Abstract
For the effective recycling of nutrients, vascular plants transport pooled inorganic ions and metabolites through the sieve tube. A novel sulfate transporter gene, Sultr1;3, was identified as an essential member contributing to this process for redistribution of sulfur source in Arabidopsis. Sultr1;3 belonged to the family of high-affinity sulfate transporters, and was able to complement the yeast sulfate transporter mutant. The fusion protein of Sultr1;3 and green fluorescent protein was expressed by the Sultr1;3 promoter in transgenic plants, which revealed phloem-specific expression of Sultr1;3 in Arabidopsis. Sultr1;3-green fluorescent protein was found in the sieve element-companion cell complexes of the phloem in cotyledons and roots. Limitation of external sulfate caused accumulation of Sultr1;3 mRNA both in leaves and roots. Movement of 35S-labeled sulfate from cotyledons to the sink organs was restricted in the T-DNA insertion mutant of Sultr1;3. These results provide evidence that Sultr1;3 transporter plays an important role in loading of sulfate to the sieve tube, initiating the source-to-sink translocation of sulfur nutrient in Arabidopsis.
Inorganic sulfate is acquired from the soil as a major source of sulfur nutrient in higher plants. Sulfate is transported to various organs through the xylem stream and used for the synthesis of sulfur-containing amino acids and numerous sulfur metabolites (Leustek and Saito, 1999). During growth and development of the young expanding organs, the sulfate reserve in the vacuoles of source organs can be remobilized through the sieve element. This long-distance transport of sulfur from the source to sink organ is conceivably mediated by the translocation of sulfate or sulfur-containing metabolites. Loading of sulfate to the sieve element is accordingly an important physiological process for the effective recycling of sulfur nutrients.
Sieve elements and companion cells are the components of the sieve tubes in higher plants linked by specialized plasmodesmata (van Bel, 1996; Haritatos et al., 2000; Oparka and Santa Cruz, 2000; van Bel et al., 2002). Loading of nutrients and metabolites to the sieve element-companion cell complexes requires the function of active transport systems that localize in the plasma membranes. Transporters present at the plasmamembranes of the enucleated-sieve elements directly carry out loading of nutrients to the phloem sap. Recent studies on Suc transporters suggest that multiple isoforms of plasmamembrane-bound Suc transporters facilitate translocation of photosynthates to the sink organs through this pathway (Lalonde et al., 1999). Nutrients can alternatively be taken up by the transporters from the apoplast to the companion cells and transported to the sieve elements through the connection of plasmodesmata. Metabolites generated in the companion cells are transferred to the sieve element through the same pathway. Glutathione and S-methyl-Met are the major sulfur compounds in the phloem sap (Bourgis et al., 1999). Furthermore, glutathione translocated in the phloem is suggested to mediate transmission of the interorgan signal of sulfur status in vascular plants (Lappartient et al., 1999). Studies with poplar more recently suggested a correlation between the demand of sulfur in shoots and the sulfate to glutathione ratio in the phloem sap (Herschbach et al., 2000).
In the past few years, numbers of genes encoding high-affinity sulfate transporters in vascular plants have been isolated and characterized (Smith et al., 1995, 1997; Vidmar et al., 1999; Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002). These high-affinity sulfate transporters are predominantly expressed in roots of sulfur-starved plants and are suggested to serve for the initial uptake of sulfate from the soil. The Arabidopsis Sultr1;1 and Sultr1;2 localize at the epidermis and cortex of roots, and are highly regulated by sulfur nutrition (Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002). Direct contribution of Sultr1;1 or Sultr1;2 to the uptake of sulfate in root was elucidated in the knockout mutant and antisense plants in Arabidopsis (Shibagaki et al., 2002; Yoshimoto et al., 2002). In the present study, the third isoform of the high-affinity sulfate transporter, Sultr1;3, was identified in Arabidopsis and was demonstrated to show specific function for the loading of sulfate into the sieve tube, facilitating retranslocation of sulfur source in plants.
