Abstract
In Arabidopsis, we previously identified two highly similar apyrases, AtAPY1 and AtAPY2. Here, T-DNA knockout (KO) mutations of each gene were isolated in a reverse genetic approach. The single KO mutants lacked a discernible phenotype. The double KO mutants, however, exhibited a complete inhibition of pollen germination, and this correlated with positive β-glucuronidase staining in the pollen of apyrase promoter:β-glucuronidase fusion transgenic lines. The vast majority of the pollen grains of these mutants were identical to wild type in size, shape, and nuclear state and were viable as assayed by metabolic activity and plasma membrane integrity. Complementation with either AtAPY1 or AtAPY2 cDNA rescued pollen germination, confirming that the phenotype was apyrase specific. Despite the redundancy of the two apyrases in rescue potential, transmission analyses suggested a greater role for AtAPY2 in male gamete success. The effect of mutant apyrase on the transmission through the female gametophyte was only marginal, and embryo development appeared normal in the absence of apyrases. The male-specific double KO mutation is fully penetrant and shows that apyrases play a crucial role in pollen germination.
Pollen germination represents a short, yet very critical event in a series of steps leading to the fertilization of the ovule. Mature pollen is shed from the anther as desiccated grains and rehydrates upon contact with the stigma. During hydration, the pollen shape changes from elongated to round, and Ca2+ enters the pollen grain, leading to rearrangement of the cytoskeleton and establishment of a Ca2+ gradient (Heslop-Harrison and Heslop-Harrison, 1992a, 1992b; for schematic illustration, see Johnson and McCormick, 2001). Germination follows rapidly, usually within minutes in Arabidopsis (Pruitt and Hülskamp, 1994), and is visually manifested by the emergence of a pollen tube. Although pollen germination in vivo is a tightly regulated process dependent on multiple cues from the stigma (Johnson and Preuss, 2002), pollen grains will germinate in vitro in aqueous medium minimally containing Suc, boric acid, and Ca2+ (Taylor and Hepler, 1997). Few genes have been identified so far that are specific for the germination stage (for reviews, see Franklin-Tong, 2002; Johnson and Preuss, 2002). Here, we introduce the enzyme apyrase as an essential player in this early gametophytic phase.
Apyrases hydrolyze nucleoside tri- and diphosphates and are highly active and most likely ubiquitous, because they have been found in all pro- and eukaryotic organisms examined for their presence. In animals, these enzymes have been shown to play important regulatory roles in the mediation of signaling events, for example the quenching of the neurotransmitter ATP (Plesner, 1995; Komoszynski and Wojtczak, 1996).
The function of plant apyrases has been studied most intensively in legumes, namely Dolichos biflorus, wild soybean (Glycine soja), Medicago truncatula, and pea (Pisum sativum). In wild soybean and D. biflorus, two apyrases each were cloned, Db-apyrase-1/Db-apyrase-2 (Roberts et al., 1999) and Gs50/Gs52 (Day et al., 2000), all of which contain a predicted transmembrane domain at the N terminus. In M. truncatula, a total of four apyrase sequences were isolated, Mtapy1 to -4, two of which exhibited an increase in transcript levels upon inoculation of roots with rhizobia (Cohn et al., 2001). A similar response was observed in wild soybean with Gs52 (Day et al., 2000). Involvement of apyrase in nodulation was further substantiated by findings that Nod signals bind to Db-apyrase-1 and stimulate its enzymatic activity (Roberts et al., 1999).
Additional physiological roles of plant apyrases have been found through studies using the pea “NTPase” (an earlier, more generic name given to the pea apyrase; Hsieh et al., 1996). Overexpression of this apyrase in Arabidopsis lines led to increased scavenging of extracellular phosphate (Thomas et al., 1999). These transgenic lines also displayed higher resistance to toxins such as cycloheximide compared with wild type (Thomas et al., 2000). In addition, the same pea apyrase was found to copurify with the cytoskeleton and was hypothesized to play a role in mRNA transport along the cytoskeleton (Shibata et al., 1999).
Despite these initial studies of apyrase function, the general importance of this enzyme in plants is not well understood. In this study, the two previously characterized apyrases AtAPY1 and AtAPY2 (Steinebrunner et al., 2000) from Arabidopsis were targeted in a T-DNA knockout (KO) screen to gain more insight into generic physiological functions of apyrases in plants. The two proteins were previously shown to share many similarities including 87% sequence identity, the same ATP to ADP hydrolysis ratios, and ubiquitous expression with only slight differences in transcript levels. However, a gel overlay assay tested only Atapy1 positive for calmodulin binding (Steinebrunner et al., 2000). Here, we report the isolation and analysis of KO lines for each apyrase individually, and we show that pollen not expressing any apyrase is unable to germinate. This reveals a novel role for apyrases in plants and identifies an unexpected enzyme constituent required for plant sexual reproduction.
