Abstract
Arbuscular mycorrhizal (AM) fungi take up photosynthetically fixed carbon from plant roots and translocate it to their external mycelium. Previous experiments have shown that fungal lipid synthesized from carbohydrate in the root is one form of exported carbon. In this study, an analysis of the labeling in storage and structural carbohydrates after 13C1 glucose was provided to AM roots shows that this is not the only pathway for the flow of carbon from the intraradical to the extraradical mycelium (ERM). Labeling patterns in glycogen, chitin, and trehalose during the development of the symbiosis are consistent with a significant flux of exported glycogen. The identification, among expressed genes, of putative sequences for glycogen synthase, glycogen branching enzyme, chitin synthase, and for the first enzyme in chitin synthesis (glutamine fructose-6-phosphate aminotransferase) is reported. The results of quantifying glycogen synthase gene expression within mycorrhizal roots, germinating spores, and ERM are consistent with labeling observations using 13C-labeled acetate and glycerol, both of which indicate that glycogen is synthesized by the fungus in germinating spores and during symbiosis. Implications of the labeling analyses and gene sequences for the regulation of carbohydrate metabolism are discussed, and a 4-fold role for glycogen in the AM symbiosis is proposed: sequestration of hexose taken from the host, long-term storage in spores, translocation from intraradical mycelium to ERM, and buffering of intracellular hexose levels throughout the life cycle.
The arbuscular mycorrhizal (AM) symbiosis is important because it benefits most land plants. AM plants show enhanced growth, increased resistance to biotic and abiotic stresses, and greater ecological diversity (for review, see Smith and Read, 1997). AMs are also responsible for directing the movement of huge quantities of photosynthate to the soil (for review, see Douds et al., 2000; Graham, 2000). Carbon in the root flows from plant to fungus in the form of sugars (Shachar-Hill et al., 1995; Solaiman and Saito, 1997), and together with the transfer of mineral nutrients from fungus to root (Koide and Schreiner, 1992; George et al., 1995; Jakobsen, 1995), this is the nutritional mainstay of what is arguably the world's most important mutualistic symbiosis.
Rather little was known about the forms and pathways through which carbon flows in the AM symbiosis until recently (Jennings, 1995; Smith and Read, 1997). The development of in vitro monoxenic AM root cultures (Mugnier and Mosse, 1987) with separate host and fungal compartments (St. Arnaud et al., 1996) has facilitated the application of stable isotope labeling (Pfeffer et al., 1999), gene expression analysis (Lammers et al., 2001; Bago et al., 2002), in vivo microscopy (Bago et al., 2002), and other methods (for review, see Fortin et al., 2002). Together with previous work on enzyme activities and analysis of metabolites, these approaches have begun to illuminate the metabolic pathways by which carbon is handled in the AM symbiosis (for review, see Bago et al., 2000).
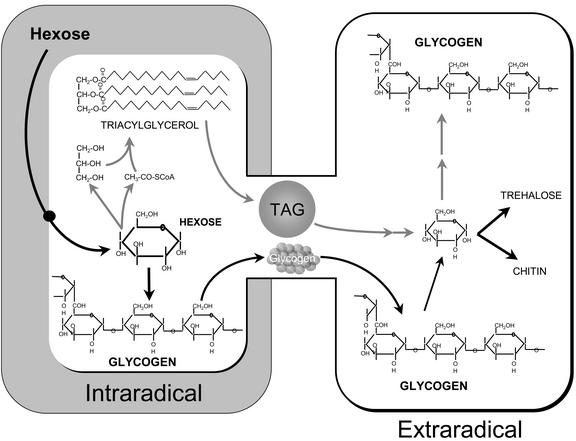
AM fungi obtain most or all of their carbon within the host root. Here, they acquire hexose and transform it into trehalose and glycogen, typical fungal carbohydrates (Shachar-Hill et al., 1995). Triacylglyceride (TAG) is the main form of stored carbon in AM fungi (Beilby and Kidby, 1980; Jabaji-Hare, 1998), and this is mostly or exclusively made in the intraradical mycelium (IRM; Pfeffer et al., 1999). Some of this storage lipid flows from the IRM to the extraradical mycelium (ERM; Pfeffer et al., 1999), and in vivo microscopic observations indicate that the rate of export is sufficient to account for the high levels of stored lipid in the ERM (Bago et al., 2002). The glyoxylate cycle is active in the ERM (Lammers et al., 2001), and this pathway appears to be important in using exported TAG to make carbohydrate in the ERM.
The goal of this investigation was to determine whether all the carbon exported from host roots is, in fact, in the form of lipid. There are two reasons to investigate this. First, the conversion of carbohydrate to lipid within the IRM followed by translocation to the ERM and reconversion into carbohydrates for structural, storage, and biosynthetic uses is inefficient (close to one-half of the carbon is lost in converting hexose into TAG and back) and a highly unusual, if not unique, transport strategy in biology. Although there may be particular reasons for this strategy that stem from AM anatomy and the high carbon and energy density of lipids (Bago et al., 2002), the cost invites a closer audit. Second, because cytoplasmic streaming along AM hyphae (Bago et al., 1998, 2002) and the conversion of hexose into glycogen and trehalose in the IRM are active (Shachar-Hill et al., 1995), the translocation of carbohydrates seems likely to occur to some degree.
RESULTS
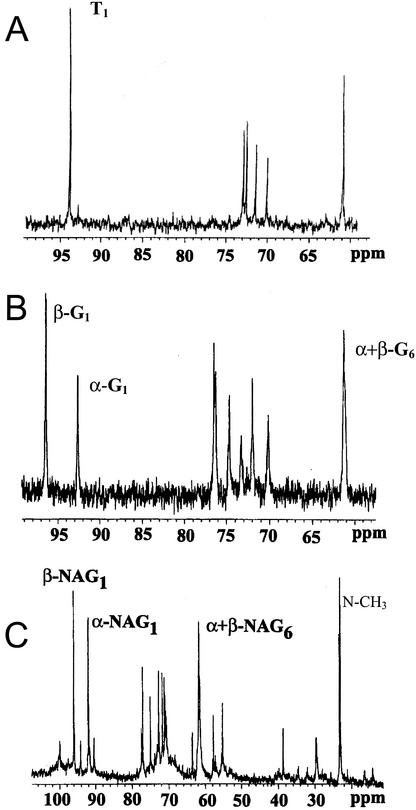
Isotopic labeling in trehalose, glycogen, and chitin in the ERM was analyzed after supplying 13C1 Glc to mycorrhizal carrot (Daucus carota) roots to track the metabolism and movement of carbohydrate from the IRM to the ERM. Figure 1 shows spectra of these carbohydrates extracted from the ERM and, for glycogen and chitin, hydrolyzed to their constituent hexose monomers. The intensity of the different peaks reflects the levels of 13C in each of the carbon positions of trehalose, Glc from glycogen, and N-acetyl glucosamine (NAG) from chitin. The identities of the hydrolysis products were confirmed by comparison with NMR and mass spectra of authentic samples. There are more peaks in the Glc and NAG spectra compared with those of trehalose because the latter compound exists in only one form in solution (a symmetric 1, 1 α,α-Glc dimer), resulting in unique signals from each of the six carbon positions of its Glc moieties, whereas Glc and NAG each exist in two anomeric forms giving two signals from each carbon position.
