Pennisi, 1998), and the Taqman assay (Livak et al., 1995). In this study, we use allele-specific oligonucleotide hybridization (Jordan et al., 2002) to detect polymorphisms among many different accessions of E. longifolia. The goal of this study is to assess the genetic relationships between and within different geographical populations of E. longifolia.
RESULTS
E. longifolia plants were collected from six geographically distinct locations in peninsular Malaysia, and genomic DNA was extracted from each of the 47 plants (Fig. 1A; Table I). Five degenerate oligonucleotide-primed (DOP)-PCR primers, each with 3′ sequences of different lengths and composition, were used to amplify fragments from genomic DNA of one of these individual plants. According to Jordan et al. (2002), increasing the length of the specific sequence at the 3′ end of the primer should decrease the complexity of the PCR product mixture. Lowering the annealing temperature and/or shortening extension times should also reduce the size of PCR products. In the present, experiment we used DOP-PCR primers with nine to 12 nucleotides at their 3′ ends and lower annealing temperature (42°C) than have been reported for DOP-PCR with Arabidopsis, mouse, and human (Jordan et al., 2002). DOP-PCR products were ligated into pCR2.1-TOPO plasmid for further analysis.
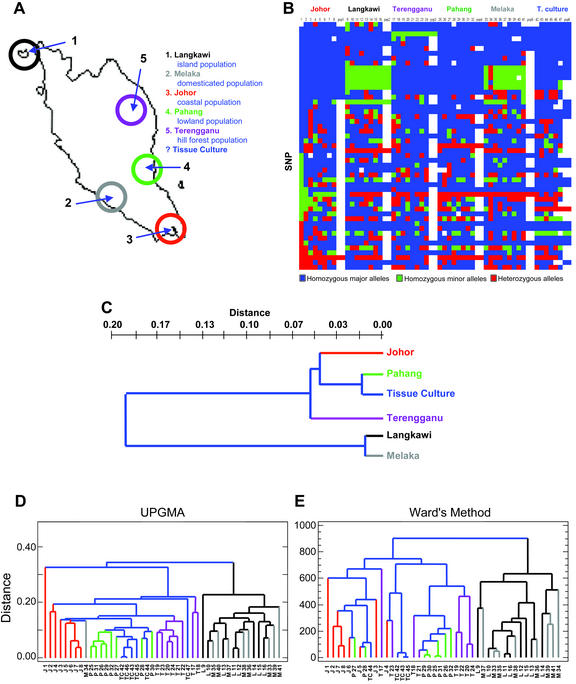
Figure 1.
A, Peninsular Malaysia depicting geographic distribution of populations from which E. longifolia accessions were collected. The geographic origin of materials propagated in tissue culture was not known. Although domesticated populations were similarly maintained on a plantation in southwest Malaysia, the geographic origins of the founders of this population were not known. B, Summary of SNP data for individual plants from six collections. Homozygosity and heterozygosity are indicated by color: blue, homozygous for the major allele; red, homozygous for the minor allele, green, heterozygous; white, no/inconclusive data, except in the final (summary) column for each population, where white indicates the presence of polymorphic loci within that population. C, Dendrogram from unweighted pair group method (UPGMA) cluster analysis based on the unbiased genetic distance in E. longifolia populations of Nei (1978). Color coding corresponds to populations as shown in A. D, Dendrogram from UPGMA cluster analysis based on the unbiased genetic distance between individuals of Nei (1978). Annotations indicate populations from which individuals originated: J, Johor, M, plantation grown; P, Pahang, TC, tissue culture collection; T, Terengganu; and L, Langkawi. E, Dendrogram from Ward's analysis of genetic distance between individuals.
Table I.
Summary of locations and samples studied
| Population | Population Code | Population Type | Latitude/Longitude | No. of Samples |
|---|---|---|---|---|
| Langkawi | L | Island | 6°13′/99°45′ | 8 |
| Melaka | M | Plantation | 2°20′/102°10′ | 9 |
| Johor | J | Coastal | 1°37′/104°15′ | 8 |
| Pahang | P | Lowland forest | 2°50′/103°20′ | 8 |
| Terengganu | T | Hill forest | 5°45′/103°10′ | 8 |
| Tissue culture | TC | Unknown | Unknown | 6 |
Locus-Specific (LS) Primer
A total of 480 clones (96 from each of the five DOP-PCRs) were characterized by end sequencing with universal primers that anneal to the common vector sequences. Terminal sequences were obtained from 263 of these clones, of which 132 (55%) were unique (Table II). DOP-PCR primer 2 produced the highest percentage of unique sequences (68%), whereas the lowest percentage of unique sequences was obtained from DOP-PCR 1 (30%).