RESULTS
Identification of KO Lines
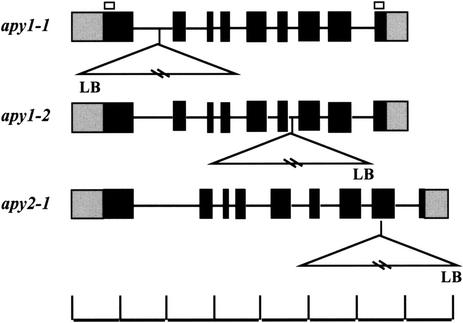
Approximately 60,500 individual T-DNA lines generated by the Arabidopsis KO Facility at the University of Wisconsin-Madison were screened. Three T-DNA insertions were found within the AtAPY1 gene, all within introns, and the three mutants were designated apy1-1, apy1-2 (Fig. 1), and apy1-3. One T-DNA insertion was localized to exon 8 of the AtAPY2 gene (apy2-1; Fig. 1). Line apy1-3 was not pursued further because it could not be associated with a specific pool in a later stage of the PCR screening procedure. For the other mutant lines, plants homozygous for the T-DNA insertion were subjected to reverse transcriptase (RT)-PCR analysis to verify true KO conditions.
Figure 1.
Genomic structure of the apyrase genes and location of the T-DNA insertions. Black and gray boxes represent translated and untranslated exons, respectively, whereas the lines represent introns. T-DNA insertions are denoted by triangles. The white boxes indicate the position of the primers ApyF and ApyR, which were used in the screen for the three T-DNA KO lines apy1-1, apy1-2, and apy2-1. LB, Left border sequence. The scale marks 500-bp distances. Exons and introns are drawn to scale.
apy1-1, apy1-2, and apy2-1 Represent True Apyrase KO Lines
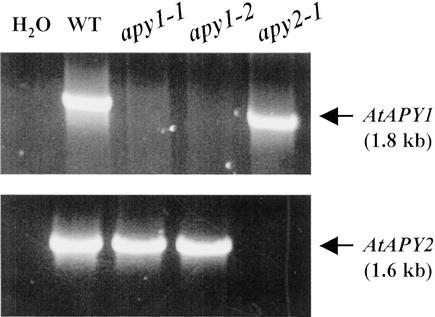
The apy1-1 and apy1-2 mutants failed to amplify AtAPY1 product, whereas the apy2-1 mutation produced no AtAPY2 cDNA (Fig. 2). The PCR products corresponded to the expected cDNA sizes, ruling out the possibility that the products stemmed from genomic DNA contamination. Therefore, all three T-DNA lines represented true apyrase KO lines.
Figure 2.
RT-PCR analysis of KO lines. RT-PCR of AtAPY1 and AtAPY2 in wild type and the three KO lines. In the lane marked H2O, water was used as template instead of cDNA as a control.
All Three Homozygous Mutant Lines Contain Two T-DNA Insertions before the Backcross
To obtain single insertion lines, individual homozygous mutant plants of each line (apy1-1, apy1-2, and apy2-1) were backcrossed once to the Wassilewskija wild type. The F1 generation was selfed and the progeny (F2) from individual F1 plants was assayed for segregation ratios of kanamycin (kan) resistance (Table I). All six analyzed F2 individuals of the apy1-2 background showed a ratio of resistant to sensitive seedlings between 3:1 and 15:1, which suggested possible linkage of two T-DNA insertions. The F2 generations of the apy1-1 background, however, segregated in a 3:1 manner except for one F2 individual. Therefore, the backcross eliminated a second linked T-DNA insertion in almost all of the F1 progeny. The apy2-1 F1 generation produced an equal number of progeny segregating in 3:1 and 15:1 ratios. This segregation pattern is indicative of a parental line with two unlinked T-DNA insertions, one of which, in this case the desired one, being homozygous and the second one being heterozygous. None of the three KO lines harbored more than two T-DNA insertions before the backcross, which is in accordance with the average number of 1.7 loci for T-DNA inserts per transformed line projected by the Arabidopsis KO facility (http://www.biotech.wisc.edu/NewServicesAndResearch/Arabidopsis). F2 individuals of the KO lines apy1-1 and apy2-1 with a kan resistance segregation ratio of 3:1 likely contained a single insertion and were screened for homozygosity by PCR for further analysis.