Figure 1.
13C NMR spectra of trehalose (A), Glc from glycogen (B), and NAG from chitin (C). The carbohydrates were obtained by sequential extraction and hydrolysis from ERM that was harvested 2 months after providing 13C1 Glc to the mycorrhizal roots. Spectroscopic conditions (see “Materials and Methods”) were such that the different intensities of the signals from different carbons accurately reflect the different labeling levels in those positions of the carbohydrates. The C1 and C6 signals are indicated for each compound. Greek symbols refer to the two anomeric forms in which Glc and NAG exist in solution.
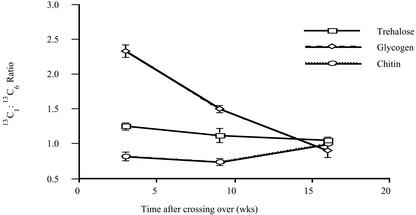
Quantitatively, there are different labeling patterns in spectra of the three different carbohydrates, although some features are qualitatively the same. Thus, the labeling is consistently higher in positions 1 and 6 and lower in 2 through 5, but the relative intensities in C1 and C6 differ among the three carbohydrates and for the same carbohydrate at different times after labeling. The ratio of labeling in C1 and C6 is important for deducing the metabolic history of carbohydrate in the ERM (see “Discussion”), and these values are plotted in Figure 2. There are no statistically significant changes in the 13C1:13C6 ratio in trehalose or chitin with time, however, a 1.5-fold decrease in that ratio was observed in glycogen from young compared with mature mycelium, and an additional 1.7-fold decrease from mature to old mycelium. When 13C6 Glc was supplied to the colonized roots for 6 weeks, the C6 of trehalose in the ERM was more labeled than the C1 position and was significantly higher than the labeling measured in C1 when 13C1 Glc was supplied (spectra not shown).
Figure 2.
The relative labeling in the C1 and C6 positions of carbohydrates from the ERM as a function of time after providing 13C1 Glc to the mycorrhizal roots. Data are derived from the intensities of C1 and C6 signals in spectra such as those shown in Figure 1. Error bars for are ± sem for at least three replicates per experiment.
Spectra such as those in Figure 1 yield relative labeling levels. Absolute percentages of 13C levels were determined from 1H NMR spectra (not shown): After supplying 13C1 Glc, labeling in C1 of trehalose was 16% ± 2%, it was 21% ± 2% in glycogen C1, and it was 16% ± 2% in C1 of chitin. (Here and elsewhere, values are quoted as mean ± se of the mean for three to six independent replicates.) Labeling was much lower, 8% ± 1%, in the methyl carbon of the acetyl moiety of chitin. There were no statistically significant trends in labeling levels as a function of labeling time in the C1 or methyl carbons of the carbohydrates. Labeling in C5 of glycogen and trehalose was 1.5 times lower than in the C6 position 3 to 12 weeks after supplying 13C1 Glc, and 1.3 times lower after 16 weeks. When 13C1,2 (double-labeled) Glc was supplied to the mycorrhizal roots, labeling in C1 positions of carbohydrates in the ERM after 10 to 13 weeks was close to 1.5 times higher than when 13C1 was used and splittings in the C1 signals in 13C NMR spectra revealed that C1 singly labeled and C1,2 doubly labeled hexose molecules were present (spectra not shown).
Because storage lipid is a precursor for carbohydrate production via the glyoxylate cycle (Lammers et al., 2001), labeling was also assessed in fungal TAG extracted from the same ERM tissues as the carbohydrates. Spectra of fungal storage lipids extracted from the ERM after providing 13C1 Glc to the mycorrhizal roots show labeling levels that do not change in the range of labeling times measured (3–16 weeks; spectra not shown). The even-numbered positions of fatty acid moieties of TAG are labeled 9.0% ± 0.8% and the C1,3 positions of the glyceryl moiety of TAG are labeled 8.1% ± 0.7%. These values are all significantly lower than the labeling levels measured in the C1 and the C6 carbons and are similar to the 13C levels in C5 of carbohydrates in the ERM after 13C1 Glc was supplied to the mycorrhizal roots. Mass spectra of fatty acids from the TAG were consistent with a single population of labeled molecules (data not shown). Labeling levels in the even-numbered positions of fatty acids are similar to the labeling in the methyl of NAG.
These results show that the origin of carbohydrates in the ERM is only partly from lipid (see “Discussion”). How far then do fluxes through gluconeogenesis and the glyoxylate cycle contribute to the different carbohydrate pools in the ERM? 13C2 acetate or 13C1,3 glycerol was supplied to the ERM for 6 to 8 weeks to address this question. Substantial labeling was observed in trehalose in the ERM, with 11% ± 2% labeling in C1 when 13C1,3 glycerol was provided, and 33% ± 6% when 13C2 acetate was used. The 13C1:13C6 ratio for trehalose was 0.6 ± 0.1 for both substrates. Labeling in C1 of glycogen was substantial but lower, being 18% ± 3% when 13C2 acetate was supplied and too low to quantify reliably by NMR in experiments with labeled glycerol. The ratio of C1:C6 labeling in glycogen and in chitin was approximately 0.5. The 13C1:13C6 ratios for trehalose and glycogen in the ERM are much lower than found when 13C1 Glc was provided to the mycorrhizal roots (Fig. 2).
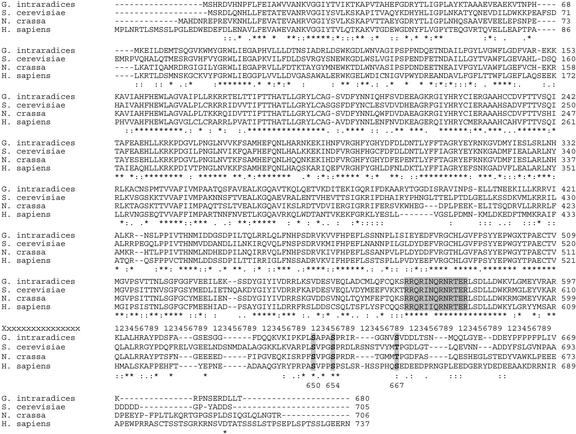
To confirm whether the glycogen and NAG are synthesized in the ERM and presymbiotic fungal tissue (hereafter referred to as germinating spores, see “Materials and Methods”) by the usual metabolic pathways, we obtained Glomus intraradices cDNA sequences for genes encoding enzymes highly similar to other fungal enzymes catalyzing unique steps in these pathways. The glycogen synthase sequence was obtained by PCR from cDNA template derived from germinating spores. The primers were degenerate and designed by reverse translation of two highly conserved regions of fungal glycogen synthase sequences. A fragment of the predicted size was cloned and sequenced and was found to share sequence similarity with the fungal genes. Specific primers were then designed from the sequence and a RACE kit was used to obtain full-length sequences (see “Materials and Methods”). Figure 3 shows the deduced amino acid sequence of the G. intraradices glycogen synthase together with multiple alignments for glycogen synthase sequences from other organisms. The highest degree of homology to a known glycogen synthase is to that from N. crassa, which has 66% identity and 78% similarity to the G. intraradices enzyme at the amino acid level. Regions of high conservation are indicated, and the known phosphorylation sites are also labeled. The G. intraradices GS protein is identical to the yeast protein sequence over a 13-amino acid region that has been shown to be responsible for the allosteric effect of Glc-6-P on GS activity (Pederson et al., 2000).