Table II.
Summary of unique sequences identified and no. of locus-specific primers used
| DOP Primer | 3′ Enda | Unique/Total Sequencesb | Uniquec | Amplified in >2 Individualsd | Amplifiede |
|---|---|---|---|---|---|
| % | % | ||||
| 1 | aagcgatgt | 26/88 | 30 | 26 | 100 |
| 2 | aagcgatgact | 17/25 | 68 | 17 | 100 |
| 3 | atcgagctgac | 34/54 | 64 | 28 | 82 |
| 4 | accttggaacg | 32/55 | 58 | 29 | 91 |
| 5 | aagcgatgactg | 23/41 | 56 | 23 | 100 |
| Total | 132/263 | 55 | 123 | 95 |
Composition of the arbitrary, specified regions of primers used in DOP-PCR.
No. of unique/total sequences encountered when sequencing cloned DOP-PCR products.
Percentage of DOP-PCR products displaying unique sequences in the cloned set.
No. of unique regions that were successfully amplified from genomic DNA of two or more discrete individuals using locus-specific primers designed according to the unique sequences identified from DOP-PCR products.
Percentage of unique sequences that could be recovered from two or more accessions of E. longifolia using locus-specific primers.
SNP Identification and Validation
One hundred and thirty-two sets of LS primers were made and used for the PCR amplification of fragments from the genomic DNA of three E. longifolia individuals representing three locations (one each from Johor [J1], Langkawi [L9], and Terengganu [T17]). One hundred and twenty-three (95%) of the LS primers produced a single product of the same length from at least two individuals (Table II). The resulting PCR products were sequenced directly (without first cloning into plasmid vectors). From the sequence data of these products, a visual comparison of aligned sequences revealed 144 putative SNPs.
Putative SNPs were validated using allele-specific oligonucleotide (ASO) hybridization. A polymorphism was considered to be an ASO-validated SNP (true SNP) if the LS products hybridized to its corresponding ASO, matching the pattern of the sequencing results. A total of 98 SNPs (71%) were validated (Table III).
Table III.
Summary of SNPs identified from five DOP-PCR
| DOP-PCR | Primer Amplified | Putative SNP | Validated SNP | Validated |
|---|---|---|---|---|
| % | ||||
| 1 | 26 | 42 | 20 | 48 |
| 2 | 17 | 28 | 20 | 69 |
| 3 | 28 | 31 | 24 | 77 |
| 4 | 29 | 24 | 19 | 79 |
| 5 | 23 | 19 | 15 | 79 |
| Total/average | 123 | 144 | 98 (68%) | 71 |
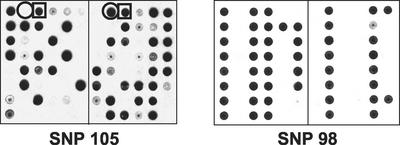
Using the same method, we then scored 47 individuals with respect to 58 ASO-validated SNPs (two of which are shown in Fig. 2). The genotyping results from seven of the SNPs were eliminated because the data were difficult to score due to weak hybridization signal or failure of the ASO to hybridize in most of the samples tested.
Figure 2.
Examples of SNP genotyping of 47 E. longifolia samples. In each case, identical blots were prepared carrying LS-PCR products from each of the different accessions; then one blot was hybridized with the ASO for the first allele, and the second blot was hybridized with the ASO for the second allele. The circle indicates an individual that is homozygous for the second allele, whereas the box adjacent to it demonstrates that another accession was heterozygous for these two alleles. The lack of heterozygosity detected with SNP98 illustrates how some SNPs reported starkly different results.
SNP Genotyping Data
Data generated from SNP genotyping showed a high degree of polymorphism between individuals (Fig. 1B). We categorized the SNP data according to the alleles observed, homozygous (minor and major) alleles, and heterozygous alleles. For each SNP, the more commonly occurring alleles were called major alleles, and the less common alleles found were categorized as minor (rare) alleles. Samples that contained both alleles were scored as heterozygous.