Table I.
Segregation of kan resistance in F2 individuals after backcrossing
| Line | F2 Individualsa
|
Total No. of F2 Individuals | Average No. of Seedlings Scored per F2 Individual | T-DNA Copy No.c | |||
|---|---|---|---|---|---|---|---|
| 3:1 | 15:1 | 63:1 | 3:1<x<15:1b | ||||
| apy1-1 | 12 | – | – | 1 | 13 | 125 | 2 |
| apy2-1 | 4 | 3 | – | – | 7 | 182 | 2 |
| apy1-2 | – | – | – | 6 | 6 | 127 | 2 |
Homozygous individuals of each mutant line (apy1-1, apy1-2, and apy2-1) were backcrossed once to wild type. The resulting F1 generation was selfed, and the F2 progeny from individual F1 plants was assayed for segregation ratios of kan resistance. Mutants segregating in a 3:1 (resistant:sensitive) ratio indicated a single insertion and were used for subsequent analyses.
No. of F2 individuals that showed the specified segregation ratios of resistant to sensitive seedlings. The chi-square goodness of fit test was used to calculate whether the population proportion agreed with the specified segregation ratio in a 95% confidence interval.
No. of F2 individuals that showed segregation ratios of resistant to sensitive seedlings between 3:1 and 15:1.
No. of T-DNA insertions in parental homozygous mutant line before backcrossing.
Phenotype of Single KO Lines Appears Identical to Wild Type
Homozygous individuals from line apy-1 and apy-2 with single insertions were evaluated for phenotypic abnormalities. apy1-1 as well as apy2-1 mutants were morphologically indistinguishable from wild-type plants. Specific tissues like roots, cotyledons, leaves, and flowers were examined, as well as the overall size and form of the plants. Earlier studies showed that the overexpression of pea apyrase (GenBank accession no. P52914) in Arabidopsis conferred enhanced growth in the presence of extracellular ATP (xATP; Thomas et al., 1999) and increased resistance to cycloheximide (Thomas et al., 2000). However KO and wild-type plants did not differ either in their ability to salvage xATP or in their resistance to cycloheximide when it was applied at concentrations from 10 to 250 ng mL−1 (data not shown).
Double KO apy1-1/apy1-1; apy2-1/apy2-1 Are Not Produced
Because the single KO lines did not display a phenotype under the conditions tested, apy1-1 and apy2-1 lines were crossed to create a double mutant line and to overcome possible functional redundancy. Homozygous apy1-1 and apy2-1 plants from the F2 generation of the kan resistance segregation analysis were crossed reciprocally. The subsequent generation was selfed, and the progeny was screened by PCR for the apy1-1/apy1-1; apy2-1/apy2-1 genotype. A total of 89 plants were analyzed, but not a single apy1-1/apy1-1; apy2-1/apy2-1 plant was among them. The probability of finding at least one apy1-1/apy1-1; apy2-1/apy2-1 in the given sample size was greater than 99.7%, provided that Mendelian laws applied. This premise should be true according to the law of independent assortment, because the two apyrase genes are located on different chromosomes.
To identify the stage of lethality of double KO apy1-1/apy1-1; apy2-1/apy2-1 plants, seed germination and embryonic development of individual self-fertilized double heterozygous plants were examined. Seeds were germinated, and siliques were inspected for the presence of aborted embryos or arrested ovules. However, neither the germination rate nor the frequency of defective seeds was reduced by the expected percentage of double KO genotypes.
The apy1-1/apy2-1 Trait Is Not Transmitted through the Male
To substantiate a defect in the apy1-1/apy2-1 gametes and to dissect the effects of mutant alleles in the male and female, the T-DNA transmission efficiencies (TEs) through each gamete were determined. The TE of a trait describes the percentage of gametes successfully transmitting the trait to the progeny relative to the number of gametes expected to carry it. In a heterozygote, the expected number of gametes harboring the trait equals the number of gametes lacking it, assuming random segregation during meiosis and the absence of post-meiotic selection (Howden et al., 1998). In this case, the T-DNA-tagged mutant alleles served as the traits of interest. The double heterozygous mutant lines were reciprocally backcrossed with wild type and the number of plants per each of the four possible genotypic outcomes was scored. If the mutant alleles were transmitted normally (TE = 100%), all four genotypes would be represented equally. When wild type was used as the pollen donor, TE of the mutant allele apy1-1 was reduced to 72.2%, whereas that of mutant allele apy2-1 was normal (Table II). This result suggested that only the loss of AtAPY1 negatively affected the female gametes. The most important observation from this cross was that the female apy1-1/apy2-1 gamete was viable, as shown by the existence of the double heterozygous progeny. The male, however, could not transmit the apy1-1/apy2-1 trait at all (Tables II and III), which was consistent with the earlier finding that no apy1-1/apy1-1; apy2-1/apy2-1 was produced by self-fertilized double heterozygous plants.
