Abstract
The lipid monogalactosyl diacylglycerol (MGD) is a major structural component of photosynthetic membranes in chloroplasts. Its formation is catalyzed by the enzyme MGD synthase. In many plants, MGD derives from two different biosynthetic pathways: the prokaryotic pathway, which operates entirely within the plastid, and the eukaryotic pathway, which involves steps in the endoplasmic reticulum. Here, we describe the identification and characterization of an Arabidopsis mutant with a defective MGD synthase gene (MGD1). The mutant was identified in a screen of T-DNA lines for individuals with defects in chloroplast biogenesis. It has a yellow-green phenotype that correlates with a ≈50% deficiency in total chlorophyll per plant. A single T-DNA insertion is located adjacent to the transcription initiation site of the MGD1 gene, and the abundance of MGD1 mRNA is reduced by 75% compared with wild type. Correlation between steady-state MGD1 transcript levels and MGD synthase activity (also reduced by 75% in mgd1) suggests that MGD1 is the most important MGD synthase in green tissues. The amount of MGD in mutant leaves is reduced by 42% compared with wild type. MGD from the mutant contains 23% less 16:3 fatty acid and 10% more 18:3 fatty acid. Because 16:3 is a characteristic feature of MGD from the prokaryotic pathway, it is possible that MGD1 operates with some preference in the prokaryotic pathway. Finally, the MGD-deficiency of mgd1 is correlated with striking defects in chloroplast ultrastructure, strongly suggesting a unique role for MGD in the structural organization of plastidic membranes.
Galactose-containing lipids are the predominant nonproteinaceous components of photosynthetic membranes in plants, algae, and a variety of bacteria. The two most common galactolipids are monogalactosyl diacylglycerol (MGD) and digalactosyl diacylglycerol (DGD). In plants, MGD and DGD occur exclusively in plastidic membranes, where they account for about 50 and 20 mol% of the lipid matrix, respectively (1). Up to 80% of all lipids in plants are associated with photosynthetic membranes, and MGD is widely considered to be the most abundant membrane lipid on earth. Most vegetables and fruits in human and animal diets are rich in galactolipids, and their breakdown products represent an important dietary source of galactose and polyunsaturated fatty acids (2, 3).
Galactolipids play an important role in the organization of photosynthetic membranes. The abundance and physical properties of MGD make it particularly important in this respect. Its small galactose head group and large unsaturated fatty acid chains give it a cone-like molecular shape and a consequent predisposition to form nonlamellar, hexagonal-phase aggregates (4). The molecular shape of MGD may be important for the structural organization of thylakoid membranes. Because much of the photosynthetic apparatus is embedded within thylakoids, the lipids that make up these membranes are of profound importance. Evidence also suggests that MGD is more directly involved in certain photosynthetic reactions (5–9). Because photosynthesis is the only significant mechanism of energy input into the living world, the importance of all factors that contribute to the efficiency of the process are inestimable.
The final step in MGD biosynthesis occurs in the plastid envelope and is catalyzed by MGD synthase (EC 2.4.1.46). This enzyme transfers d-galactose from UDP-galactose to sn-1,2-diacylglycerol (DAG) (10). Cucumber and spinach MGD synthases have been described in detail (11, 12), and similar Arabidopsis sequences are present in the GenBank database (12). The DAG used in MGD synthesis is thought to be derived, in Arabidopsis, from two different biosynthetic pathways (13, 14). The prokaryotic pathway operates exclusively in the plastid and produces DAG containing an 18-carbon fatty acid at the sn-1 position of the glycerol backbone, and a 16-carbon fatty acid at the sn-2 position. The eukaryotic pathway involves steps in the endoplasmic reticulum and produces predominantly DAG with 18-carbon fatty acids at both positions. Differing substrate specificities of acyltransferases in the plastid and endoplasmic reticulum are thought to account for the differences between the DAG species produced by each pathway. As a result of this DAG heterogeneity, MGD from the prokaryotic pathway contains 16:3 (hexadecatrienoic acid) at the sn-2 position, whereas MGD from the eukaryotic pathway contains 18-carbon fatty acids at the sn-2 position; both forms contain mostly 18-carbon fatty acids at the sn-1 position. Whereas both pathways operate in plants such as Arabidopsis (designated 16:3 plants), more advanced species such as pea (18:3 plants) have dispensed with the prokaryotic pathway and produce predominantly MGD with 18-carbon fatty acids at both positions only (15).