RESULTS
Identification of a Novel High-Affinity Sulfate Transporter, Sultr1;3 in Arabidopsis
The Arabidopsis genome (Arabidopsis Genome Initiative, 2000) contains 14 members of sulfate transporter genes that are assumed to function independently for the uptake and distribution of sulfate in various cell types. We have recently characterized the function of two distinct high-affinity sulfate transporters, Sultr1;1 and Sultr1;2, that facilitate the initial uptake of sulfate at the root epidermis and cortex in Arabidopsis (Shibagaki et al., 2002; Yoshimoto et al., 2002). In the present study, a putative open reading frame, At1g22150, that potentially encodes the third isoform of the high-affinity sulfate transporter in Arabidopsis was identified on the BAC clone F2E2 (accession no. AC069252) and designated Sultr1;3.
The Sultr1;3 cDNA (accession no. AB049624) was isolated by reverse transcriptase (RT)-PCR from the root RNA of sulfur-starved Arabidopsis plants. The open reading frame of Sultr1;3 encoded a polypeptide of 656 amino acids that shows 70.0% and 83.8% identities to Sultr1;1 and Sultr1;2, respectively (Fig. 1A). Sultr1;3 was able to complement the lesion of sulfate uptake capacity of the yeast mutant, CP154-7A that lacks two sulfate transporter genes, SUL1 and SUL2 (Fig. 1B). The growth of yeast mutant cells expressing the Sultr1;3 cDNA on low-sulfur medium was comparable with those containing the Sultr1;1 or Sultr1;2 cDNAs, suggesting that Sultr1;3 encodes a functional sulfate transporter (Fig. 1B). The phylogenic relationships of plant sulfate transporters indicated that Sultr1;3 falls into the group of high-affinity sulfate transporters in the vascular plants (Smith et al., 1995, 1997; Vidmar et al., 1999; Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002; Fig. 2). As a consequence, Sultr1;1, Sultr1;2 and Sultr1;3 were the three members of high-affinity sulfate transporters encoded by the Arabidopsis genome.
Figure 1.
Comparison of Sultr1;1, Sultr1;2, and Sultr1;3 sulfate transporters. A, Protein sequence alignment of Sultr1;1, Sultr1;2, and Sultr1;3. The alignment was performed using the ClustalW program. Black shading indicates identical amino acid residues. Gray shading indicates similar amino acid residues. B, Complementation of yeast mutant CP154-7A. Yeast mutant cells expressing Sultr1;1, Sultr1;2, and Sultr1;3 cDNAs or the empty vector pYE22m were grown at 30°C for 2 d on SD medium containing 0.1 mm of sulfate as a sole sulfur source.
Figure 2.
Phylogenic tree of plant sulfate transporters. The neighbor-joining tree was produced based on the alignment of the full-length sequences using ClustalW program. Munich Information Center for Protein Sequences code numbers (http://mips.gsf.de/proj/thal/) of the Arabidopsis transporters and GenBank/EMBL/DNA data bank of Japan accession numbers are indicated in the parentheses. Arabidopsis transporters are indicated in bold letters.
Sultr1;3 Is Inducible by Sulfur Limitation
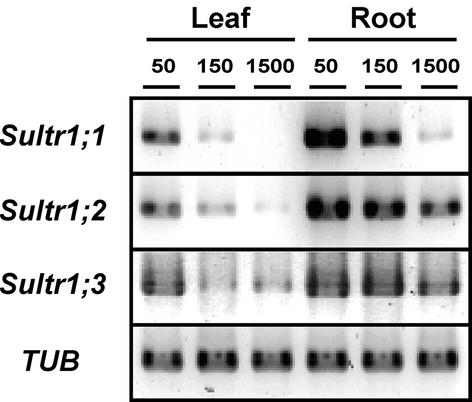
The effect of sulfur limitation on the mRNA accumulation of Sultr1;3 was investigated by RT-PCR. Arabidopsis plants were grown continuously for 2 weeks on GM (Germination Medium) (Valvekens et al., 1988) containing 50, 150, or 1,500 μm sulfate, respectively. Sultr1;3 mRNA was expressed both in leaves and roots and was abundantly expressed under sulfur-deficient conditions particularly in leaves (Fig. 3). The increased accumulation of Sultr1;3 mRNA by sulfur limitation was comparable with those observed in Sultr1;1 and Sultr1;2 expression. It is suggested that these three high-affinity sulfate transporters are strictly regulated by the changes of the sulfur status. The inducible expression of mRNA by sulfur limitation is one of the general features of the high-affinity sulfate transporter genes in plants.
Figure 3.