Table II.
Genetic analysis of T-DNA transmissions
| Transmission | Cross | Gamete Frequencya
|
Totalb | TEc (a) | TEc (b) | |||
|---|---|---|---|---|---|---|---|---|
| ab | AB | aB | Ab | |||||
| Female | ♀ AaBb × wt ♂ | 27 | 46 | 43 | 51 | 167 | 72.2 (<0.01*) | 87.6 (<0.25) |
| Male | ♀ wt × AaBb ♂ | 0 | 30 | 37 | 23 | 90 | 69.8 (<0.005*) | 34.3 (<0.001*) |
A, AtAPY1; a, apy1-1; B, AtAPY2, b; apy2-1. Genotypes of progeny from reciprocal crosses were determined by PCR and used to infer the genotype of the gamete contributed by the mutant parent in each cross. The no. of inferred gametes for each genotype is shown.
Total no. of genotyped progeny.
TE of the indicated T-DNA calculated as follows: TE (%) = (observed no. of mutant allele/observed number of wild-type allele) × 100. P values are given in parentheses and are indicated with an asterisk if significantly different from 100% as determined by the chi-squared test.
Table III.
Transmission of the double KO alleles
These results implied that the defect in the male transmission was severe and possibly entailed dysfunctional or nonviable male gametophytes. The TE of apy1-1 was reduced through the male to the same extent as through the female. A striking difference, however, was the strong reduction in TEmale of apy2-1. Without an intact AtAPY2 gene, only approximately one-third of the male gametes led to viable offspring. In general, the apyrases seemed to play a larger role in the male because the mutation in AtAPY1 and particularly in AtAPY2 diminished the chance of being passed on. If both functional apyrase genes were absent, transmission was completely abolished.
Apyrase Promoters Are Active in Pollen
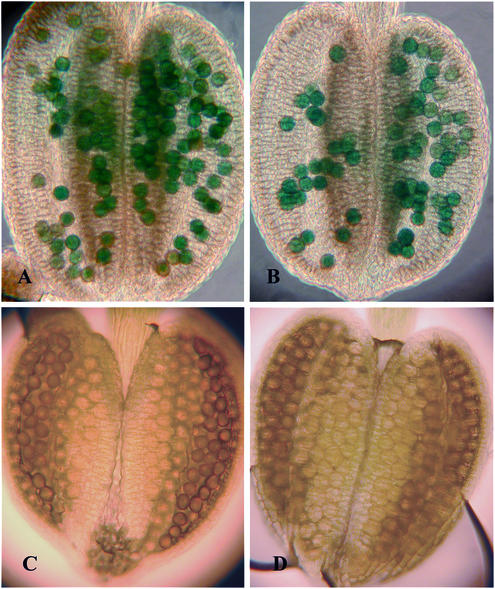
The genetic analyses (screen for apy1-1/apy1-1; apy2-1/apy2-1 and T-DNA transmissions) suggested a crucial role of apyrase in the male gametophyte. To show that the apyrase promoters are active in pollen, the expression of the two apyrases was analyzed by using promoter:β-glucuronidase (GUS) fusion transgenic lines. Anthers from mature flowers of stages 11 to 13 from four independent lines were each stained either with AtAPY1:GUS or AtAPY2:GUS construct for AtAPY1:GUS and AtAPY2:GUS expression, respectively. Expression was clearly detectable in mature pollen grains for either apyrase promoter (Fig. 3, A and B), but not in immature pollen (Fig. 3, C and D).
Figure 3.
AtAPY1:GUS and AtAPY2:GUS expression analysis in pollen. Anther from flowers at anthesis (stage 13) with mature pollen (A and B) and anther from flowers at stage 11 with immature pollen (C and D) were stained for expression of AtAPY1:GUS and AtAPY2:GUS. A and C, Anthers from plants transformed with AtAPY1:GUS; B and D, anthers from plants transformed with AtAPY2:GUS.