Figure 3.
Multiple alignments of glycogen synthase enzymes. The organisms are G. intraradices, Neurospora crassa, Saccharomyces cerevisiae, and human (Homo sapiens). Conservation of phosphorylation sites at the C terminus are shown in bold: Ser(650), Ser(654), and Thr(667) (numbering is according to the S. cerevisiae protein). Thr 667 is conserved between S. cerevisiae and N. crassa, but substituted by Ser in G. intraradices and human. The region defining the Glc-6-P allosteric binding site known in yeast (Pedersen et al., 2000) is also highlighted.
A putative glycogen-branching enzyme (BE) cDNA was discovered in random expressed sequence tag (EST) sequencing efforts. A full-length cDNA was obtained by RACE methodology and was sequenced in full. Figure 4 shows the deduced amino acid sequence of this clone in alignment with other glycogen BEs. BE from Aspergillus oryzae was very similar to the G. intraradices enzyme at the amino acid level (69% identity and 81% similarity).
Figure 4.
Multiple alignments of BE sequences. The organisms are G. intraradices, A. oryzae, Emericella nidulans, N. crassa, and humans. Two Arg residues and a Tyr in the human protein (Y329, R515, and R524) whose mutation is associated with glycogen storage disease (Ziemssen et al., 2000; Lossos et al., 1998) are also highlighted.
Random sequencing of a cDNA library from germinating spores also yielded cDNA with homology to Gln fructose-6-P aminotransferase (GFAT), which catalyzes the first committed metabolic step in glucosamine synthesis. The EST fragment had 59% identity over a 202-amino acid region to the Saccharomyces cerevisiae protein. The nucleotide sequences of these and other putative metabolic genes have been deposited in GenBank (GFAT, BE603749; BE, AF503447; and GS, BE603748), and may also be seen at http://darwin.nmsu.edu/∼plammers/glomus/.
We used quantitative real-time PCR to measure the number of copies of mRNA for GS, β-tubulin, and rRNA to analyze the expression of glycogen synthase in different tissues of G. intraradices (Table I). The results from control samples without reverse transcriptase (RT) showed that there was no significant contamination of RNA with fungal DNA. Uncolonized roots gave no detectable signals from fungal mRNAs and counts for fungal rRNA that were at least 10,000 times smaller than for colonized roots. Thus, this approach is very selective as well as very sensitive for measuring fungal gene expression within host roots as well as in ERM and germinating spores. The levels of tubulin expression are different in the different fungal tissues, raising doubts as to its suitability as a control housekeeping gene for studies of gene expression. Relative to fungal rRNA levels, the number of mRNA molecules encoding the putative GS is very similar in IRM and ERM and about an order of magnitude lower in germinating spores than in both of these tissues.
The detection of significant GS expression in the ERM, and the labeling of glycogen and trehalose when 13C-labeled permeant metabolic precursors were provided, demonstrate significant glycogen synthesis in the ERM. This suggests the possibility that carbohydrates synthesized in the ERM might be translocated back to the IRM as was shown for TAG (Bago et al., 2002). In accordance with this, labeling in extracts of mycorrhizal roots was examined after exposure to 13C1,3 glycerol and 13C2 acetate to the ERM. No labeling was detected in C1 of trehalose (NMR spectra not shown) with a detection limit of approximately 3%. It was not possible to assess labeling in glycogen and chitin from IRM.
Levels of stored carbohydrate decrease during spore germination (Bécard et al., 1991), but the detection of significant GS expression in germinating spores suggests that glycogen is also being synthesized. To confirm whether this is so, we incubated germinating spores with 13C2 acetate or 13C1,3 glycerol. Labeling in C1 of trehalose was 55% ± 4% when 13C2 acetate was supplied and was 49% ± 2% when spores were germinated in the presence of 13C1,3 glycerol. The ratio of 13C in C1 and C6 was 0.78 ± 0.02 and 0.72 ± 0.02 for labeling with acetate and glycerol, respectively. Labeling in the C1 of glycogen was substantial although lower than in trehalose, being 33% ± 2% and 33% ± 4% for samples labeled with acetate and glycerol, respectively; the corresponding C1:C6 ratios were 0.63 ± 0.02 and 0.62 ± 0.05. Labeling was also detected in chitin, although NAG yields were low from germinating spores, preventing reliable quantitation of percentage of 13C levels.
DISCUSSION
Evidence for the Export of Carbohydrate as well as Lipid to the ERM
The spectra in Figure 1 show that supplying AM colonized roots with Glc labeled in C1 results in labeling of trehalose, glycogen, and chitin in the ERM. This labeling is consistent with the previous report of labeling in trehalose in the ERM (Bago et al., 1999) and reflects the fact that AM fungi depend on the host plant for carbon, and export substantial amounts of this carbon from mycorrhizal roots to the ERM (for review, see Douds et al., 2000). Previous studies have shown that hexose is taken up directly by the fungus within the root (Shachar-Hill et al., 1995; Solaiman and Saito, 1997), but not by the ERM (Pfeffer et al., 1999), and that this is converted directly into trehalose and glycogen (Shachar-Hill et al., 1995), as well as lipid in the IRM (Pfeffer et al., 1999). Other experiments showed the export of lipid from the root (Pfeffer et al., 1999; Bago et al., 2002) and its conversion by the ERM into carbohydrate via the glyoxylate cycle and gluconeogenesis (Lammers et al., 2001). The sequential operation of these processes forms a mechanism for the export of carbon from mycorrhizal roots (for review, see Bago et al., 2000).
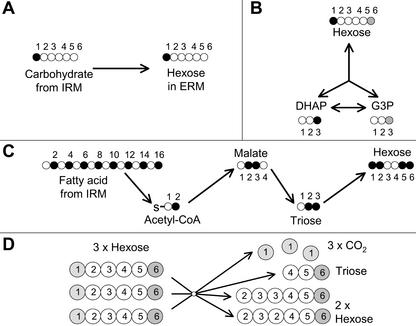
The finding that carbohydrates in the ERM are labeled in C5 is consistent with a significant flux through this route. C5 labeling is particularly indicative of the glyoxylate cycle and gluconeogenesis operating from 13C2-labeled acetyl-coenzyme A (CoA) as it is not expected to become labeled via the other pathways shown to operate in this tissue (Lammers et al., 2001; Fig. 5). The labeling levels in C5 are similar to the labeling levels in the even-numbered positions of the fatty acid moieties of the lipid, which is consistent with lipid as the origin of a significant proportion of the carbohydrate (Fig. 5C). The acetyl moiety of chitin is labeled to the same extent as the fatty acids of lipid, suggesting that this too originates from acetyl-CoA generated by lipid breakdown. The synthesis of glycogen in the ERM as part of this route necessitates the presence of glycogen synthase in the ERM, and the results of Table I demonstrate that it is expressed in ERM as well as the IRM.