Genetic Diversity
Overall, of all the loci tested 49% to 75% (average = 64%) were polymorphic within a population (Fig. 1B; Table IV). The lowest number of polymorphic loci (and therefore presumably the least genetic diversity) was observed in tissue culture samples, and the highest frequency of polymorphic loci was observed in the Melaka (plantation) population. Nei's (1978) mean expected heterozygosity was higher than observed with an average of He = 0.216 (±0.029) and Ho = 0.182 (±0.035), respectively. The Pahang population had the lowest heterozygosity, He = 0.177 (±0.028), whereas the highest heterozygosity was from the Johor population, He = 0.246 (±0.028). In four of the populations, the value of Ho was lower than He, indicating an excess of homozygotes in these groups. However, in the cases of the Pahang and tissue culture populations, the differences between the observed and the expected heterozygosities were not significantly different.
Table IV.
Summary of genetic diversity in E. longifolia populations
| Population | Na | Aa | P | Ho | He |
|---|---|---|---|---|---|
| Johor | 7.9 (0.0) | 1.7 (0.1) | 70.6 | 0.177 (0.032) | 0.246 (0.028) |
| Langkawi | 8.0 (0.0) | 1.6 (0.1) | 64.7 | 0.189 (0.035) | 0.227 (0.030) |
| Terengganu | 8.0 (0.0) | 1.7 (0.1) | 66.7 | 0.177 (0.029) | 0.233 (0.029) |
| Pahang | 7.9 (0.1) | 1.6 (0.1) | 56.9 | 0.172 (0.038) | 0.177 (0.028) |
| Melaka | 8.9 (0.1) | 1.7 (0.1) | 74.5 | 0.198 (0.035) | 0.230 (0.027) |
| Tissue culture | 5.7 (0.1) | 1.5 (0.1) | 49.0 | 0.178 (0.039) | 0.182 (0.030) |
| Mean | 7.7 (0.05) | 1.6 (0.1) | 63.7 | 0.182 (0.03) | 0.216 (0.03) |
Na, Mean sample size per locus; Aa, average no. of alleles per polymorphic locus; P, percentage of polymorphic loci (95% criterion); Ho, observed heterozygosity; and He, expected heterozygosity. Values in parentheses are ses.
Total genetic diversity (Ht) and gene diversity within populations at polymorphic loci averaged 0.288 and 0.219, respectively (Table V). The mean genetic differentiation between populations (Gst) was 0.240 indicating that about 24% of the observed genetic diversity was due to variation between E. longifolia populations and the remaining 76% was a function of genetic differentiation among plants within populations.
Table V.
Summary of G-statistic (Nei, 1978) calculated from 51 polymorphic loci over six populations
| Locus | Ho | Hs | Ht | Dst | Gst |
|---|---|---|---|---|---|
| SNP01 | 0.060 | 0.058 | 0.059 | 0.001 | 0.025 |
| SNP02 | 0.021 | 0.021 | 0.021 | 0.000 | −0.002 |
| SNP03 | 0.400 | 0.351 | 0.384 | 0.033 | 0.086 |
| SNP04 | 0.042 | 0.078 | 0.081 | 0.003 | 0.042 |
| SNP05 | 0.282 | 0.240 | 0.245 | 0.005 | 0.020 |
| SNP06 | 0.000 | 0.000 | 0.278 | 0.278 | 1.000 |
| SNP07 | 0.206 | 0.160 | 0.186 | 0.026 | 0.142 |
| SNP08 | 0.736 | 0.452 | 0.501 | 0.049 | 0.098 |
| SNP09 | 0.188 | 0.175 | 0.172 | −0.003 | −0.018 |
| SNP10 | 0.208 | 0.242 | 0.252 | 0.010 | 0.041 |
| SNP11 | 0.424 | 0.324 | 0.336 | 0.012 | 0.036 |
| SNP12 | 0.083 | 0.219 | 0.246 | 0.028 | 0.113 |