Effect of sulfur limitation on mRNA levels of Sultr1;1, Sultr1;2, and Sultr1;3. Arabidopsis plants were grown on GM medium containing 50, 150, or 1,500 μm sulfate for 2 weeks. RT-PCR analysis of Sultr1;1, Sultr1;2, Sultr1;3, and α-tubulin (TUB) was carried out with gene-specific primers as described in “Materials and Methods.”
Sultr1;3 Is a Phloem-Specific Sulfate Transporter
The cell type-specific expression of Sultr1;3 was studied by introducing a fusion gene construct of Sultr1;3 and green fluorescent protein (GFP; Chiu et al., 1996) in Arabidopsis. A DNA fragment of Sultr1;3 gene that starts from the 5′-region 2,541 bp upstream of the translation initiation site and terminates before the stop codon of Sultr1;3 transporter (+2,981) was amplified from the Arabidopsis genomic DNA by PCR and fused to the coding sequence of GFP. This fusion gene construct enables the expression of the Sultr1;3-GFP fusion protein under the control of the Sultr1;3 promoter. The Sultr1;3-GFP fusion gene construct was stably integrated into the Arabidopsis genome by Agrobacterium-mediated transformation (Clough and Bent, 1998). The expression of Sultr1;3-GFP fusion protein was analyzed in 16 independent transgenic lines grown for 10 d on GM medium (Valvekens et al., 1988).
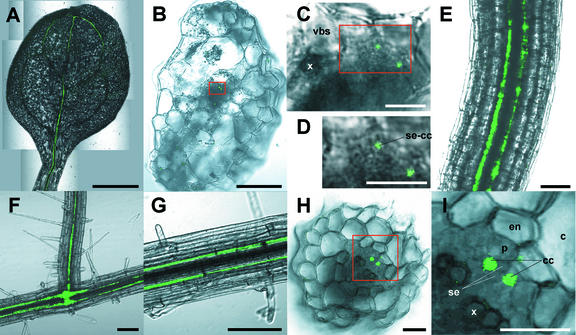
In transgenic Arabidopsis, fluorescence of Sultr1;3-GFP was detected in the phloem of cotyledons (Fig. 4, A–D), hypocotyls (Fig. 4E), and roots (Fig. 4, F–I). Expression of Sultr1;3-GFP in the cotyledon was confined within the sieve element-companion cell complexes (Fig. 4, C and D). Sultr1;3-GFP was mainly found in the source organs, and no green fluorescence was detected in the sink organs such as young rosette leaves. In roots, the level of green fluorescence in the phloem was most remarkable in the mature part of the primary roots and at the branching point of the lateral roots (Fig. 4F). More precisely, the fluorescence was detected in the companion cells in roots (Fig. 4, H and I). The patterns of the cell type-specific expression of Sultr1;3 was completely different from those of Sultr1;1 and Sultr1;2 (Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002). These results suggest that Sultr1;3 may have specific function in the transport of sulfate through the sieve element in Arabidopsis.
Figure 4.
Phloem-specific localization of Sultr1;3. Sultr1;3 promoter-coding sequence-GFP fusion gene construct was expressed in transgenic Arabidopsis. Ten-day-old plants grown on GM agar medium were analyzed. A, Cotyledon (bar = 500 μm). B, Cross section of the petiole of cotyledon (bar = 100 μm). C, Enlarged image corresponding to the red square in B (bar = 10 μm). D, Enlarged image corresponding to the red square in C (bar = 10 μm). E, Hypocotyls (bar = 100 μm). F, Junction of lateral root (bar = 100 μm). G, Root (bar = 100 μm). H, Cross section of the mature part of root (bar = 10 μm). I, Enlarged image corresponding to the red square in H (bar = 10 μm). c, Cortex; cc, companion cell; en, endodermis; p, pericycle; se, sieve element; se-cc, sieve element-companion cell complex; vbs, vascular bundle sheath; and x, xylem.