Figure 5.
Illustration of the labeling patterns in the hexose units of carbohydrates in the ERM that result from the activity of different pathways after providing 13C1 Glc to the mycorrhizal roots. A, The direct export of carbohydrate from the IRM; Glc uptake in the IRM results in labeling in C1 of carbohydrate in the IRM that is then exported to the ERM. B, Hexose triose cycling: Hexose initially labeled in C1 is broken down to dihydroxyacetone phosphate labeled in the C3 position, this then labels the C3 of glyceraldehyde-3-P through the action of triose phosphate isomerase, subsequently, gluconeogenic flux results in the labeling of C6 of carbohydrates. C, Synthesis of carbohydrates from lipids made in the IRM: The fatty acids (mainly C16) of lipid made in the IRM from 13C1 Glc are labeled in the even-numbered positions, export of this lipid to the ERM followed by β-oxidation produces acetyl-CoA units labeled in C2, the glyoxylate cycle produces malate labeled in C2 and C3, which after decarboxylation labels triose in C2 and C3, this triose labels carbohydrates in C1, C2, C5, and C6. D, The oxidative pentose phosphate pathway operating in cyclic mode: Three hexose molecules are converted to two hexose molecules, three molecules of CO2, and one of triose. The fate of each of the six carbons of hexose is shown. The light and dark shading of C1 and C6 carbons in C is used to emphasize the loss of label from the C1 positions and the retention of label in the C6 positions. This acts to lower the ratio of 13C1:13C6 in carbohydrates.
However, synthesis from lipid cannot fully account for the labeling pattern in carbohydrates. Thus labeling in C3 and C4 positions, as previously observed (Bago et al., 1999; Lammers et al., 2001), indicates refixation of 13CO2 released by respiration. This is consistent with the finding that supplying 13CO2 resulted in labeling selectively in C3 and C4 (spectra not shown). More importantly, labeling was at all time points higher in C1 of carbohydrates than in any C position of the lipids in the ERM after supplying 13C1 Glc to the colonized roots. In principle, carbohydrate could be made in the ERM from a subpopulation of more highly labeled TAG, thereby explaining the higher labeling in the C1 of carbohydrates. However, this possibility is not supported by gas chromatography-mass spectrometry results because the profile of fatty acid molecules of different masses is consistent with a single population of modestly labeled fatty acid molecules in lipids in the ERM (data and mass isotopomer distribution analysis not shown).
The observation of greater labeling in C1 than in C6 in trehalose and glycogen at shorter and intermediate times when 13C1 Glc is provided (Fig. 2) also indicates that a significant proportion of carbohydrate in the ERM is derived from a nonlipid source. Higher labeling in C6 than in C1 of trehalose in the ERM when 13C6 Glc is provided is consistent with this. Label from lipids that reaches hexose via the formation of labeled triose in the ERM and gluconeogenesis from triose would label C1 and C6 equally (Fig. 5C). The subsequent action on hexose of other pathways such as recycling between hexose and triose pools or cyclical flux through the pentose phosphate pathway cannot account for greater labeling in C1 than in C6 starting from 13C1 hexose (Fig. 5, B and D).
A 13C1:13C6 ratio greater than one in extraradical fungal carbohydrates suggests the export of 13C1-labeled carbohydrate from the IRM to the ERM because carbohydrate in the IRM is labeled almost exclusively in C1 when 13C1 Glc is provided (Shachar-Hill et al., 1995; this study). There is evidence from enzymatic (Saito, 1995) and labeling (Bago et al., 1999; Lammers et al., 2001) studies for pentose phosphate activity in the ERM and germinating spores. Flux through the pentose phosphate pathway reduces labeling in C1 relative to C6 and accounts for the 13C1:13C6 ratios being less than 1 when labeled glycerol or acetate were used (Fig. 5D). None of the other pathways of primary metabolism for which there is evidence in AM fungi (for review, see Bago et al., 2000) can account for the elevated 13C1:13C6 ratio, and the direct export of carbohydrate from the mycorrhizal root appears to be the only straightforward explanation.
The Identity of Exported Carbohydrate
What form(s) of carbohydrate is involved in the export from IRM to ERM? Glycogen is the most likely candidate because it is found in significant amounts in AM fungi (Bonfante et al., 1994; Bago et al., 1998); it is actively made in the IRM from hexose taken up by the fungus in the root and reaches high fractional enrichments in C1 but not C6 when exogenous 13C1 hexose is supplied (Shachar-Hill et al., 1995); and the fractional labeling in glycogen C1 in the ERM is greater than in trehalose and the 13C1:13C6 ratio is also much greater indicating that it is not made from trehalose in the ERM. The labeling observed is consistent with the possibility that the breakdown of exported glycogen in the ERM is the cause of the 13C1:13C6 ratio in trehalose being greater than one. Also consistent with this inference is the fact that trehalose becomes more highly labeled than glycogen when glycerol or acetate is provided to the ERM, which indicates that more of the trehalose is synthesized in the ERM than glycogen.
Because chitin is not water soluble, and because its C1 percentage labeling and 13C1:13C6 ratio are lower than those of glycogen or trehalose, it is likely that the glucosamine from which NAG and then chitin are made derive from hexose in the ERM rather than being exported from the IRM. The putative gene sequences for GFAT and chitin synthase suggest that the usual pathway of chitin synthesis is active in the ERM. Ubalijoro et al. (2001) have also reported the expression of chitin synthase in the ERM. The labeling in the acetyl moieties of chitin is lower than in the C1 and C6 of the glucosamine moieties, which suggests that the acetyl group is made from lipid-derived acetyl-CoA in the ERM.
The 13C-labeling levels in C1 of glycogen, trehalose, and chitin are two to three times higher at all time points than expected in hexose made from lipid; and with the exception of glycogen at the first time point, the same is true of C6 labeling. This indicates that carbohydrate movement from IRM to ERM plays a significant role in AM fungal metabolism in the symbiosis. Quantifying the relative contributions of carbohydrate and lipid to total carbon export would require additional information on the labeling in ERM carbohydrates and on the relative proportions of carbohydrate and lipid that are directed to other metabolic fates (protein, nucleic acids, respiration, and secondary metabolites) in the ERM.