| SNP13 | 0.153 | 0.190 | 0.268 | 0.077 | 0.289 |
| SNP14 | 0.407 | 0.386 | 0.374 | −0.012 | −0.031 |
| SNP15 | 0.021 | 0.021 | 0.438 | 0.417 | 0.952 |
| SNP16 | 0.104 | 0.098 | 0.100 | 0.001 | 0.013 |
| SNP17 | 0.104 | 0.227 | 0.248 | 0.021 | 0.083 |
| SNP18 | 0.813 | 0.481 | 0.484 | 0.003 | 0.006 |
| SNP19 | 0.060 | 0.061 | 0.059 | −0.002 | −0.033 |
| SNP20 | 0.019 | 0.019 | 0.438 | 0.420 | 0.957 |
| SNP21 | 0.329 | 0.325 | 0.352 | 0.027 | 0.075 |
| SNP22 | 0.063 | 0.063 | 0.061 | −0.001 | −0.024 |
| SNP23 | 0.039 | 0.040 | 0.039 | −0.001 | −0.015 |
| SNP24 | 0.271 | 0.358 | 0.351 | −0.007 | −0.019 |
| SNP25 | 0.123 | 0.129 | 0.186 | 0.057 | 0.307 |
| SNP26 | 0.000 | 0.200 | 0.453 | 0.253 | 0.559 |
| SNP27 | 0.000 | 0.072 | 0.082 | 0.009 | 0.111 |
| SNP28 | 0.366 | 0.339 | 0.371 | 0.032 | 0.087 |
| SNP29 | 0.000 | 0.091 | 0.452 | 0.361 | 0.799 |
| SNP30 | 0.019 | 0.072 | 0.457 | 0.386 | 0.843 |
| SNP31 | 0.083 | 0.435 | 0.497 | 0.062 | 0.125 |
| SNP32 | 0.195 | 0.410 | 0.461 | 0.051 | 0.110 |
| SNP33 | 0.049 | 0.120 | 0.125 | 0.005 | 0.043 |
| SNP34 | 0.060 | 0.326 | 0.342 | 0.016 | 0.047 |
| SNP35 | 0.096 | 0.189 | 0.178 | −0.011 | −0.062 |
| SNP36 | 0.451 | 0.476 | 0.496 | 0.021 | 0.041 |
| SNP37 | 0.188 | 0.206 | 0.233 | 0.027 | 0.115 |
| SNP38 | 0.495 | 0.364 | 0.402 | 0.037 | 0.093 |
| SNP39 | 0.019 | 0.019 | 0.438 | 0.420 | 0.957 |
| SNP40 | 0.102 | 0.157 | 0.170 | 0.013 | 0.078 |
| SNP41 | 0.021 | 0.057 | 0.062 | 0.005 | 0.077 |
| SNP42 | 0.421 | 0.356 | 0.499 | 0.143 | 0.286 |
| SNP43 | 0.097 | 0.200 | 0.256 | 0.055 | 0.216 |
| SNP44 | 0.160 | 0.445 | 0.507 | 0.062 | 0.123 |
| SNP45 | 0.502 | 0.424 | 0.485 | 0.061 | 0.126 |
| SNP46 | 0.113 | 0.162 | 0.212 | 0.051 | 0.239 |
| SNP47 | 0.132 | 0.232 | 0.246 | 0.014 | 0.059 |
| SNP48 | 0.417 | 0.461 | 0.505 | 0.045 | 0.088 |
| SNP49 | 0.021 | 0.142 | 0.134 | −0.008 | −0.063 |
| SNP50 | 0.021 | 0.063 | 0.062 | −0.001 | −0.018 |
| SNP51 | 0.083 | 0.415 | 0.385 | −0.030 | −0.078 |
| Overall | 0.182 | 0.219 | 0.288 | 0.069 | 0.240 |
Hs, Within population genetic diversity. Dst, Average gene diversity among populations.
Genetic Distance
The mean genetic distances D (Nei, 1978) between populations was 0.119, with a range of 0.012 to 0.229 (Table VI). UPGMA cluster analysis between populations using Nei's unbiased genetic distance revealed two distinct clusters (Fig. 1C). The first cluster comprised Johor, Pahang, tissue culture samples, and the more distantly related individuals from the Terengganu population. The Langkawi and Melaka populations clustered in a second group, suggesting that the plantation material was derived from the island population. Similar results were obtained when cluster analysis was performed on the individual plants (Fig. 1, D and E). With very few exceptions (e.g. M34), these plants clustered into cohesive groups corresponding to their distinct geographic origins. When Ward's method was used for cluster analysis, all of the plantation-grown individuals clustered with the Langkawi population.
Table VI.