Sultr1;3 Mediates Interorgan Transport of Sulfate
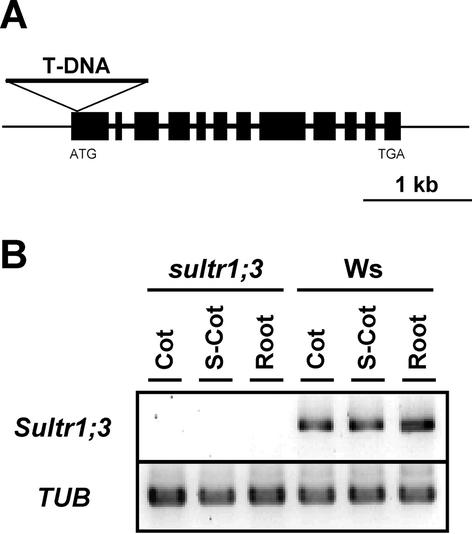
Arabidopsis T-DNA insertion mutant of Sultr1;3 was isolated from the T-DNA tagged population by reverse genetic strategy (Krysan et al., 1999). From 60,480 T-DNA transformed lines generated at the University of Wisconsin, we have identified a mutant line containing a single insertion in the coding region of Sultr1;3. T-DNA was integrated in the first exon of Sultr1;3 between the position +6 to +38 of the translation initiation site, generating a 31 bp deletion (Fig. 5A). The insertion site of T-DNA was determined by sequencing DNA fragments amplified with specific primers for Sultr1;3 and the border regions of T-DNA. Progenies containing a single homozygous insertion of T-DNA was selected through Southern hybridization, and propagated for further experiments. Expression of Sultr1;3 mRNA was entirely eliminated in the homozygous mutant (Fig. 5B).
Figure 5.
Disruption of Sultr1;3 gene by T-DNA insertion. A, Location of the T-DNA within the Sultr1;3 gene. Thick bars and lines indicate exons and introns, respectively. T-DNA is not drawn to scale. B, RT-PCR analysis of Sultr1;3 and α-tubulin (TUB) in the sultr1;3 mutant and Ws wild type. RNA was extracted from cotyledons (Cot), shoot without cotyledons (S-Cot), and roots of plants grown on GM medium for 10 d.
The movement of 35S-labeled sulfate from the source to sink organs was determined in the sultr1;3 and the background Wassilewskija (Ws) wild-type plants. To determine the rate of phloem-mediated translocation of sulfate to the sink organs, plants were grown for 10 d on GM medium, and the cotyledons were fed with 35SO42−. In these young seedlings, sulfate accumulated in the cotyledons can be reused in the expanding immature organs. After 1 h of labeling, plants were rinsed and further incubated for 1 h in non-labeled nutrient solution. Plants were excised into three parts—cotyledon, shoot without cotyledon, and roots—and the accumulation of radioactivities in each organ was counted. In the wild-type plants, 12% to 15% of labeled sulfate were transported out of the cotyledons (Table I). It is suggested that the labeled sulfate rapidly moved out from the cotyledons and accumulated in the shoot apical region. However, in the sultr1;3 mutant, most of the radioactive sulfate was still present in the cotyledons. Less than 5% of the labeled sulfate was transported from the cotyledon to distant sink organs in the mutant (Table I). These results strongly indicates that disruption of Sultr1;3 sulfate transporter can attenuate the source-to-sink transport of sulfate. It is suggested that Sultr1;3 high-affinity sulfate transporter participates in the loading of sulfate to the sieve tube particularly in the source organs, controlling the flux of sulfur on the stream of phloem sap.
Table I.
Movement of 35S-labeled sulfate in sultr1;3 and wild-type plants
| Distribution of 35S
|
|||
|---|---|---|---|
| Cotyledon | Shoot without Cotyledon | Root | |
| % | |||
| Experiment 1 (n = 9) | |||
| Ws | 84.76 ± 8.58 | 12.66 ± 8.72 | 2.58 ± 1.86 |
| sultr1;3 | 95.27 ± 1.56 | 3.28 ± 1.59 | 1.45 ± 0.58 |
| (P = 0.006) | (P = 0.012) | (P = 0.113) | |
| Experiment 2 (n = 8) | |||
| Ws | 88.08 ± 7.93 | 9.99 ± 7.34 | 1.93 ± 2.03 |
| sultr1;3 | 96.22 ± 0.67 | 2.62 ± 0.51 | 1.16 ± 0.27 |
| (P = 0.023) | (P = 0.025) | (P = 0.317) | |
Cotyledons of 10-d-old plants grown on GM medium were labeled with 35SO42−. Translocation of the radioactivity to the distal organs was determined in two independent experiments. The values indicate distribution of the radioactivity detected in each organ after 1 h of incubation (means ± sd). Statistical significance of the difference between the Ws wild type and sultr1;3 mutant is shown in parentheses.