Synthesis, Turnover, and Regulation of Glycogen through the Fungal Life Cycle
Small-scale random sequencing of cDNAs from germinating spores (Jun et al., 2002) generated a sequence to a putative glycogen BE (Fig. 3), which suggested significant expression of this gene. Expression levels of BE correlate with glycogen levels in yeast (Thon et al., 1992), although it does not appear to exert significant control over flux in mammalian cells (Skurat et al., 1996).
The quantification of significant expression levels for glycogen synthase (Table I), together with the substantial levels of labeling in glycogen observed after adding labeled glycerol or acetate, confirm active glycogen synthesis at this stage of the life cycle. Labeling data indicate that glycogen synthesis is active in IRM (Shachar-Hill et al., 1995), ERM, and germinating spores (this study), and the expression of glycogen synthase in all these tissues is consistent in this conclusion. Carbohydrate is broken down during spore germination in AM (Bécard et al., 1991) and other fungi (Thevelein et al., 1982), and in vivo experiments have shown that glycogen can also be rapidly turned over in the IRM (Shachar-Hill et al., 1995).
The finding that the deduced amino acid sequence of glycogen synthase contains putative phosphorylation and Glc-6-P allosteric regulatory sites (Fig. 4), together with the absence of dramatic differences in expression levels in the different tissues during symbiosis, is consistent with the idea that posttranslational regulation governs glycogen turnover in G. intraradices as in other organisms (Skurat et al., 1996). This conclusion is also consistent with the fact that glycogen labeling after exposure to labeled acetate or glycerol is higher in germinating spores than in ERM despite the lower expression levels of glycogen synthase in germinating spores (Table I).
Fungal lipid within the root becomes labeled when 13C-glycerol was supplied to the ERM (Bago et al., 2002), which was interpreted as representing recirculation of lipid from the ERM to the IRM because the lipid in the ERM became modestly labeled under these conditions. By contrast, no labeling was detected in the trehalose of colonized roots when labeled acetate or glycerol was supplied to the ERM even though labeling in trehalose and glycogen in the ERM is much higher than in the glyceryl moiety of TAG. This suggests that no significant flux of carbohydrate occurs from ERM to IRM or at least that the rate of turnover of trehalose in the IRM is high compared with any carbohydrate recirculation from ERM to IRM. In vivo NMR experiments support the idea of rapid turnover of carbohydrates in the IRM from absorbed hexose (Shachar-Hill et al., 1995). If as these results indicate lipid but not carbohydrate recirculation from ERM to IRM is significant, any plant-to-plant transfer of carbon mediated by the fungus is more likely to be mediated by translocation of lipid than of carbohydrate.
The fact that labeling is higher in C6 of glycogen and trehalose than in C5 in the ERM when 13C1 Glc was supplied to the colonized roots suggests that there is also a significant bidirectional flux between hexose and triose in the ERM that increases the labeling in C6 by transferring label between C1 and C6 (Fig. 5B). Together with glycogen turnover, triose/hexose recycling is consistent with the observed drop in the 13C1:13C6 ratio at longer time points (Fig. 2). This hexose/triose recycling is in contrast to the situation in the IRM, where no scrambling of label was observed between C1 and C6 (Shachar-Hill et al., 1995).
We conclude that the flux of carbon from within mycorrhizal roots to the ERM is in the form of carbohydrate and lipid, and that glycogen is probably the predominant carbohydrate moved within the fungus. The dual pathways of carbon movement are illustrated in Figure 6. Turnover of glycogen and the expression of glycogen synthase are consistent with a 4-fold role for glycogen in the AM symbiosis: sequestration of hexose taken from the host, long-term storage in spores, translocation from IRM to ERM, and buffering of intracellular hexose levels throughout the life cycle.
Figure 6.
Illustration of the two routes by which we propose that carbon is moved from the IRM to the ERM in the AM symbiosis. Carbon taken up from the host in the form of hexose is converted to carbohydrates and storage lipids in the IRM. Lipid and glycogen are translocated from IRM to ERM. Storage (glycogen and trehalose) and structural (chitin) carbohydrates are synthesized in the ERM from hexose that is derived from exported carbohydrate as well as from lipid.
MATERIALS AND METHODS
Production and Labeling of Fungal Material
Fungal material for NMR experiments was obtained as previously published (Bago et al., 1999; Pfeffer et al., 1999; Lammers et al., 2001). In brief, Ri T-DNA-transformed carrot (Daucus carota clone DC1) roots colonized by Glomus intraradices (DAOM 197198, Biosystematic Research Center, Ottawa) were grown in petri plates with two compartments as described by St-Arnaud et al. (1996). The mycorrhizal roots were confined to one compartment, but the extraradical fungus was allowed to grow over the divider and into the other compartment. After 3 weeks, the ERM had extensively developed in the root-free medium and it mainly consisted of absorptive hyphae. In the following 6 to 13 weeks (9–20 weeks after the crossing over), the fungus sporulated extensively in this fungal compartment.
To label the fungal material, 99% (atom%) 13C-enriched substrates dissolved in 0.5 to 1 mL of water were added (through a sterile 0.2-μm filter) to the compartment containing the mycorrhizal roots or the ERM (approximately 12 mL of media in each compartment) 1 to 2 weeks after the fungus had crossed the plastic barrier (Pfeffer et al., 1999). The substrates and final concentrations used were as follows: 13C1-Glc, 25 mm; 13C1,3-Gly, 10 mm; and 13C2-acetate, 4 mm. Depending on the time treatment, the ERM was collected 3 weeks (“young mycelium”), 9 to 13 weeks (“mature mycelium”), or 16 weeks (“old mycelium”) after labeling. Fungal tissue was recovered by blending the solidified medium (in a commercial blender; Waring) in sodium-citrate buffer (10 mm, pH 6; Doner and Bécard, 1991) at high speed for 5 s, followed by intermittent blending at low speed for 5 min (Pfeffer et al., 1999). The fungal compartments of three plates were collected and combined for each sample, except for young mycelium experiments in which six to nine compartments were pooled. The ERM was then collected on a 38-μm sieve, rinsed with water, and frozen at −80°C.
To perform germinating spores experiments, fungal tissue from 12-week-old (after crossing over) ERM was recovered as explained above except for blending for 45 s at high speed (Bago et al., 1999). Spores were germinated in the dark in liquid M medium without Suc for 14 d (at 32°C with 2% [v/v] CO2; Bago et al., 1999). 13C2-Acetate and 13C1,3-Gly were added as filter-sterilized solutions to the medium for a final concentration of 4 and 10 mm, respectively. In all cases, more than 80% of the spores germinated within 3 d and they formed a macroscopically visible diffuse mycelium during the incubation period. After incubation, the fungal material was recovered and frozen at −80°C.
Carbohydrate Extraction, Enzymatic Digestions, and NMR Sample Preparation
To obtain the different fractions of fungal carbon molecules studied, sequential extraction of soluble carbohydrates, neutral lipids, glycogen, and chitin were performed.