DGD synthase catalyzes the transfer of galactose from one molecule of MGD to another, producing DGD and DAG in equimolar amounts. The majority of Arabidopsis DGD contains 18:3 in the sn-2 position, suggesting that it is largely derived from the eukaryotic pathway. An Arabidopsis mutant (dgd1) with a pronounced deficiency in DGD lipids (containing only 10% of the wild-type level) was identified by screening for plants with altered leaf lipid composition (16). The DGD1 locus was recently cloned and found to encode a galactosyltransferase-like protein (17). Simultaneous heterologous expression of the DGD1 and a cucumber MGD synthase gene resulted in the reconstitution of the plant galactolipid biosynthetic pathway in Escherichia coli.
In this report, we describe the identification and characterization of an Arabidopsis MGD synthase mutant. The data provide important new information on the operation of galactolipid biosynthetic pathways in 16:3 plants, and demonstrate the significance of MGD lipids for normal chloroplast biogenesis. Possible roles for the different Arabidopsis MGD synthase species are discussed.
Materials and Methods
Plant Materials and Growth Conditions.
All plants described are Arabidopsis thaliana of the Columbia ecotype. The T-DNA construction present in the mgd1 mutant has been described (18). Plants were grown on 1× Murashige and Skoog medium (GIBCO/BRL) containing 1% sucrose, or on soil, in continuous white light. Incubator temperatures were maintained at a constant 20°C. For electron microscopy, plants were grown in vitro in the dark or the light; sample preparation and analysis was carried out according to described procedures (19).
Molecular Analyses.
Total RNA was extracted from plants according to described procedures (20). Reverse transcription was carried out with oligo(dT) primers and Superscript II reverse transcriptase (GIBCO/BRL).
Rapid amplification of cDNA ends (RACE) PCR was carried out with the switch mechanism at the 5′ end of RNA templates RACE cDNA amplification kit (CLONTECH). MGD1 primers used in RACE experiments were 5′-AATCTCTCCCACGACGCGCTTCATC-3′ and 5′-ATGCAAAACCCTTCAACGGTAACCC-3′ at the 5′ end of the gene, and 5′-ATTAGGCAGTGCAAGAGAGTTGAGG-3′ and 5′-TACGGACAAGCTCGTGCATATCATG-3′ at the 3′ end. RACE PCR products were cloned by using the pGEM-T Easy Vector System (Promega) before sequencing.
For reverse transcription–PCR, primers that amplify a product of 967 bp corresponding to the MGD1 mRNA (5′-ATGGTGTTGAAGCTGATCGG-3′ and 5′-TCTTGACCAGCGATGTAACC-3′), and others that amplify a product of 728 bp corresponding to translation initiation factor eIF4E mRNA (5′-AAACCATGGCGGTAGAAGACACTC-3′ and 5′-AAGATCTAGAAGGTTTCAAGCGGTGTAAG-3′) (21) were used. Both sets of primers were included in each PCR, and amplification was performed for 20 cycles. PCR products were analyzed by Southern blotting by using gene-specific radiolabeled probes. Bands were visualized and quantified by using a PhosphorImager (Molecular Dynamics). Presented values for MGD1 expression are means ± SE from four independent experiments.
Database searches were performed at the U.S. National Center for Biotechnology Information by using the blast program (22). The sequence and annotation of bacterial artificial chromosome clone F28M20 was provided by the EU Arabidopsis sequencing project. Amino acid sequence alignment was performed by using the megalign program (DNAstar, Madison, WI) by the Clustal method.
Biochemical Analyses.
Lipids were extracted from leaves of 5-week-old plants and separated by TLC according to previously described procedures (16). Bands corresponding to each lipid class were isolated and used to prepare fatty acid methyl esters by incubating in 1 M HCl in methanol at 80°C for 30 min. Methyl esters were quantified by gas chromatography by using myristic acid as an internal standard (23).