Vast Majority of Pollen Carrying the Mutation Display a Normal Nuclear Phenotype and Appear Viable
Pollen grains from double heterozygous plants were analyzed for morphological and nuclear aberrations as well as viability. Roughly one-fourth of this pollen was expected to carry the apy1-1/apy2-1 genotype. The mean from 10 independent flowers, 94% ± 3.1%, was identical to the normal wild-type pollen in size, shape, color, and nuclear composition (data not shown). Pollen grains that were not considered identical to wild type were smaller, were irregular in shape, and contained no nuclei. The percentage of such misformed pollen in wild type amounted to 2.4% ± 2.8% (mean from three independent flowers). The fraction of the pollen that looked normal in double heterozyous plants and wild type was viable, as assayed by the presence of active cellular esterases and intact plasma membranes (Heslop-Harrison and Heslop-Harrison, 1970).
Germination Rate Is Reduced by the Predicted Percentage of Double KO Pollen
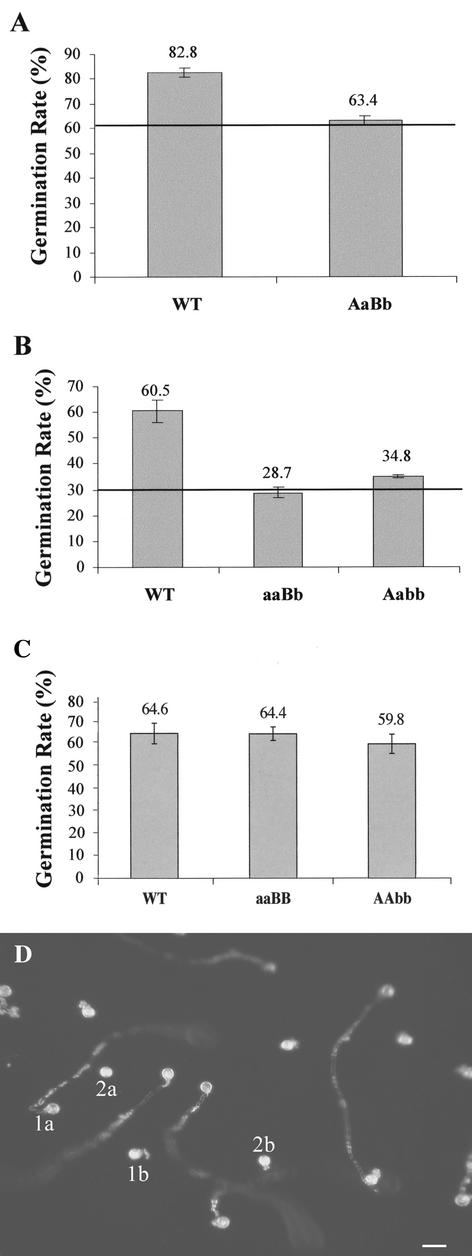
As part of an effort to identify a functional defect of apy1-1/apy2-1 pollen, the in vitro germination rates of pollen from double heterozygous plants and from mutant plants harboring only one wild-type apyrase gene were compared with that of wild type. Because the absolute germination rates varied greatly from one experiment to another (Fig. 4, A–C), data from independent experiments were not pooled. Environmental and handling conditions are difficult to reproduce exactly for each experiment, but are evidently more similar for pollen of the same experiment. Therefore, only germination rates from pollen germinated in parallel in the same 12-h time period were compared. In addition, only those experiments were considered in which the wild-type germination was at least 60% (Fig. 4D).
Figure 4.
Reduction in pollen germination rate by the percentage of expected double KO genotypes, and maintenance of wild-type germination rates in single KO pollen. The germination rates were calculated as the number of germinating pollen divided by the number of pollen sown multiplied by 100. The horizontal line marks 75% (A) and 50% (B) of the average wild-type germination rate, respectively. A and B, One representative experiment out of five each; C, one representative experiment of two done. A, AtAPY1; a, apy1-1; B, AtAPY2; and b, apy2-1. Bar graphs represent means with sds from triplicate assays (wild-type pollen) and from quadruplicate assays (pollen from double heterozygous plants; A) and from triplicate assays (B and C). For each assay the number of pollen grains counted was more than 300. D, Dark-field image of a representative section from a completed germination assay in which approximately 70% of the pollen germinated. Pollen with long tubes (e.g. 1a) and short tubes (1b) were counted as germinated. Pollen with no (2a) or barely visible and undefined (2b) protrusions was regarded ungerminated. Scale bar = 50 μm.