Soluble carbohydrate and neutral lipid extracts were prepared as previously described (Pfeffer et al., 1999). In brief, samples were lyophilized and ground with mortar and pestle with acid-washed sand at −20°C in 3 to 5 mL of methanol/water (MeOH/H2O; 70/30, v/v). After filtration (using no. 2 filter paper; Whatman, Clifton, NJ), the methanol was removed by evaporation under reduced pressure and the aqueous solution was freeze dried. The solid residue remaining after MeOH/H2O extraction was freeze-dried and re-extracted in 30 to 40 mL of boiling isopropyl alcohol for 20 min to extract neutral lipids. After filtering, the solvent was removed by evaporation under a stream of nitrogen.
Solid residues from lipid extraction were treated enzymatically for glycogen breakdown to Glc. This was accomplished by resuspending samples in a sodium acetate buffer (50 mm, pH 4.5) and adding five units of an amyloglucosidase (EC 3.2.1.3; reference no. 10115; Fluka, Buchs, Switzerland) solution (in deionized water) per sample. Samples were then incubated at 50°C with continuous stirring for 3 h. After decanting, supernatants were acquired with a Pasteur pipette and were further centrifuged to completely remove solid particles. The aqueous solution was then freeze dried and kept at 4°C until NMR analysis.
Labeling patterns in chitin were characterized in solids obtained from glycogen digestions. First samples were pretreated for 20 min in boiling water with a KOH solution (1 m) to remove proteins and other material coating the fungal cell wall (Ruiz-Herrera, personal communication). After decanting, KOH was removed and solids were washed in deionized water, and then soaked in a potassium phosphate buffer (200 mm, pH 6.0). One unit of chitinase (EC 3.2.1.14; reference no. C-1525; Sigma, St. Louis), 1 unit of chitosanase (EC 3.2.1.132; reference no. C-9830; Sigma), and 0.1 unit of β 1–3 glucanase (EC 3.2.1.6; reference no. L-9259; Sigma; each prepared as a stock solution in the potassium phosphate buffer) were added jointly to the samples, which were then incubated at 37°C for 7 d in continuous agitation. Sodium azide (0.01%, w/v) was added per sample to prevent growth of any contaminant during the digestion process. After digestion, samples were decanted and supernatants were centrifuged, freeze-dried, and kept at 4°C until NMR analysis.
For NMR analysis, carbohydrate extracts as well as glycogen and chitin digestions were dissolved in 750 μL of deuterated water. Lipid extracts were dissolved in 750 μL of deuterated chloroform. In all cases, insoluble matter was removed by centrifugation.
NMR Spectroscopy and Quantification of 13C Labeling
Spectra were obtained using a spectrometer (UnityPlus 400 MHz; Varian, Palo Alto, CA) with an superconducting magnet (9.4T; Oxford Instruments, Oxford), although several 1H extract spectra were acquired at 750 MHz on spectrometers (Bruker, Billerica, MA). A 5-mm broadband probe was used for 13C spectra, whereas a 5-mm 1H, inverse detection probe with Z gradient and broadband decoupling coil was used for 1H spectra. 13C spectra were accumulated with 80° pulse angles, WALTZ-1H decoupling, and with recycle times of 4.2 s for aqueous samples and 13.2 s for samples dissolved in chloroform. For 1H spectra, 80° pulses and recycle times of 4.5 s were used, and when necessary, 12-s recycle times were used to prevent distortion of the relative intensities of the 1H-12C and 1H-13C signals (London, 1988). Total acquisition times for 13C and 1H extract were between 12 and 36 h, depending on the concentration of each sample.
The identification of peaks in 13C and 1H spectra was made from literature values (Fan, 1995) or via comparison of spectra of purified compounds (Bago et al., 1999; Pfeffer et al., 1999). 13C chemical shifts were referenced to the signals of 13C1-trehalose (94.1 ppm) in carbohydrate extracts, 13C1-β-Glc (96.8 ppm) in glycogen digestions or the NAG-CH3 group in cell wall enzyme-treated samples. 1H signals were referenced to water (4.67 ppm at 35°C). Carbon and proton shifts were expressed in parts per million with respect to tetramethylsilane at 0 ppm.
C1:C6 and C6:C5 ratios were obtained from integrals of the respective peaks. 13C-isotopic abundance (atom percentage of 13C) of the labeled positions of a given compound was calculated by measurement of the 13C-1H satellites of 1H signals in proton spectra. To be specific, the satellites of the 1H signals coupled to (i) the C1 and 1′ of trehalose, 1H signal at 5.18 ppm for MeOH/H2O extracts; (ii) C1 of Glc, 1H signal at 5.22 ppm for digested glycogen samples, and (iii) the methyl carbon of NAG, 1H signal for chitin digestions were used.
Gene Cloning, Sequencing, and Quantification of Gene Expression
Glycogen Synthase and Glycogen BE Gene Isolation
The glycogen synthase gene from G. intraradices was isolated using degenerate PCR primers designed from conserved sequence motifs in a series of fungal glycogen synthase amino acid sequences. These were aligned using ClustalW program, and degenerate PCR primers were designed based on regions of high sequence conservation as follows: forward: 5′-CAYGARTTYCARAAYYTNCA-3′; and reverse: 5′-GTRTANCCCCANGGYTCRTA-3′.
cDNA from 11-d germinating spores of G. intraradices served as template along with no-template negative control. The PCR product of the expected size was gel isolated and cloned into Topo vector (Invitrogen, Carlsbad, CA). Plasmid DNA from random clones was isolated and sequenced as described previously (Lammers et al., 2001).
The glycogen BE was discovered in a random EST sequencing screen of clones from a germinating spore cDNA library from G. intraradices (Lammers et al., 2001).
Obtaining Full-Length Sequences by RACE
Full-length cDNA sequences were obtained using the SMART RACE cDNA amplification kit (CLONTECH, Palo Alto, CA). Two hundred-fifty nanograms of total RNA was used to synthesize each 5′- and 3′-RACE-ready cDNA. The sequences of gene-specific primers used for RACE were based on the sequences of gene fragments obtained from PCR-based amplification (GS) or from random EST sequence data (glycogen BE). Primers were synthesized by IDT (Coralville, IA). Primer sequences were as follows: GS 5′-RACE gene-specific primer (GSP): 5′-GCCCCAAGGTTCATAATAACTTGGGAACACGCC-3′; nested GSP: 5′-TGAAATATCACCACCCGTATAGCGTGCAGC-3′; GS 3′-RACE GSP: 5′-GCGGCAACTCAATCATTTGCTGTGGAAGC-3′; NGSP2: 5′-TTTGTTCGTGGTTGCCATCTTGGCGTGTTCC-3′; RACE primers for glycogen BE: 5′-RACE GSP1: 5′-CCAGGCTGGAAGACGATCGATACATTCG-3′; NGSP1: 5′-CAATCAAACTTGCAGCAACGGCATTAGGTGCC-3′; 3′-RACE GSP: 5′-ATATACCGCGAATGGGCACCTAATGCC-3′; NGSP2: 5′-GACTACACCAGAAGGCGAATGTATCGATCG-3′. The resulting RACE fragments for each gene were cloned into the pGEM-T Easy vector (Promega, Madison, WI) and were sequenced with M13 forward and reverse primers.