DISCUSSION
Long-distance transport of nutrients and metabolites from the source to sink organs is mediated by the sieve element-companion cell (SE-CC) complexes of the phloem in the vasculature. Import of nutrients from the apoplastic space of vasculature to the SE-CC complex requires the function of transporters localizing at the plasmamembrane of the sieve element or the companion cells. In this study, we identified a novel phloem-localizing sulfate transporter, Sultr1;3, and elucidated its specific function in the SE-CC complex in Arabidopsis.
The high sequence similarity of Sultr1;3 with the other group 1 sulfate transporters in vascular plants indicated that Sultr1;3 is a member of the high-affinity sulfate transporters (Figs. 1A and 2). Over 70% of the identities were found among the protein sequences of Sultr1;1, Sultr1;2, and Sultr1;3. Genetic complementation of the yeast sulfate transporter mutant by the expression of Sultr1;3 clearly indicates that this transporter protein can function as sulfate transporter (Fig. 1B). In addition, Sultr1;3 mRNA was abundantly accumulated in sulfur-starved plants, which was comparable with the induced expression of Sultr1;1 and Sultr1;2 (Fig. 3). These results suggested that Sultr1;1, Sultr1;2, and Sultr1;3 may have closely related properties for sulfate transport in Arabidopsis under sulfur deficiency.
However, the spatial expression pattern of Sultr1;3 was completely different from those characterized for the other two high-affinity sulfate transporters, Sultr1;1 and Sultr1;2 in Arabidopsis. Sultr1;1 and Sultr1;2 are suggested to carry out the initial acquisition of sulfate at the root surface of Arabidopsis. These transporters were mainly expressed in the root epidermis and cortex, and their mRNA levels increased by limitation of external sulfate (Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002). Transgenic plants expressing the Sultr1;3-GFP fusion protein under the control of Sultr1;3 promoter displayed specific expression of GFP in the phloem of cotyledons, hypocotyls, and roots (Fig. 4). Expression of Sultr1;3-GFP was restricted to the SE-CC complexes in the mature organs such as cotyledons (Fig. 4, A–D) but not in the young developing rosette leaves. In roots, Sultr1;3-GFP was localized in the companion cells (Fig. 4, H and I). These expression patterns of Sultr1;3-GFP suggested that Sultr1;3 may function for the phloem-mediated transport of sulfate in Arabidopsis. Phloems have three functional parts for the source-to-sink transport of nutrients. They consist of collection phloems in the minor veins of source leaves, transport phloems, and release phloems in the sink organs for unloading (van Bel, 1996). Our results presented here indicate that Sultr1;3 mainly localizes within the transport phloem, suggesting its specific function for retrieval of sulfate leaked out from the sieve tube during the long-distance transport. This may partly contribute to retain the sulfur flux of source-to-sink transport recovering the leakage of sulfate from the sieve tubes within the transport phloem.
Analysis on the T-DNA mutant provided direct evidence for the contribution of Sultr1;3 transporter to the long-distance transport of sulfate in Arabidopsis. Feeding of 35SO42− to the cotyledons and measurements of its distribution to the distant organs revealed that transport of sulfate to the sink tissue is restricted in the sultr1;3 mutant (Table I). In the mutant, the efficiency of movement of the labeled sulfate from the cotyledons to sink organs was approximately 30% of the wild type. These results indicate that Sultr1;3 plays an important role in source-to-sink transport of sulfate. In general, sulfate ions pooled in the vacuoles or degraded from the organic compounds in the source tissues are transported to the apoplastic space of the vasculature of minor veins. Sulfate is subsequently imported into the sieve elements of collection phloems, initiating the flow of long-distance transport of sulfate toward the sink organs. The result of Sultr1;3-GFP localization suggests that this initial loading process in the collection phloems requires the functions of other transport systems independent from those associated with Sultr1;3. It is suggested that Sultr1;3 transporter is more likely responsible for the retrieval of sulfate within the transport phloem in Arabidopsis. The analysis of the sultr1;3 mutant suggests that recovery or retrieval of sulfate within the transport phloem significantly promotes the interorgan translocation of sulfate (Table I). Overaccumulation of Sultr1;3 mRNA under sulfur limitation may secure retention of sulfate in the sieve tube, which can facilitate the transport of limiting amount of sulfate from the cotyledon to the young developing sink organs under sulfur-stressed conditions (Fig. 3). Furthermore, our results suggest that Sultr1;3 in the root phloem carries out uptake of sulfate directly to the companion cells. The exact pathways for loading of sulfate in the transport phloems of leaves are yet to be investigated. The present study demonstrated the first identification of a phloem-specific sulfate transporter that participates in the interorgan movement of sulfur nutrient in vascular plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis plants were grown on GM medium (Valvekens et al., 1988) at 22°C under 16 h/8 h light and dark cycles. Sulfate-deficient GM medium was prepared by replacing sulfate salts contained in Murashige and Skoog (1962) salts with equivalent chloride salts as described previously (Takahashi et al., 2000). Agar was rinsed in deionized water to remove the contamination of sulfate.