According to Mendelian laws, double heterozygous plants and plants with only one wild-type apyrase gene are expected to produce 25% and 50% of pollen with the apy1-1/apy2-1 genotype, respectively. Pollen germination from several wild-type and double heterozygous apy1-1/+; apy2-1/+ plants was compared. The average germination rate of pollen from double heterozygous plants was reduced to 75% of the wild-type value, namely 63.4% ± 1.7% (Fig. 4A). This 25% reduction coincided with the expected percentage of double KO pollen. In single heterozygous plants, apy1-1/apy1-1; apy2-1/+ and apy1-1/+; apy2-1/apy2-1, the germination rate was reduced to 28.7% ± 2.0% and 34.8% ± 0.5%, respectively (Fig. 4B). This represented a 50% lower germination rate compared with the wild-type value of 60.5% ± 4.5%. This reduced germination rate again matched the expected value if double KO pollen did not germinate.
As a further control, the germination rate of pollen from single KO lines was determined in comparison with pollen from wild type, and no significant difference was found (Fig. 4C). These results showed that single KO male gametes were functional, whereas apy1-1/apy2-1 ones were not as suggested by the genetic transmission data (Table II). Hydration is a prerequisite for germination and immediately precedes the latter event. The in vitro germination assays showed that pollen grains, regardless of their genotypic background, rapidly changed their shape from elliptical to almost spherical upon incubation in germination medium or when placed on wild-type stigmas (data not shown). Therefore, water uptake was not visibly affected in mutant pollen.
Wild-Type Copies of the Apyrase Genes Rescue the Pollen Phenotype
To establish unequivocally that the disruption of the apyrase genes AtAPY1 and AtAPY2 caused nonfunctional male gametes, double heterozygous plants were transformed with wild-type AtAPY1 and AtAPY2 cDNA, respectively, to rescue the phenotype. Transformants were selected on hygromycin and were grown in the presence of dexamethasone (DEX), because the introduced cDNAs were placed under a DEX-inducible promoter (Aoyama and Chua, 1997). Plants were also transformed with vector alone as a control. Hygromycin-resistant plants were screened by PCR for the double heterozygous genotype and then used as pollinators for wild-type ovules as described in the crosses for the T-DNA transmission analyses (Table II, male transmission). If either AtAPY1 or AtAPY2 transgene was able to complement the pollen phenotype, double heterozygous progeny should be recovered. The progeny from this cross was grown in the presence of kan to eliminate wild-type seedlings. The remaining plants were screened by PCR for simultaneous presence of apy1-1 and apy2-1. Several double heterozygous plants all of which contained transgenic apyrase, either AtAPY1 or AtAPY2, were identified (Table IV). Control pollen with empty vector alone did not result in any double heterozygous progeny in 20 plants analyzed (Table IV).
Table IV.
Rescue of male transmission of double KO mutant by complementation with wild-type apyrase
Wild-type stigmas were pollinated with pollen from double heterozygous plants transformed with DEX-inducible AtAPY1 and AtAPY2, respectively. The genotypes of the progeny were determined. Retrieval of genotype AaBba demonstrated that transmission of the double mutant pollen (aba) was restored (compare with Table II, male transmission).
A, AtAPY1; a, apy1-1; B, AtAPY2; b, apy2-1. No. of plants with the complemented genotype are given.
No. of genotyped plants.
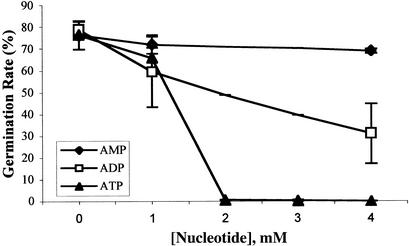
Apyrase Substrates Inhibit Wild-Type Pollen Germination
To further investigate the role of apyrase in pollen germination, the apyrase substrates ATP and ADP were added to the germination medium of wild-type pollen. ATP abolished pollen germination at concentrations of 2 mm and higher (Fig. 5). The addition of ADP also led to inhibition, but to a lesser degree. At 4 mm ADP, the germination rate was still 40% of the untreated control (Fig. 5). AMP, which cannot be cleaved by apyrase, reduced the germination rate to only 90% of untreated levels at the highest concentration of 4 mm (Fig. 5). The germination medium contains 2.7 mm CaCl2, and all three nucleotides chelate Ca2+, so at 4 mm ATP, the free Ca2+ concentration of the medium was reduced to 0.236 mm (Bers et al., 1994). To test for a possible effect on germination by this reduction, pollen was germinated in medium without adding Ca2+. The germination rate was found not to be significantly affected, demonstrating that even the background levels of Ca2+ were sufficient for germination.
Figure 5.
Effect of ATP, ADP, and AMP on wild-type pollen germination. Wild-type pollen was germinated in the presence of various concentrations of ATP, ADP, or AMP. The data points represent means with sds of three independent experiments, each performed in duplicate. An average of 354 pollen grains was counted per sample.