Estimates of mean genetic distance (Nei, 1978) between six populations of E. longifolia
| Population | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1. Johor | – | |||||
| 2. Langkawi | 0.229 | – | ||||
| 3. Terengganu | 0.058 | 0.222 | – | |||
| 4. Pahang | 0.031 | 0.190 | 0.038 | – | ||
| 5. Melaka | 0.179 | 0.012 | 0.171 | 0.156 | – | |
| 6. Tissue culture | 0.058 | 0.201 | 0.059 | 0.014 | 0.170 | – |
DISCUSSION
In general, the present study shows that populations of E. longifolia possess a high level of genetic diversity (P = 64%, He = 0.216 ± 0.029). The mean heterozygosity between populations of E. longifolia was higher compared with other regional, tropical, long-lived trees reported from isozyme studies (He = 0.125+0.012; Hamrick et al., 1992), conifers (He = 0.145+0.008; Hamrick et al., 1992), eucalyptus (He = 0.182; Moran and Hopper, 1987), and Acacia auriculformis (He = 0.081, Wickneswari and Norwati, 1993). However, the value for E. longifolia is lower than for timber species such as Shorea leprosula (He = 0.369+0.025; Lee et al., 2000b), Stemonoporous oblingifolius (He = 0.297+0.039; Murawski and Bawa, 1994), and Dryobalanops aromatica (He = 0.459+0.117; Lee et al., 2000a). The validity of this comparison may be questioned because the studies were made using different molecular markers, i.e. SNPs versus isozymes. In isozyme studies, the level of genetic diversity between populations is determined by allele frequency, mean number of alleles, heterozygosity, percentage of polymorphic loci, etc. In the present study, the level of genetic diversity was determined by the mean of heterozygosity and percentage of polymorphic loci. Allele frequency and mean number of polymorphic alleles were excluded from our calculation because the technique was developed to detect only two different alleles.
The perceived diversity within E. longifolia might also be affected by the small number of individuals per population used in the study. According to electrophoretic surveys by Nei and Roychoudhury (1974), Nei (1978), and Gorman and Renzi (1979), a large number of loci should be examined if the number of individuals per locus is small. In fact, a few individuals are sufficient for estimating heterozygosity if a sufficiently large number of loci are examined (Gorman and Renzi, 1979). Although it is difficult to compare the data from electrophoretic and SNP-based studies (the former being a protein-based assay and the latter focusing on DNA), preliminary data from recent electrophoretic assays (Norfiza et al., 2001) support the conclusion that SNP markers provide more sensitive assays of genetic diversity.
The mean degree of population differentiation in E. longifolia (Gst = 0.24) is higher than wind-pollinated species such as conifers (Gst = 0.05; Matheson et al., 1989) and S. leprosula (Gst = 0.085; Lee et al., 2000b), and insect-pollinated eucalyptus (Gst = 0.10–0.12; Moran and Hopper, 1987). Low values for population differentiation suggest that effective and extensive seed migration in these species minimizes fragmentation of these populations. In contrast, a high Gst value in E. longifolia might reflect poor pollen movement and the limitations of gravity-based seed dispersal in this species, both of which would be expected to genetically isolate individual populations.
Cluster analysis between populations of E. longifolia revealed two major groups, with the plantation population being closely related to that from Langkawi, suggesting that individuals or seedlings from plantation might have originated from Langkawi. The populations from Pahang and the tissue culture collection are closely related to the population from Johor, whereas the population from Terengganu forms a somewhat more distantly related subpopulation within the second group. The distinction between the population from Langkawi and the other two groups could be explained by its geographical isolation (it is an island population), which is expected to limit gene exchange between populations.
Similar results were observed in cluster analysis among individuals (Fig. 1, D and E). An interesting finding was observed with tissue culture samples, where callus samples that are capable of producing somatic embryos cluster together (TC42, TC43, TC44, TC45, and TC46), whereas a nonembryogenic callus line (TC47) did not (data on regenerability not shown). Although many more individuals should be tested in this regard before a firm correlation can be drawn, this observation suggests that SNPs could be employed as molecular markers for predicting whether a particular accession of E. longifolia will be amenable to regeneration via somatic embryogenesis. This result would be very valuable for micropropagation of this sought-after forest species.