Chloroplasts used in the labeling experiments to determine MGD and DGD synthase activities were isolated as described (16, 24). Aliquots equivalent to 250 μg of chlorophyll were resuspended in 0.3 M sorbitol/20 mM Tricine-KOH (pH 7.6)/5 mM MgCl2/2.5 mM EDTA, and incubated with 0.5 μCi UDP-[14C]galactose (329 mCi/mmol; Amersham International) in a total volume of 130 μl. Lipids were extracted from aliquots of 25 μl, removed after increasing incubation periods, and separated by TLC (23). The radioactivities of bands corresponding to MGD and DGD were measured by scintillation counting. These data were used to determine the rate of [14C]galactose incorporation into each lipid by regression analysis.
Results
Identification of the mgd1 Mutant.
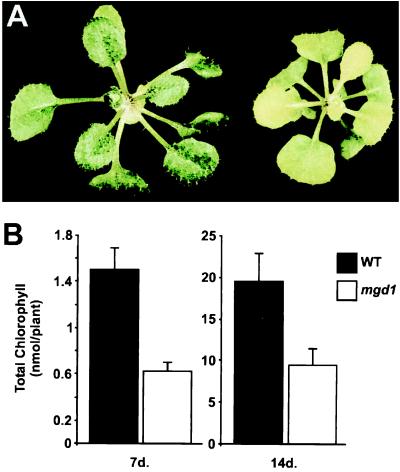
The Arabidopsis MGD synthase 1 (mgd1) mutant was identified in a screen of T-DNA-mutagenized plants for individuals with defects in chloroplast biogenesis (25). Mutant mgd1 plants have a chlorotic phenotype (Fig. 1A) that is inherited in a Mendelian fashion consistent with the presence of a single recessive mutation (data not shown). The mutation is associated with reductions in the amount of chlorophyll (Fig. 1B). Total chlorophyll contents for wild-type and mutant plants grown in vitro for 1 or 2 weeks were determined. Mutant plants were found to contain ≈50% less chlorophyll than wild-type plants at each time point (Fig. 1B) and the chlorotic phenotype of the mutant was observed to persist throughout development. However, there is some leaf heterogeneity in older plants with younger leaves being more pale.
Figure 1.
Visible phenotype of the mgd1 mutant. (A) Photograph showing 23-day-old seedlings. Plants were grown in vitro in continuous white light. The wild type is shown on the left and the mgd1 mutant is shown on the right. (B) Chlorophyll measurements. Plants were grown in vitro in continuous white light for either 7 or 14 days. Chlorophyll was extracted by using N,N′-dimethylformamide and determined photometrically according to described procedures (26). Presented values are means from six measurements of 10 seedlings (7 days) or 13 measurements of 2 seedlings (14 days) ± SE.
Characterization of the mgd1 Locus.
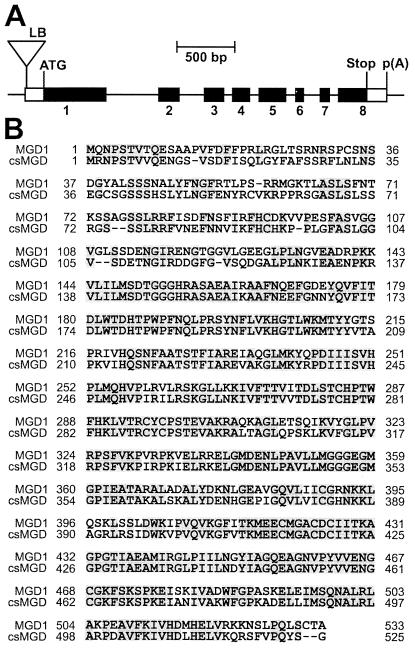
The mgd1 mutation cosegregated with a single T-DNA-associated herbicide-resistance marker over a backcross-derived F2 population of 100 plants (data not shown). This demonstrated tight genetic linkage of the two loci and indicated that the T-DNA insertion is most likely responsible for the mgd1 phenotype. Arabidopsis DNA flanking the T-DNA left border was isolated by using thermal asymmetric interlaced PCR (TAIL-PCR) (27) and found to correspond to a sequenced region of the genome on chromosome 4 corresponding to bacterial artificial chromosome clone F28M20 (GenBank accession no. AL031004). Bacterial artificial chromosome sequence annotation revealed the T-DNA insertion to be upstream (at position −148 relative to the translation initiation codon) of a predicted gene encoding a putative MGD synthase (Fig. 2A). Cosegregation of the mutant MGD synthase gene (mgd1) and the chlorotic phenotype over a population of 50 plants was demonstrated by using PCR, strongly suggesting a causal relationship between the T-DNA insertion and the mutant phenotype.