The sultr1;3 mutant in the Ws background was screened from 60,480 random T-DNA insertion population generated at the University of Wisconsin (http://www.biotech.wisc.edu/Arabidopsis/default.htm). PCR screening (Krysan et al., 1999) was carried out following the user guidelines. Oligonucleotide primers, 1;3-F (5′-CGGCAAGCAAATACACCGTATGTCCACAA-3′) and 1;3-R-W (5′-TTACACTTGACCTCTACGTCACACGATTG-3′) were designed according to the nucleotide sequence of BAC clone, F2E2 (accession no. AC069252) to screen T-DNA insertions in the coding sequence of Sultr1;3. Single insertion of T-DNA was confirmed by Southern hybridization analysis. The integration site of the T-DNA was determined by sequencing PCR fragments amplified with the Sultr1;3 and T-DNA-specific primers.
Cloning of Sultr1;3 cDNA
Molecular biological experiments were carried out according to the standard protocols (Sambrook et al., 1989). The Sultr1;3 cDNA was isolated by RT-PCR. Oligonucleotide primers, Sultr1;3-FE (5′-CAGTGAATTCATGTCGGCTAGAGCTC-3′) and Sultr1;3-RE (5′-TAGTGAATTCTCAGACCTCGTCGGAC-3′) were designed to amplify the coding sequence of Sultr1;3 according to the nucleotide sequence of BAC clone, F2E2. Total RNA was extracted from the roots of 2-week-old Arabidopsis ecotype Columbia plants grown vertically on sulfur-deficient media containing 100 μm of sulfate using RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Reverse transcription was carried out using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) as described previously (Yoshimoto et al., 2002). PCR was carried out on the first-strand cDNA using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). The amplified EcoRI-ended fragment was cloned into the EcoRI site of pBluescriptII SK- (Stratagene) and fully sequenced on both strands.
Expression of Sultr1;3 cDNA in Yeast
The EcoRI-ended fragment of Sultr1;3 cDNA described above was cloned into the EcoRI site of the yeast expression vector, pYE22m (Ashikari et al., 1989). The resulting plasmid was transferred into the Brewer's yeast (Saccharomyces cerevisiae) mutant strain CP154-7A (Matα, his3, leu2, ura3, ade2, trp1, sul1::LEU2, and sul2::URA3; Cherest et al., 1997) by the lithium acetate method (Gietz et al., 1992), and the transformants were selected on synthetic dextrose (SD) minimal medium (Sherman, 1991) containing 20 g L−1 Glc, 0.25 mm of homo-Cys, and required amino acids. Complementation of the mutant was tested on sulfur-deficient SD medium containing 0.1 mm of sulfate as a sulfur source.
RT-PCR
Preparation of total RNA and reverse transcription was carried out as described for the isolation of Sultr1;3 cDNA. First-strand cDNA that derives from 10 ng of total RNA was used for the amplification of Sultr1;3. PCR was carried out by ExTaq DNA polymerase (Takara, Tokyo) using gene-specific primers, 1;3G-FSac (5′-CATAGCAATGTCGGCTAGAGCTCATC-3′) and 1;3-R (5′-AGATTTTGTCGTGTCCTATCAAGTCCGCA-3′). PCR was carried out for 24 cycles where cDNAs were exponentially amplified. Amplification of Sultr1;1, Sultr1;2, and α-tubulin (Ludwig et al., 1987) was carried out as described previously (Yoshimoto et al., 2002). PCR products were separated in agarose gels and stained with SYBR green (Takara). Signals were detected and quantified using FluorImager 595 (Molecular Dynamics, Sunnyvale, CA) with a 515 to 545 nm band-pass filter.