Real-Time RT-PCR Quantification of Gene Expression
RNA was extracted from the ERM, germinating spores, and colonized and uncolonized root tissue using the method of Lammers et al. (2001). Gene expression was monitored using an PRISM 7700 instrument (Applied Biosystems, Foster City, CA) and “Taqman” assays designed for β-tubulin, isocitrate lyase, and 18S ribosomal RNA. The amplification and probe sequences for each assay are shown below along with amplicon sizes. Primers were used at 500 nm and probes at 100 nm final concentrations. Duplicate 5-ng samples of total RNA from ERM and germinating spores were analyzed by Taq-Man assays as described in Lammers et al. (2001). Assays of RNA from colonized (IRM) and uncolonized roots used duplicate 100 ng of total RNA. The critical threshold cycle values for unknowns were converted into absolute copy numbers by comparison against standard curves of critical threshold cycle versus absolute copy number using plasmid DNA template containing each gene. Standard DNAs were prepared using Qiagen (Valencia, CA) kits and were quantified by UV absorbance spectroscopy. All standard curves were determined in duplicate at 102, 103, 104, and 106 copies for each assay (not shown). Goodness of fit R values for all curves was equal or greater than 0.990. Results obtained from no RT control assays demonstrate the absence of any significant DNA contamination of the RNA. Likewise, quadruplicate control measurements using no added template for each assay were negative over 45° cycles (not shown). Absolute quantification was based on standard curves for each assay.
To prepare template for Taq-Man RT-PCR assays, 100 ng of total RNA was treated with RNase-free DNase-I (DNA free; Ambion, Austin, TX) for 1 h followed by DNase-I removal as specified by the manufacturer. Duplicate assays used 5-ng aliquots of the DNase-treated RNA preincubated for 15 min at 95°C then placed on ice to remove any potential interfering secondary structures. The reverse amplification primer served as the primer for reverse transcription. Each RT-PCR assay was run in 50 mL of total volume using One-Step RT-PCR Master mix containing AmpliTaq Gold DNA polymerase to which 12.5 units of MultiScribe enzyme was added (all from Applied Biosystems, Foster City, CA). The reactions were incubated at 48°C for 60 min for reverse transcription, followed by a 10-min incubation at 95°C to activate the AmpliTaq Gold polymerase and 45 cycles of 15 s at 95°C, and 1 min at 60°C. MultiScribe enzyme was omitted from the no-RT control reactions.
GS Amplicon 74-bp Amplicon
Forward primer: 5′-AACAGCTTGACCTTTCAGTGCTT-3′; reverse primer: CCGTTGTTGCATTTATTGTCATG; Taq-Man Probe: 5′-FAM-CACAGCAAATGATTGAGTTGCCGCA-3′-TAMRA-3′.
β-Tubulin 82-bp Amplicon
Forward primer: 5′-AGAAAGTCTACCACGGAAAATAGTAGCT-3′; reverse primer: 5′-TTCACGTAATATGATGGCTGCAT-3′; Taq-Man Probe: 5′-FAM-CGGTCAAATATCTTCCATGACGAGGATCG-3′-TAMRA-3′.
18S rRNA 76-bp Amplicon
Forward primer: 5′-CCGTGAATCATCGAATCTTTGAA-3′; reverse primer: 5′-CACTGACCCTCAAACAGGCATA-3′; Taq-Man Probe: 5′-FAM-TGCACTCTCTGGCAACCCGGG-3′-TAMRA-3′
Table 1.
. Transcript copy numbers for fungal genes expressed in colonized and uncolonized roots, extraradical mycelium, and germinating spores of G. intraradices
| Gene | Colonized Roots (per 100 ng total RNA) | Uncolonized Roots (per 100 ng total RNA) | Extraradical Mycelium (per 5 ng total RNA) | Germinating Spores (per 5 ng total RNA) | ||||
|---|---|---|---|---|---|---|---|---|
| +RT | −RT | +RT | −RT | +RT | −RT | +RT | −RT | |
| Glycogen synthase | 1.7 ± 0.6 × 103 | <50 | <50 | <50 | 3 ± 0.7 × 104 | <50 | 3.2 ± 2.6 × 103 | <50 |
| Tubulin | 4.0 ± 0.2 × 103 | <50 | <50 | <50 | 6.7 ± 2.6 × 103 | <50 | 2.8 ± 0.7 × 103 | <50 |
| rRNA | 2.9 ± 0.4 × 107 | <50 | 2 ± 0.5 × 103 | <50 | 6.6 ± 2.0 × 108 | 55 ±3 | 5.6 ± 0.74 × 108 | 102 ± 6 |
Real-time RT-PCR experiments were performed on DNAse I-treated total RNA isolated from the indicated tissues. Table values are the mean transcript copy numbers (n = 2) ± half the full range of values.
ACKNOWLEDGMENTS
We wish to thank Aisha Abdul-Wakeel and Dr. Daniel Schwartz (U.S. Department of Agriculture-Agricultural Research Service) for technical assistance.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.007765.