Figure 2.
Structure of the MGD1 gene. (A) Schematic representation of the gene disrupted in the mgd1 mutant. The depicted gene encodes the MGD synthase enzyme, MGD1. Filled boxes correspond to translated regions of the MGD1 transcript; open boxes correspond to untranslated regions of the MGD1 transcript. Exons are numbered from 1 to 8. The T-DNA insertion is represented by a triangle. Indicated features include the T-DNA left border (LB), the translation initiation codon (ATG), the translation termination codon (Stop), and the polyadenylation site [p(A)]. (B) Alignment of the MGD1 and csMGD1 amino acid sequences. Dashes indicate gaps introduced to maximize alignment. Residues conserved between both sequences are shaded.
To accurately determine the structure of the MGD1 ORF, a full-length MGD1 cDNA (GenBank accession no. AF241797) was isolated by using 5′ and 3′ RACE-PCR. The ORF encoded within the MGD1 cDNA matched exactly the predicted ORF (F28M20.30) found within bacterial artificial chromosome clone F28M20. Three different 5′ RACE-PCR products corresponding to three different transcription initiation sites were identified. The transcription initiation sites are located at positions −159, −113, and −58 relative to the translation initiation codon. The T-DNA insertion lies downstream of the transcription initiating site at position −159, but just upstream of those at positions −113 and −58. Two different polyadenylation sites were identified; these are located 133 bp and 168 bp downstream of the translation termination codon. The MGD1 schematic shown in Fig. 2A corresponds to the longest possible transcript.
The sequence of the protein encoded by the MGD1 cDNA (GenBank accession no. CAA19745) is shown in Fig. 2B. It has been aligned with the sequence of a well-characterized MGD synthase from cucumber (11) (GenBank accession no. AAC49624; referred to here as csMGD1). Two additional Arabidopsis MGD synthase sequences (MGD2 and MGD3) are present in the GenBank database; MGD2 (GenBank accession no. CAA04005) and MGD3 (GenBank accession no. AAD28678) have previously been referred to as atMGD type B and atMGD type C, respectively (12). MGD1 shares 71.4% amino acid sequence identity with csMGD1, but only 53.2% identity with the MGD2 and 55.4% identity with the MGD3.
Analysis of MGD1 Expression in the Mutant.
To assess the severity of the mgd1 mutation directly, MGD1 mRNA levels in wild-type and mutant plants were compared by using reverse transcription–PCR; MGD1 expression could not be detected on Northern blots (data not shown). PCR products were analyzed by Southern blotting after 20 cycles of amplification, conditions shown previously to produce quantitative data (28). MGD1 expression data were normalized by using translation initiation factor eIF4E (21) expression data. Expression of MGD1 mRNA in 10-day-old plants was determined to be 100.0 ± 19.9 for wild-type plants, and 25.1 ± 4.6 for mgd1 mutant plants (arbitrary units).
Characterization of the mgd1 Biochemical Phenotype.
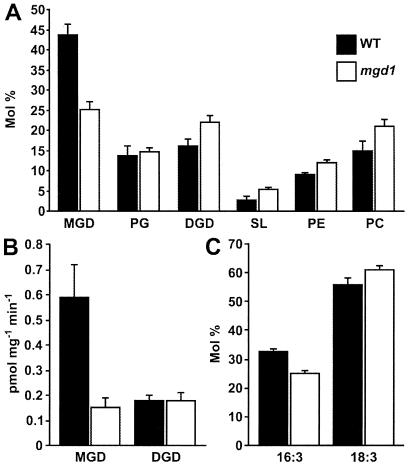
To confirm the identity of mgd1 as a MGD synthase-defective mutant, the effect of the mutation on lipid composition was investigated. Total leaf lipids extracted from wild-type and mutant plants were separated by TLC and quantified (Fig. 3A). The data indicate that mgd1 plants contain 42% less MGD than wild-type plants. Reduced abundance of MGD in mgd1 was found to be accompanied by an increased abundance of the other major plastid and leaf lipids, DGD, sulfolipid, phosphatidylethanolamine, and phosphatidylcholine; the abundance of phosphatidylglycerol was not significantly altered in the mutant. The total lipid content of leaves was not found to be significantly altered by the mutation (fatty acid per fresh weight values (mg/g) for wild type and mgd1 are 3.78 ± 0.54 and 4.40 ± 0.36, respectively) indicating that the major effect of mgd1 is MGD loss.