Sultr1;3-GFP
The fusion gene construct of Sultr1;3 and GFP (Chiu et al., 1996) for plant transformation was constructed as follows. Oligonucleotide primers, 1;3P-FHd (5′-AAGCTTGAGGTTTAATCTTCGTGCTTG-3′) and 1;3G-RSac (5′-GATGAGCTCTAGCCGACATTGCTATG-3′) were designed according to the nucleotide sequence of BAC clone F2E2 to amplify a fragment that starts from the 5′-promoter region 2,541 bp upstream of the translation initiation site and terminates at the SacI site at the position 11 bp downstream of the translation initiation site. Oligonucleotide primers, 1;3G-FSac (5′-CATAGCAATGTCGGCTAGAGCTCATC-3′) and 1;3c-RXb (5′-TCTAGAGACCTCGTCGGACAGTTTAG-3′) were designed to amplify the rest of the coding sequence of Sultr1;3 by PCR. PCR was carried out on genomic DNA prepared from Arabidopsis ecotype Columbia using Pfu Turbo DNA polymerase (Stratagene). BamHI-EcoRI fragment of the 35S-omega-sGFP(S65T) (Chiu et al., 1996) containing the GFP coding sequence and the nopaline synthase terminator was placed into the position of β-glucuronidase and the nopaline synthase terminator in the binary plasmid, pBI101 (BD Biosciences Clontech, Palo Alto, CA). The HindIII-SacI and SacI-XbaI fragments of Sultr1;3 were inserted between the HindIII and XbaI site of this promoter-less GFP binary vector constructed in pBI101. The binary plasmid containing the Sultr1;3 promoter and Sultr1;3 coding region-GFP fusion in-frame was transferred to Agrobacterium tumefaciens GV3101 (pMP90; Koncz and Schell, 1986) by the freeze-thaw method (Chen et al., 1994). Arabidopsis plants were transformed according to the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on GM medium (Valvekens et al., 1988) containing 50 mg L−1 kanamycin sulfate. Kanamycin-resistant T2 progenies of 16 independent lines were analyzed. Tissues were embedded in 5% (w/v) agar and cut into 150-μm cross sections with a microslicer DTK-1000 (Dosaka, Kyoto). Fluorescence of GFP in transgenic plants was observed under a BX61 microscope equipped with a FV500 confocal laser scanning system and a 505- to 525-nm band-pass filter (Olympus, Tokyo).
35S Feeding Experiment
One-microliter drop of 5 mm Na235SO4 solution (18.5 kBq; Amersham Biosciences UK, Ltd., Buckinghamshire, UK) containing 0.1% (v/v) Triton X-100 was fed to cotyledons of 10-d-old plants grown on GM medium (Valvekens et al., 1988). After 1 h of incubation, plants were rinsed three times in water and left 1 h in non-labeled GM nutrient solution. Plants were excised into three parts—cotyledon, shoot without cotyledon, and roots—and digested by adding 20 μL of 100 mm HCl to one mg fresh weight of plant tissue. Tissues were extracted for 1 h in 100 mm HCl, and the radioactivity was determined in a scintillation counter (Aloka, Tokyo).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Dr. Y. Surdin-Kerjan (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France) for the yeast mutant strain CP154-7A; Dr. Y. Tanaka (Suntory Ltd., Osaka, Japan) for the yeast expression vector pYE22m; Dr. Y. Niwa (University of Shizuoka, Japan) for the GFP expression vector 35S-omega-sGFP(S65T). We thank the Arabidopsis Biological Resource Center and the Arabidopsis Knockout Facility of University of Wisconsin Biotech Center for providing the pools of T-DNA insertion mutants. We are grateful to all colleagues in the laboratory for valuable suggestions and discussions.
Footnotes
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan, by the Japan Society for the Promotion of Science, and by Core Research for Evolutional Science and Technology of Japan Science and Technology.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.014712.