Another strategy to identify SNPs among the genomes of undercharacterized species might be to collect sequence information from the 3′ ends of cloned expressed sequence tags (ESTs; e.g. Ching and Rafalski, 2002). SNPs identified in this manner would offer the advantages of (a) the sequences not being anonymous (i.e. the expressed genes with which they are associated may be identified by a BLAST search [Altschul et al., 1997]); and (b) a higher likelihood that the SNPs initially identified are allelic. Although such a method might be useful for identifying SNPs in E. longifolia, the lack of a suitable EST library—or for that matter a reliable RNA extraction protocol—made the DOP-PCR strategy preferable because it requires only vanishingly small amounts of genomic DNA. Other advantages of the DOP-PCR method include (a) the fact that it provides a simple method for PCR amplification of all of the markers in a single batch, thus providing a template for genotyping; and (b) the fact that DOP-PCR may also allow for identification of SNPs from regions of the genome that are not particularly gene rich, enabling more complete coverage of the genome. Although an EST-based strategy might have the benefit of permitting gene identification, this would still not allow the assignment of the newly discovered SNPs to a map location in E. longifolia because gene locations are completely uncataloged in this species, and very few assumptions could be made about synteny between E. longifolia and other species. Nonetheless, some caveats associated with the DOP-PCR technique should be acknowledged. Because no controlled segregating populations of these primarily wild-grown plants were available at the time of this study, we were unable to carry out allelism tests for the SNPs identified by the DOP-PCR method. We hope to work with controlled populations in the future to address this issue and to begin constructing genetic maps for this species. From previous results with human, mouse, and Arabidopsis DNA (Jordan et al., 2002), we found the DOP-PCR technique favors the identification of DNA fragments and SNPs that are present in single copies. These studies also showed that the SNPs identified through DOP-PCR segregated in a Mendelian manner. However, the assumption that these trends hold true in E. longifolia has not yet been tested.
From this study, we have identified 51 SNPs that can be used as genetic markers in E. longifolia. Cluster analysis showed that diversity among the different accessions of E. longifolia corresponds well with the geographic origins of each population. These markers should prove useful in preserving genetic diversity among domesticated populations of E. longifolia. These SNPS may also be developed as predictive markers for useful phenotypes such as regenerability.
MATERIALS AND METHODS
Plant Material
A total of 47 Eurycoma longifolia Jack. individuals representing four natural populations, one domesticated population, and materials that had been propagated in tissue culture were collected in Peninsular Malaysia. Leaf materials from six to nine trees were collected from each population as shown in Figure 1A. The locations of each population are listed in Table I. Total genomic DNA from leaf tissue was extracted using a modified cetyl-trimethyl-ammonium bromide method (Doyle and Doyle, 1987).
DOP-PCR Reactions
DOP-PCR primers for whole genome amplification used in the experiments were designed as described by Telenius et al. (1992). The primer includes a C/G rich 5′ anchor (CTCGAG), six “N” where N is A, C, G, or T (4,096-fold degeneracy) and nine to 12 arbitrary, specified nucleotides at the 3′ end of each primer. The DOP-PCR reaction mix was as follows: 50 ng of genomic DNA, 0.2 mm dNTPs, 2.5 units of Amplitaq DNA polymerase (Applied Biosystems, Foster City, CA), 3.0 μm degenerate primer, and 10× PCR buffer in a total of 50-μL reaction volume. The cycling profile was as follows: 94°C for 1 min; five cycles of 94°C for 30 s, 42°C for 45 s, and 72°C for 1.5 min; 35 cycles of 94°C for 30 s, 58°C for 45 s, and 72°C for 1.5 min; 72°C for 10 min; and hold at 4°C. The sequences of the DOP-PCR primers are given in Table II. All degenerate primers were purchased from Invitrogen (Carlsbad, CA). Fifty microliters (250–1,500 bp) of five DOP-PCR mixtures were gel purified using Qiaquick PCR purification kit (Qiagen USA, Valencia, CA) and shotgun cloned according to the protocol of the TOPO XL cloning kit (Invitrogen).
End Sequencing of Cloned DOP-PCR Products
The termini of 480 cloned DOP-PCR products inserts were sequenced with the universal primers (M13 Forward and M13 reverse) in 50-μL reactions: 0.2 mm dNTPs, 2.0 μL of overnight culture, 1.25 units of Amplitaq DNA polymerase, 0.4 μm each primer, and 5.0 μL of 10× buffer. The PCR cycles were 94°C for 5 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; 72°C for 10 min; and hold at 4°C. The entire products were loaded onto 1.1% (w/v) agarose gel, electrophoresed, and gel purified using Qiaquick 96 PCR purification kit (Qiagen USA). The purified PCR products were sent for automated sequencing at MWG Biotech Inc. (High Point, NC).
LS Primer Design
After sequencing of the cloned DOP-PCR products, the 20 bp of sequence immediately adjacent to the DOP-PCR primer sequences at either end of all clones were tabulated and sorted. Duplicate clones were eliminated, producing a list of unique loci. Pairs of LS primers were designed based on these unique sequences such that one primer of each pair contained a 5′ M13 forward tag (TGT AAA ACG ACG GCC AGT) and the other primer contained a 5′ M13 reverse tag (CAG GAA ACA GCT ATG ACC). Primers were synthesized by MWG Biotech Inc.