Figure 3.
Biochemical phenotype of the mgd1 mutant. (A) Lipid composition of wild-type and mutant leaves. Total leaf lipids were extracted and the different lipid classes were separated by TLC and quantified. Indicated lipids are monogalactosyl diacylglycerol (MGD), phosphatidylglycerol (PG), digalactosyl diacylglycerol (DGD), sulfolipid (SL), phosphatidylethanolamine (PE), and phosphatidylcholine (PC). (B) MGD synthase and DGD synthase activities of wild-type and mutant chloroplasts. The rate of incorporation of [14C]galactose into galactolipids in isolated chloroplasts was determined. Units are pmol UDP-[14C]galactose/mg chlorophyll/min. (C) Fatty acid composition of MGD from wild-type and mutant leaves. MGD isolated in A above was subjected to fatty acid methyl ester quantification. (A–C) Presented values are means from three (wild type) or six (mgd1) independent measurements ± SE.
MGD synthase and DGD synthase activities were investigated directly by measuring the rate of incorporation of [14C]galactose into MGD and DGD by using isolated wild-type and mutant chloroplasts (Fig. 3B). The rate of incorporation of [14C]galactose into MGD was 75% less in the mutant than in wild type. In contrast, the rate of incorporation into DGD was the same in both genotypes. These data demonstrate that mgd1 is indeed an MGD synthase-defective mutant.
To investigate the role played by MGD1 in the two-pathway scheme for the biosynthesis of plastidic galactolipids, the fatty acid composition of MGD and DGD lipids isolated in Fig. 3A was investigated. Fig. 3C shows that MGD from the mutant contained 23% less 16:3 (a characteristic constituent of MGD derived from the prokaryotic pathway) and 10% more 18:3 (the principal fatty acid component of MGD derived from the eukaryotic pathway) than MGD from wild-type plants.
Ultrastructural Analyses of mgd1 Plastids.
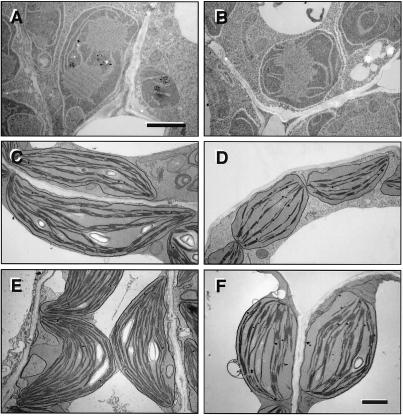
To determine the effect of the mgd1 biochemical defect on plastid development, and to assess the relative importance of MGD1 at different stages of development, wild-type and mgd1 plastids were analyzed and compared by using transmission electron microscopy. The etioplasts of 5-day-old dark grown plants and the chloroplasts of 5-day-old and 23-day-old light-grown plants were examined (Fig. 4). Etioplasts appeared essentially the same in both genotypes (Fig. 4 A and B), suggesting that MGD1 is not important for plastid development in the dark. In contrast, the chloroplasts of 5-day-old plants (in cotyledons) and 23-day-old mgd1 plants (in midsized leaves) were found to be severely underdeveloped (Fig. 4 C–F). At both developmental timepoints, mgd1 chloroplasts were found to be smaller in size than those in the wild type, have a more spherical shape, and contain fewer internal thylakoid membranes. Starch grains were either absent or reduced in size and/or number. Mutant chloroplasts also appeared to contain more plastoglobuli than wild-type chloroplasts.
Figure 4.
Ultrastructure of mgd1 plastids. Electron micrographs of representative plastids from (A) 5-day-old etiolated wild-type cotyledons, (B) 5-day-old etiolated mgd1 cotyledons, (C) 5-day-old light-grown wild-type cotyledons, (D) 5-day-old light-grown mgd1 cotyledons, (E) 23-day-old light-grown wild-type leaves, and (F) 23-day-old light-grown mgd1 leaves. A and B are at higher magnification than C–F. (Bars = 1 μm.)