Overexpression of apyrase enhanced phosphate uptake in Arabidopsis (Thomas et al., 1999), so it was possible that the lack of apyrase depressed phosphate nutrition in Arabidopsis pollen. However, addition of 2 mm, 6 mm, or 10 mm phosphate to the medium did not induce germination in mutant pollen (data not shown).
To test whether apyrase activity was necessary for pollen germination, wild-type pollen was treated with NGXT1913, a compound shown to be a strong inhibitor of apyrase activity (Windsor et al., 2002). The pooled result of four separate experiments (each performed in duplicate) showed that an inhibitor concentration of 2.5 μg mL−1 completely blocked germination, 1.0 μg mL−1 significantly (P < 0.05) reduced germination to 75.3% ± 11% of control levels, and 0.5 μg mL−1 did not affect the germination rate in a statistically significant way compared with untreated pollen.
DISCUSSION
Apyrases perform a very generic enzymatic activity, yet they clearly do not qualify as housekeeping genes. Their promoters are not ubiquitously active (Y. Sun and S.J. Roux, unpublished data), and their expression is very sensitively regulated by light (Hsieh et al., 1996).
As an approach to elucidate the physiological functions of apyrases in general and to investigate functional differences or similarities between the two Arabidopsis apyrases AtAPY1 and AtAPY2, T-DNA lines were screened for disrupted gene function. The isolated AtAPY1 mutant apy1-1 as well as the AtAPY2 mutant apy2-1 displayed no discernible phenotype. This could be due to insufficiently comprehensive histochemical and physiological analyses or to overlapping gene function. Gene redundancy and possibly, compensatory increases in protein, or transcript of the undisrupted gene could also explain why the mutants apy1-1 and apy2-1 did not demonstrate higher sensitivity to cycloheximide and did not show a decrease in efficiency of xATP salvage.
During the attempt to overcome potential gene redundancy by generating double KO lines, the absence of the apy1-1/apy1-1; apy2-1/apy2-1 genotype was discovered. Transmission analyses revealed that the apy1-1/apy2-1 genotype was not passed on through the male gamete. A functionally defective gametophyte was discovered, because the germination rate of pollen from double heterozygous mutant plants and from mutant plants containing only one wild-type apyrase gene was reduced by a percentage that correlated with the fraction of pollen genetically expected to bear the double KO genotype. Introduction of a wild-type copy of either apyrase into the apy1-1/apy1-1; apy2-1/apy2-1 background rescued the apy1-1/apy2-1 pollen grains. These complementation studies unequivocally showed that the pollen phenotype was caused by lack of apyrases and not by positional effects of the T-DNA insertion or secondary mutations. In addition, the inhibition of pollen germination by inhibiting apyrase activity demonstrated that the activity of the enzyme, not just the activity of the gene, is necessary for pollen germination. Although the apyrase inhibitor used was originally characterized only in vitro (Windsor et al., 2002) and could exhibit toxic effects on a cell unrelated to apyrase, viability assays indicated the pollen remained metabolically active, although ungerminated, after it was treated with the inhibitor (data not shown).
Apyrases Not Only Important for Pollen
Transmission analyses of the gamete apy1-1/apy2-1 demonstrated a male-specific role of apyrases. Activity of AtAPY1 and AtAPY2 promoters in pollen itself supports their functional relevance in this structure and suggests a direct mode of action of apyrases in pollen as opposed to an indirect effect by activity in surrounding tissue.
Besides the importance of apyrases in pollen, the mRNA expression profile extends to many other plant organs (Steinebrunner et al., 2000). Therefore, apyrase is likely to have additional functions. It is not unique for enzymes needed for pollen germination to have important functions beyond that stage of development. Rho-related GTPases, for example, perform an essential role during pollen germination, but are also required in later stages of plant development (Li et al., 1998).
The reduced TE of apy1-1 through the female gamete proposes an involvement of AtAPY1 in the female gametophyte; however, female gametophytes and embryos stained negative for apyrase:GUS expression (Y. Sun and S.J. Roux, unpublished data). This could be explained by apyrase expression levels below the detection limit of this method. Although statistically significant, this particular TE was not pronounced and would need to be confirmed by scoring high numbers of progeny over several consecutive generations.