SNP Identification
The LS primer pairs were used for PCR amplification of fragments from three individuals representing different E. longifolia populations. PCR was carried out in 50-μL reactions: 1 μL of appropriate DOP-PCR products as a template DNA, 0.2 mm dNTPs, 2.5 units of Amplitaq DNA polymerase, 0.4 μm each primer, and 5 μL of 10× buffer. A touch down cycling profile was as follows: 94°C for 1 min; 40 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 1 min; 10 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min; 72°C for 10 min; and hold at 4°C. The entire PCR products were separated on 1.1% (w/v) agarose gels and purified with Qiaquick 96 PCR purification kit (Qiagen USA). The purified PCR products were sequenced by MWG Biotech Inc. using the M13 forward and reverse primers. Sequence data was analyzed with the Lasergene Seq-Man II program (DNASTAR, Inc., Madison, WI) to find putative SNPs.
SNP Validation and Genotyping
For each allele of each putative SNP, a 17-mer oligonucleotide (MWG Biotech, Inc.) centered on the putative polymorphic nucleotide was made. Each putative SNP was validated by ASO hybridization to the LS-PCR product and DOP-PCR product mixtures from three individuals following the method of Jordan et al. (2002). These ASO-validated SNPs were used to genotype 47 individuals of E. longifolia (supplemental data, available at www.plantphysiol.org).
Data Analysis: SNP Genotyping
Data generated from 47 individuals were entered in an Excel spreadsheet (Microsoft, Redmond, WA) and were categorized as homozygous (minor and major) and heterozygous alleles. These are depicted in Figure 1B. In a small number of cases, data for individual SNPs were inconclusive.
Genetic Diversity
Data were scored as presence and absence of major and minor alleles (homozygous) or presence of both alleles (heterozygous) for each sample. The level of genetic diversity and cluster analysis was conducted with the program GENEPOP v.3.2a (Raymond and Rousset, 1995). Percentage polymorphic loci and mean heterozygosity between populations were estimated using Biosys-1 (Swofford and Selander, 1981). The Ht at the polymorphic loci and the Gst were determined following the G-statistic of Nei (1978) with the assistance of the FSTAT computer program (v2.1.9; http://www.unil.ch/izea/softwares/fstat.html). Nei's (1978) unbiased distance (D) was estimated between populations and individuals to generate average linkage clustering using the UPGMA. A phenogram of Nei's (1978) genetic distance was prepared using Biosys-1. Cluster analysis using Ward's method was also carried out on data from individual plants using the StatGraphics Software package (Manugistics, Rockville, MD).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Fadhilah Zainudin and Nor Hasnida for providing samples of cultured E. longifolia and Dr. Azizol Abdul Kadir, Dr. Mohd. Ilham Adenan, and colleagues at the Forest Research Institute of Malaysia for supplying plant material for this study and for technical assistance.
Footnotes
This work was supported by the Government of Malaysia and the Malaysia-Massachusetts Institute of Technology Biotechnology Partnership Program.
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.012492.
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, O'Neill MJ, Phillipson JD, Warhurst DC. Plants as source of antimalarial drugs: Part 3. Eurycoma longifolia. Planta Med Apr. 1986;2:105–107. doi: 10.1055/s-2007-969091. [DOI] [PubMed] [Google Scholar]
- Ching A, Rafalski A. Rapid genetic mapping of ESTs using SNP pyrosequencing and indel analysis. Cell Mol Biol Lett. 2002;7:803–810. [PubMed] [Google Scholar]
- Creswell A, Sackville Hamilton NR, Roy AK, Viegas BMF. Use of amplified fragment length polymorphism markers to assess genetic diversity of Lolium sp. from Portugal. Mol Ecol. 2001;10:229–241. doi: 10.1046/j.1365-294x.2001.01170.x. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Gomory S, Yakovlev I, Zhelev P, Jedinakova J, Paule L. Genetic differentiation of oak populations within the Quercus robor/Quercus petraea complex in Central and Eastern Europe. Heredity. 2001;86:557–563. doi: 10.1046/j.1365-2540.2001.00874.x. [DOI] [PubMed] [Google Scholar]