Discussion
This paper describes the identification and characterization of an Arabidopsis mutant with a T-DNA insertion in the MGD synthase gene, MGD1. The T-DNA is located 12 bp downstream of the first MGD1 transcription initiation site (Fig. 2A), and 101 bp upstream of the third transcription initiation site (RACE analysis identified three different MGD1 transcription initiation sites). MGD1 mRNA abundance in the mutant was estimated to be reduced by 75% compared with wild type by using reverse transcription–PCR. The basis for the residual expression of MGD1 in the mutant is uncertain. One possibility is that the 101-bp region which lies between the T-DNA border and the most downstream transcription initiation site contains sufficient promoter activity to drive basal levels of expression of the shortest transcript. Another possibility is that cryptic promoter sequences exist within the left border region of the T-DNA. In both cases, it is possible that the performance of minimal or cryptic promoters are under the influence of the cauliflower mosaic virus 35S enhancer sequences which are located adjacent to the right border of the T-DNA construct (18).
The abundance of MGD lipid in mature mutant leaves was found to be reduced by 42% compared with wild type (Fig. 3A). As one would expect, this MGD deficiency was associated with an increase in the relative abundance of the other major plastid and leaf lipids (Fig. 3A). The increased relative abundance of nonchloroplastic lipids (phosphatidylethanolamine and phosphatidylcholine) in the mutant suggests that the chloroplasts of mgd1 plants contribute less to the total leaf lipid pool than the chloroplasts of wild-type plants, implying that mutant chloroplasts are smaller in size and/or contain fewer thylakoid membranes (as was indeed observed; Fig. 4 C–F). Confirmation of the role played by the MGD1 locus was obtained by analyzing MGD synthase and DGD synthase activities associated with isolated mutant and wild-type chloroplasts (Fig. 3B). The rate of incorporation of [14C]galactose into MGD was found to be 75% less by using mgd1 chloroplasts. DGD incorporation rates, in contrast, were exactly the same in both genotypes. These data clearly demonstrate that the mgd1 mutant has a substantial and specific MGD synthase deficiency. The remaining MGD lipid and MGD synthase activity in mgd1 may be the result of residual MGD1 expression, or alternatively, of other MGD synthases (such as MGD2 and MGD3; see below). Given the apparent correlation between steady-state MGD1 transcript levels and the rate of incorporation of [14C]galactose into MGD (both reduced by 75% in mgd1), we conclude that the former is the case, and therefore that MGD1 is the predominant MGD synthase in chloroplasts.
Previous studies led to the identification of cDNAs encoding MGD synthases from cucumber (11) and spinach (12). In addition to these cDNAs, and to the Arabidopsis MGD1 gene described in this report, two MGD synthase-related Arabidopsis sequences can be found in the GenBank database (12). MGD1 and the cucumber and spinach MGD synthases (referred to here as csMGD1 and soMGD1) are very closely related, each sharing at least 70% amino acid sequence identity. The other Arabidopsis sequences, MGD2 and MGD3, are far more divergent, sharing only 53.2 and 55.4% amino acid sequence identity with MGD1, respectively. The relatedness of MGD1, csMGD1, and soMGD1 prompted Miège et al. (12) to group them together and refer to them as members of a type A family of MGD synthases; the Arabidopsis MGD2 and MGD3 sequences were referred to as type B and type C enzymes, respectively.
We investigated the role played by MGD1 in galactolipid metabolism by analyzing the fatty acid content of MGD from mgd1 and wild-type plants. MGD from the mutant was found to contain 23% less 16:3 and 10% more 18:3 than MGD from the wild type (Fig. 3C). Because 16:3 is a characteristic feature of MGD derived from the prokaryotic pathway, these data suggest that MGD1 operates in the prokaryotic pathway more than in the eukaryotic pathway. However, the fact that the changes in MGD fatty acid profile are only small (when compared with the change in MGD lipid abundance) indicates that MGD1 (a type A enzyme) is nevertheless the predominant MGD synthase of both pathways. These data are therefore consistent with our observation that MGD synthase activity correlates closely with MGD1 mRNA abundance in Arabidopsis, and with results obtained previously by using heterologously expressed soMGD1. Recombinant soMGD1 (also a type A enzyme) was shown to use both prokaryotic and eukaryotic DAG substrates (18/16 and 18/18 configurations, respectively) efficiently (12). The recombinant enzyme was, however, also found to have a higher affinity for eukaryotic DAG. The apparent difference between the results of the previous in vitro study (12) and the present in vivo study (Fig. 3C) perhaps reflect substrate availability to the type A enzyme in vivo. MGD1 is presumably localized at the inner envelope membrane like soMGD1 (12), and therefore, is more easily accessible to prokaryotic DAG than eukaryotic DAG. In this scenario, a mutation in MGD1 would lead to a reduction in prokaryotic MGD synthesis (as observed in this study) even though the enzyme itself might be slightly more active with eukaryotic DAG.