LITERATURE CITED
- Ashikari T, Kiuchi-Goto N, Tanaka Y, Shibano Y, Amachi T, Yoshizumi H. High expression, and efficient secretion of Rhizopus oryzae glucoamylase in the yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1989;30:515–520. [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, Fisher DB, Tarczynski MC, Li C, Herschbach C, Rennenberg H, Pimenta MJ, Shen TL et al. S-Methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell. 1999;11:1485–1498. doi: 10.1105/tpc.11.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Nelson RS, Sherwood JL. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques. 1994;16:664–668. , 670. [PubMed] [Google Scholar]
- Cherest H, Davidian J-C, Thomas D, Benes V, Ansorge W, Surdin-Kerjan Y. Molecular characterization of two high affinity sulfate transporters in Saccharomyces cerevisiae. Genetics. 1997;145:627–635. doi: 10.1093/genetics/145.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Gietz D, Jean AS, Woods DA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haritatos E, Medville R, Turgeon R. Minor vein structure and sugar transport in Arabidopsis thaliana. Planta. 2000;211:105–111. doi: 10.1007/s004250000268. [DOI] [PubMed] [Google Scholar]
- Herschbach C, van der Zalm E, Schneider A, Jouanin L, De Kok LJ, Rennenberg H. Regulation of sulfur nutrition in wild-type and transgenic poplar over-expressing γ-glutamylcysteine synthetase in the cytosol as affected by atmospheric H2S. Plant Physiol. 2000;124:461–473. doi: 10.1104/pp.124.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of T-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel types of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Boles E, Hellmann H, Barker L, Patrick JW, Frommer WB, Ward JM. The dual function of sugar carriers: transport and sugar sensing. Plant Cell. 1999;11:707–726. doi: 10.1105/tpc.11.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappartient AG, Vidmar JJ, Leustek T, Glass AD, Touraine B. Inter-organ signaling in plants: regulation of ATP sulfurylase and sulfate transporter genes expression in roots mediated by phloem-translocated compound. Plant J. 1999;18:89–95. doi: 10.1046/j.1365-313x.1999.00416.x. [DOI] [PubMed] [Google Scholar]
- Leustek T, Saito K. Sulfate transport and assimilation in plants. Plant Physiol. 1999;120:637–643. doi: 10.1104/pp.120.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Oppenheimer DG, Silflow CD, Snustad DP. Characterization of the α-tubulin gene family of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1987;84:5833–5837. doi: 10.1073/pnas.84.16.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Oparka KJ, Santa Cruz S. The great escape: phloem transport and unloading of macromolecules. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:323–347. doi: 10.1146/annurev.arplant.51.1.323. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:4–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, Davies JP. Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 2002;29:475–486. doi: 10.1046/j.0960-7412.2001.01232.x. [DOI] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT. Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA. 1995;92:9373–9377. doi: 10.1073/pnas.92.20.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Hawkesford MJ, Ealing PM, Clarkson DT, Vanden Berg PJ, Belcher AR, Warrilow AGS. Regulation of expression of a cDNA from barley roots encoding a high affinity sulfate transporter. Plant J. 1997;12:875–884. doi: 10.1046/j.1365-313x.1997.12040875.x. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K. The role of three functional sulfate transporters involved in uptake and translocation of sulfate in Arabidopsis thaliana. Plant J. 2000;23:171–182. doi: 10.1046/j.1365-313x.2000.00768.x. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel AJE. Interactions between sieve element and companion cell and the consequences for photoassimilate distribution: two structural hardware frames with associated physiological software packages in dicotyledons. J Exp Bot. 1996;47:1129–1140. doi: 10.1093/jxb/47.Special_Issue.1129. [DOI] [PubMed] [Google Scholar]
- van Bel AJE, Ehlers K, Knoblauch M. Sieve elements caught in the act. Trend Plant Sci. 2002;7:126–132. doi: 10.1016/s1360-1385(01)02225-7. [DOI] [PubMed] [Google Scholar]
- Vidmar JJ, Schjoerring JK, Touraine B, Glass ADM. Regulation of the hvst1 gene encoding a high-affinity sulfate transporter from Hordeum vulgare. Plant Mol Biol. 1999;40:883–892. doi: 10.1023/a:1006230131841. [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Takahashi H, Smith FW, Yamaya T, Saito K. Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J. 2002;29:465–473. doi: 10.1046/j.0960-7412.2001.01231.x. [DOI] [PubMed] [Google Scholar]