LITERATURE CITED
- Bago B, Azcón-Aguilar C, Goulet A, Piché Y. Branched absorbing structures (BAS): a feature of the extraradical mycelium of symbiotic arbuscular mycorrhizal fungi. New Phytol. 1998;139:375–388. [Google Scholar]
- Bago B, Pfeffer PE, Douds DD, Brouillette J, Bécard G, Shachar-Hill Y. Carbon metabolism in spores of the arbuscular mycorrhizal fungus Glomus intraradices as revealed by nuclear magnetic resonance spectroscopy. Plant Physiol. 1999;121:263–271. doi: 10.1104/pp.121.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago B, Pfeffer PE, Shachar-Hill Y. Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol. 2000;124:949–957. doi: 10.1104/pp.124.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago B, Zipfel W, Williams R, Jun J, Arreola R, Lammers P, Pfeffer PE, Shachar-Hill Y. Translocation and utilization of fungal lipid in the arbuscular mycorrhizal symbiosis. Plant Physiol. 2002;128:108–124. [PMC free article] [PubMed] [Google Scholar]
- Bécard G, Doner LW, Rolin DB, Douds DD, Pfeffer PE. Identification and quantification of trehalose in vesicular-arbuscular mycorrhizal fungi by in vivo 13C NMR and HPLC analyses. New Phytol. 1991;118:547–552. [Google Scholar]
- Beilby JP, Kidby DK. Biochemistry of ungerminated and germinated spores of the vesicular-arbuscular mycorrhizal fungus, Glomus caledonium: changes in neutral and polar lipids. J Lipid Res. 1980;21:739–750. [PubMed] [Google Scholar]
- Bonfante P, Balestrini R, Mendgen K. Storage and secretion processes in the spore of Gigaspora margarita Becker and Hall as revealed by high-pressure freezing and freeze substitution. New Phytol. 1994;128:93–101. doi: 10.1111/j.1469-8137.1994.tb03991.x. [DOI] [PubMed] [Google Scholar]
- Doner LW, Bécard G. Solubilization of gellan gells by chelation of cations. BioTechniques. 1991;5:25–29. [Google Scholar]
- Douds DD, Pfeffer PE, Shachar-Hill Y. Carbon partitioning, cost and metabolism of Arbuscular Mycorrhizae. In: Douds DD, Kapulnik Y, editors. Arbuscular Mycorrizas Physiology and Function. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. pp. 107–130. [Google Scholar]
- Fan TW-M. Recent advances in profiling plant metabolites by multi-nuclear and multidimensional NMR. In: Shachar-Hill Y, Pfeffer PE, editors. Nuclear Magnetic Resonance in Plant Biology. Rockville Maryland: American Society of Plant Physiologists; 1995. pp. 181–254. [Google Scholar]
- Fortin JA, Bécard G, Declerck S, Dalpe Y, St-Arnaud M, Coughlan AP, Piche Y. Arbuscular mycorrhiza on root-organ cultures. Can J Bot. 2002;80:1–20. [Google Scholar]
- George E, Marschner H, Jakobsen I. Role of arbuscular mycorrhizal fungi in uptake of phosphorus and nitrogen from soil. Crit Rev Biotechnol. 1995;15:257–270. [Google Scholar]
- Graham JH. Assessing costs of arbuscular mycorrhizal symbiosis in agroecosystems. In: Podila GK, Douds Jr DD, editors. Current Advances in Mycorrhizae Research. St. Paul, MN: American Phytopathological Society Press; 2000. pp. 127–140. [Google Scholar]
- Jabaji-Hare S. Lipid and fatty acid profiles of some vesicular-arbuscular mycorrhizal fungi: contribution to taxonomy. Mycologia. 1998;80:622–629. [Google Scholar]
- Jakobsen I. Transport of phosphorus and carbon in VA mycorrhizas. In: Varma A, Hock B, editors. Mycorrhiza: Structure, Function, Molecular Biology and Biotechnology. Berlin: Springer-Verlag; 1995. pp. 297–323. [Google Scholar]
- Jennings DH. The Physiology of Fungal Nutrition. Cambridge, UK: Cambridge University Press; 1995. [Google Scholar]
- Jun J, Abubaker J, Rehrer C, Pfeffer PE, Shachar-Hill Y, Lammers PJ. Expression in an arbuscular mycorrhizal fungus of genes involved in metabolism, transport, the cytoskeleton and the cell cycle. Plant Soil. 2002;224:141–148. [Google Scholar]
- Koide RT, Schreiner RP. Regulation of the vesicular-arbuscular mycorrhizal symbiosis. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:557–581. [Google Scholar]
- Lammers PJ, Jun J, Abubaker J, Arreola R, Gopalan A, Bago B, Hernandez-Sebastia C, Allen JW, Douds DD, Pfeffer PE et al. The glyoxylate cycle in an arbuscular mycorrhizal fungus: gene expression and carbon flow. Plant Physiol. 2001;127:1287–1298. [PMC free article] [PubMed] [Google Scholar]
- London R. 13C-Labeling in studies of metabolic regulation. Progress NMR Spectr. 1988;20:337–383. [Google Scholar]
- Lossos A, Meiner Z, Barash V, Soffer D, Schlesinger I, Abramsky O, Argov Z, Shpitzen S, Meiner V. Adult polyglucosan body disease in Ashkenazi Jewish patients carrying the Tyr(329) Ser mutation in the glycogen-branching enzyme gene. Ann Neurol. 1998;44:867–872. doi: 10.1002/ana.410440604. [DOI] [PubMed] [Google Scholar]
- Mugnier J, Mosse B. Vesicular-arbuscular mycorrhizal infection in transformed root-inducing T-DNA roots grown axenically. Phytopathology. 1987;77:1045–1050. [Google Scholar]
- Pederson BA, Cheng C, Wilson WA, Roach PJ. Regulation of glycogen synthase: identification of residues involved in regulation by the allosteric ligand glucose-6-P and by phosphorylation. J Biol Chem. 2000;275:27753–27761. doi: 10.1074/jbc.M003342200. [DOI] [PubMed] [Google Scholar]
- Pfeffer PE, Douds DD, Bécard G, Shachar-Hill Y. Carbon uptake and the metabolism and transport of lipids in and arbuscular mycorrhiza. Plant Physiol. 1999;120:587–598. doi: 10.1104/pp.120.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M. Enzyme activities of the internal hyphae and germinated spores of an arbuscular mycorrhizal fungus, Gigaspora margarita Becker and Hall. New Phytol. 1995;129:425–431. [Google Scholar]
- Skurat AV, Peng HL, Chang HY, Cannon JF, Roach PJ. Rate-determining steps in the biosynthesis of glycogen in COS cells. Arch Biochem Biophys. 1996;328:283–288. doi: 10.1006/abbi.1996.0174. [DOI] [PubMed] [Google Scholar]
- Shachar-Hill Y, Pfeffer PE, Douds D, Osman SF, Doner LW, Ratcliffe RG. Partitioning of intermediate carbon metabolism in VAM colonized leek. Plant Physiol. 1995;108:7–15. doi: 10.1104/pp.108.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Read DJ. Mycorrhizal Symbiosis. London: Academic Press; 1997. [Google Scholar]
- Solaiman MD, Saito M. Use of sugars by intraradical hyphae of arbuscular mycorrhizal fungi revealed by radiorespirometry. New Phytol. 1997;136:533–538. doi: 10.1046/j.1469-8137.1997.00757.x. [DOI] [PubMed] [Google Scholar]
- St-Arnaud M, Hamel C, Vimard B, Caron M, Fortin JA. Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol Res. 1996;100:328–332. [Google Scholar]
- Thevelein JM, Hollander JA, Shulman RG. Changes in the activity and properties of trehalase during early germination of yeast ascospores: correlations with trehalose breakdown as studied by in vivo 13C NMR. Proc Natl Acad Sci USA. 1982;79:3503–3507. doi: 10.1073/pnas.79.11.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon VJ, Vigneronlesens C, Mariannepepin T, Montreuil J, Decq A, Rachez C, Ball SG, Cannon JF. Coordinate regulation of glycogen-metabolism in the yeast Saccharomyces cerevisiae: induction of glycogen-branching enzyme. J Biol Chem. 1992;267:15224–15228. [PubMed] [Google Scholar]
- Ubalijoro E, Chantal C, McClung CR, Smith DL. Detection of chitin synthase class I and II type sequences in six different arbuscular mycorrhizal fungi and gene expression in Glomus intraradices. Mycol Res. 2001;105:470–476. [Google Scholar]
- Ziemssen F, Sindern E, Schroder JM, Shin YS, Zange JH, Kilimann MW, Malin JP, Vorgerd M. Novel missense mutations in the glycogen-branching enzyme gene in adult polyglucosan body disease. Ann Neurol. 2000;47:536–540. [PubMed] [Google Scholar]