The ultrastructure of mgd1 plastids was analyzed by using transmission electron microscopy. Etioplasts in the mutant appeared essentially the same as in wild type (Fig. 4 A and B), indicating that MGD1 is not important for plastid development in the dark. This result was somewhat unexpected because previous studies have shown that etioplast membranes contain high levels of MGD (29, 30). One possible explanation is that the MGD synthase function of dark-grown Arabidopsis plants is fulfilled primarily by MGD2 and/or MGD3, and not to any significant degree by MGD1.
The MGD deficiency of light-grown mgd1 plants was found to correlate with severe defects in chloroplast structure (Fig. 4 C–F). Mutant chloroplasts were substantially smaller in size and more spherical in shape than those in wild type. Interestingly, mgd1 chloroplasts contained far fewer internal thylakoid membranes and grana than wild-type chloroplasts, demonstrating an important role for MGD (which normally constitutes upwards of 50% of the thylakoid lipid matrix) in their structural organization and biogenesis. MGD has a small galactose head group and splayed fatty acid tails which together give the molecule a cone-like shape, a predisposition to form nonlamellar, hexagonal-phase aggregates, and the ability to induce curvature in lamellar membranes (4). It is possible that these unique characteristics of MGD are particularly important for the organization of thylakoids and the formation of granal stacks.
The observed structural abnormalities of mgd1 chloroplasts would indeed be predicted to impact significantly on their functional capabilities. Apart from indirect structural effects on photosynthetic capacity, the MGD deficiency of the mutant might also be predicted to affect photosynthesis more directly. MGD is highly enriched in the photosystem II reaction center/core complex (5) and a single MGD molecule is tightly associated with the photosystem II reaction center (6). It has been shown to stimulate the activity of chloroplastic ATP synthase in vitro (7), and was found to be the exclusive lipid associated with the xanthophyll cycle enzyme, violaxanthin de-epoxidase (8). The mgd1 mutant therefore represents a unique resource for studying the role of MGD in different photosynthetic processes in vivo. It will also prove invaluable as attempts are made to assess the role of MGD in other important processes in which it has been proposed to play a role, such as the translocation of nucleus-encoded proteins across the plastid envelope (31–33).
Acknowledgments
This work was supported by grants from the U.S. Department of Energy (ER13993 to J.C. and ER20305 to C.B.) and the University of Leicester (to P.J.), and by a Human Frontier Science Program fellowship (to P.J.). J.C. is an associate investigator of the Howard Hughes Medical Institute.
Abbreviations
- MGD
monogalactosyl diacylglycerol
- DGD
digalactosyl diacylglycerol
- DAG
sn-1,2-diacylglycerol
- RACE
rapid amplification of cDNA ends
- mgd1
MGD synthase 1
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100132197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100132197
References
- 1.Douce R, Joyard J. Annu Rev Cell Biol. 1990;6:173–216. doi: 10.1146/annurev.cb.06.110190.001133. [DOI] [PubMed] [Google Scholar]
- 2.Ohlsson L, Blom M, Bohlinder K, Carlsson A, Nilsson J. J Nutr. 1998;128:239–245. doi: 10.1093/jn/128.2.239. [DOI] [PubMed] [Google Scholar]
- 3.Andersson L, Bratt C, Arnoldsson K C, Herslof B, Olsson N U, Sternby B, Nilsson A. J Lipid Res. 1995;36:1392–1400. [PubMed] [Google Scholar]
- 4.Webb M S, Green B R. Biochim Biophys Acta. 1991;1060:133–158. [Google Scholar]