- Gorman GC, Renzi J., Jr Genetic distance and heterozygosity estimates in electrophoretic studies: effects of sample size. Copeia. 1979;1979:242–249. [Google Scholar]
- Hamrick JL, Godt MJW, Sherman-Broyles SL. Factor influencing levels of genetic diversity in woody plant species. New For. 1992;6:96–124. [Google Scholar]
- Jordan B, Charest A, Dowd JF, Blumenstiel JP, Yeh RF, Osman A, Housman DE, Landers JE. Genome complexity reduction for SNP genotyping analysis. Proc Nat Acad Sci USA. 2002;99:2924–2927. doi: 10.1073/pnas.261710699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SL, Ang KC, Norwati M. Genetic diversity of Dryobalanops aromatica gaertn.F. (Dipterocarpaceae) in peninsular Malaysia and its pertinence to genetic conservation and tree improvement. For Genet. 2000a;7:209–217. [Google Scholar]
- Lee SL, Wickneswari R, Mahani MC, Zakri AH. Genetic diversity of a tropical tree species, Shorea leprosula miq. (Dipterocarpaceae), in Malaysia: implications for conservation of genetic resources and tree improvement. Biotropica. 2000b;32:213–224. [Google Scholar]
- Livak KJ, Marmaro J, Todd JA. Towards fully automated genome-wide polymorphism screening. Nat Genet. 1995;9:341–342. doi: 10.1038/ng0495-341. [DOI] [PubMed] [Google Scholar]
- Maguire TL, Saenger P, Baverstocks P, Henry R. Microsatellites analysis of genetic structure in the mangrove sp. Avicennia marina (Forsk.) Vierh. (Avicenniaceae) Mol Ecol. 2000;9:1853–1862. doi: 10.1046/j.1365-294x.2000.01089.x. [DOI] [PubMed] [Google Scholar]
- Matheson AC, Bell JC, Barnes RD. Breeding systems and genetic structure in some central American pine populations. Silvae Genet. 1989;38:107–113. [Google Scholar]
- Moran GF, Hopper SD. Conservation of the genetic resources of rare and widespread eucalyptus in remnant vegetation. In: Saunders DA, Arnold GW, Burbidge AA, Hopkins AJM, editors. Nature Conservation: The Role of Remnants of Native Vegetation. Sydney: Surrey Beatty and Sons; 1987. pp. 151–162. [Google Scholar]
- Murawski DA, Bawa KS. Genetic structure and mating system of Stemonoporus oblongifolius (Dipterocarpaceae) in Sri Lanka. Am J Bot. 1994;81:155–160. [Google Scholar]
- Nassar JM, Hamricks JL, Fleming TH. Genetic variation and population structure of the mixed-mating cactus, Melacactus curvispinus (Cactaceae) Heredity. 2001;87:69–79. doi: 10.1046/j.1365-2540.2001.00910.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Roychoudhury AK. Sampling variance of heterozygosity and genetic distance. Genetics. 1974;76:379–390. doi: 10.1093/genetics/76.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norfiza MR, Norwati M, Mohd Rosli H, Mohd Faizal K, Osman A. Genetic diversity of Eurycoma longifolia in peninsular Malaysia. In: Latif Ibrahim A, editor. Proceedings of the Second Malaysia-Massachusetts Institute of Technology Biotechnology Partnership Programme Symposium, 5–6 November 2001, Kuala Lumpur, Malaysia. Kuala Lumpur: The National Biotechnology Directorate; 2001. p. 1. [Google Scholar]
- Pennisi E. Sifting through and making sense of genome sequences. Science. 1998;280:1692–1693. doi: 10.1126/science.280.5370.1692. [DOI] [PubMed] [Google Scholar]
- Quagliaro GM, Vischr M, Tyrka M, Olivieri AM. Identification of wild and cultivated sunflower for breeding purposes by AFLP markers. J Hered. 2001;92:38–42. doi: 10.1093/jhered/92.1.38. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2) population genetic software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- Swofford DL, Selander RB. BIOSYS-1: a FORTRAN program for the comprehensive analysis of electrophoretic data in population genetics and systematics. J Hered. 1981;72:281–283. [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Norderskjold M, Ponder BA, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- Walter R, Epperson BK. Geographic pattern of genetic variation in Pinus resinosa: Area of greatest diversity is not the origin of postglacial populations. Mol Ecol. 2001;10:103–111. doi: 10.1046/j.1365-294x.2001.01177.x. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Gandour G, Perkins N, Winchester E, Lander ES. Large-scale identification, mapping, & genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Wickneswari R, Norwati M. Genetic diversity of natural populations of Acacia auriculiformis. Aust J Bot. 1993;41:65–77. [Google Scholar]