- 5.Tremolières A, Dainese P, Bassi R. Eur J Biochem. 1994;221:721–730. doi: 10.1111/j.1432-1033.1994.tb18785.x. [DOI] [PubMed] [Google Scholar]
- 6.Murata N, Fujimura Y, Higashi S. Biochim Biophys Acta. 1990;1019:261–268. [Google Scholar]
- 7.Pick U, Weiss M, Gounaris K, Barber J. Biochim Biophys Acta. 1987;891:28–39. [Google Scholar]
- 8.Yamamoto H Y, Higashi R M. Arch Biochem Biophys. 1978;190:514–522. doi: 10.1016/0003-9861(78)90305-3. [DOI] [PubMed] [Google Scholar]
- 9.Siefermann-Harms D, Ross J W, Kaneshiro K H, Yamamoto H Y. FEBS Lett. 1982;149:191–196. [Google Scholar]
- 10.Joyard J, Douce R. In: Biochemistry of Plants. Stumpf P K, editor. New York: Academic; 1987. pp. 215–274. [Google Scholar]
- 11.Shimojima M, Ohta H, Iwamatsu A, Masuda T, Shioi Y, Takamiya K. Proc Natl Acad Sci USA. 1997;94:333–337. doi: 10.1073/pnas.94.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miège C, Maréchal E, Shimojima M, Awai K, Block M A, Ohta H, Takamiya K I, Douce R, Joyard J. Eur J Biochem. 1999;265:990–1001. doi: 10.1046/j.1432-1327.1999.00801.x. [DOI] [PubMed] [Google Scholar]
- 13.Browse J, Somerville C. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:467–506. [Google Scholar]
- 14.Heinz E, Roughan P G. Plant Physiol. (1983) 1983;72:273–279. doi: 10.1104/pp.72.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mongrand S, Bessoule J J, Cabantous F, Cassagne C. Phytochemistry. 1998;49:1049–1064. [Google Scholar]
- 16.Dörmann P, Hoffmann-Benning S, Balbo I, Benning C. Plant Cell. 1995;7:1801–1810. doi: 10.1105/tpc.7.11.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dörmann P, Balbo I, Benning C. Science. 1999;284:2181–2184. doi: 10.1126/science.284.5423.2181. [DOI] [PubMed] [Google Scholar]
- 18.Weigel D, Ahn J, Blazquez M, Borevitz J, Christensen S, Fankhauser C, Ferrandiz C, Kardailsky I, Neff M, Nguyen J, et al. Plant Physiol. 2000;122:1003–1014. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H-m, Culligan K, Dixon R A, Chory J. Plant Cell. 1995;7:1599–1610. doi: 10.1105/tpc.7.10.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Napoli C, Lemieux C, Jorgensen R. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez C M, Freire M A, Camilleri C, Robaglia C. Plant J. 1998;13:465–473. doi: 10.1046/j.1365-313x.1998.00047.x. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Rossak M, Schafer A, Xu N, Gage D A, Benning C. Arch Biochem Biophys. 1997;340:219–230. doi: 10.1006/abbi.1997.9931. [DOI] [PubMed] [Google Scholar]
- 24.Price C A, Hadjeb N, Newman L, Reardon E M. In: Plant Molecular Biology Manual. Gelvin S B, Schilperoort R A, editors. Dordrecht: Kluwer; 1994. pp. 1–15. [Google Scholar]
- 25.Jarvis P, Chen L-J, Li H-M, Peto C, Fankhauser C, Chory J. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- 26.Porra R J, Thompsin W A, Kriedman P E. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- 27.Liu Y-G, Mitsukawa N, Oosumi T, Whittier R F. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 28.Jarvis P, Belzile F, Dean C. Plant J. 1997;11:921–931. doi: 10.1046/j.1365-313x.1997.11050921.x. [DOI] [PubMed] [Google Scholar]
- 29.Ryberg M, Sandelius A S, Selstam E. Physiol Plant. 1983;57:555–560. [Google Scholar]
- 30.Selstam E, Sandelius A S. Plant Physiol. 1984;76:1036–1040. doi: 10.1104/pp.76.4.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keegstra K. In: Photosynthesis. Briggs W, editor. New York: Liss; 1989. pp. 347–357. [Google Scholar]
- 32.van 't Hof R, Demel R A, Keegstra K, de Kruijff B. FEBS Lett. 1991;291:350–354. doi: 10.1016/0014-5793(91)81318-3. [DOI] [PubMed] [Google Scholar]
- 33.Bruce B. Plant Mol Biol. 1998;38:223–246. [PubMed] [Google Scholar]