Other possible functions are suggested by the segregation ratios. The segregation ratios of resistant:sensitive F2 individuals should have been approximately 5:1 for the apy1-1 line and 7:1 for the apy2-1 line according to the TE obtained in Table II. This discrepancy in TEs can be explained by secondary effects influencing transmission. The backcross of double heterozygous plants with wild type represents a competitive environment for ovules and pollen, respectively, as opposed to a backcross with a single KO. Ovules without AtAPY1, for example, may be less efficient in attracting pollen tubes than ovules without AtAPY2 as suggested by the corresponding TEs in Table II. Pollen tubes without AtAPY2 may similarly be slower in reaching ovules than competing pollen tubes with a different genotype. Therefore, the difference in TE depending on the type of backcross and depending on the sex of the gamete support the notion of specific functions of each apyrase, despite the redundancy of gene function in pollen germination.
Mutations Affecting Early Stages of Pollen Germination
So-called “fully-penetrant male gametophytic mutants” have rarely been found, and this term has been generally used to refer to single mutations causing a lethal effect in the gametophyte. An example is the mutant T-DNA transmission defect (Ttd) line 38 (Procissi et al., 2001), whose mutant pollen grains seemed to contain traces of chromatin but never the three characteristic nuclei of mature wild-type pollen. Ttd38 and apy1-1/apy2-1 represent two of the few early mutations identified to date that affect stages either before or at germination.
eceriferum mutants (Hülskamp et al., 1995) are also blocked at the germination stage, at least in vivo, but germinate normally in vitro in contrast to ttd38 and apy1-1/apy2-1. To our knowledge, the only other mutation leading to a complete failure to produce a pollen tube in vitro and in vivo except for occasional swelling at the germination pore was found in petunia (Petunia hybrida) and maize (Zea mays) mutants that lacked chalcone synthase activity, necessary for flavonoid bioysynthesis (Coe et al., 1981; Taylor and Jorgensen, 1992). Germination of these flavonoid-deficient pollen could be fully restored by providing flavonoids in the form of wild-type stigmas (in vivo) or in the germination medium (in vitro; Mo et al., 1992).
Eliminating Reasons for Lack of Germination
The apyrase mutant apy1-1/apy2-1 could not be rescued by any compounds in the wild-type stigma exudate as shown in the cross of mutant pollen to female wild type (Table II). The same cross and microscopic analyses confirm functional hydration. The percentage of misformed pollen was slightly higher in mutant plants, but it did not match the expected percentage of double KO pollen. The inhibition of germination of wild-type shaped pollen could not be simply accounted for by death. The viability assay performed tested for plasma membrane integrity and metabolic activity (Heslop-Harrison and Heslop-Harrison, 1970), both evidently signs of vital cells.
The lack of germination of apyrase-deprived pollen, alternatively, could be explained by a very slow germination process. Pollen tubes of the tip1-2 mutant, for example, grow much more slowly than wild type (Ryan et al., 1998). If the pollen tubes of apy1-1/apy2-1 grains emerged at a much later time point than wild type, the pollen would appear not to have germinated. However, in wild-type pollen, germination is usually visible within 15 min of initiation (Pruitt and Hülskamp, 1994), and germination studies are generally evaluated after a 4- to 6-h time period. In this study, the window of germination was extended to a minimum of 12 h, which makes lack of time a very unlikely reason for the absence of germination.
Possible Roles of Apyrase in Pollen
Hydropathy analysis (Steinebrunner et al., 2000) predicts that the Arabidopsis apyrases AtAPY1 and AtAPY2, like most animal apyrases, are probably inserted into the plasma membrane with the catalytic site facing the exterior. This prediction has been experimentally verified for apyrase homologs in pea and D. biflorus (Thomas et al., 1999; Kalsi and Etzler, 2000). In this role as an ectoapyrase, three possible functions potentially relevant for pollen germination could be predicted based on literature reports: one as a facilitator of phosphate uptake (Thomas et al., 1999), one as a quencher of an xATP signal (Lew and Dearnaley, 2000), and one as an agent needed to maintain a steep ATP gradient across the plasma membrane and thus to facilitate a symport process critical for pollen germination (Thomas et al., 2000).
Phosphate ions are not a needed component of pollen germination medium, and addition of phosphate to the medium does not rescue mutant pollen, so it would seem unlikely that the ability of apyrase to facilitate phosphate uptake would be relevant to its role in pollen germination. Could xATP and xADP, both of which can depolarize the membrane potential of Arabidopsis root hairs (Lew and Dearnaley, 2000), represent an inhibitory signal to pollen germination, thus creating a need for ectoapyrase activity to overcome the inhibition? Consistent with this idea, the addition of ATP and ADP inhibited the germination of wild-type pollen severely (Fig. 5). However, the millimolar concentrations of these compounds needed for inhibition have uncertain relevance to levels that might ever be found in the coat of mature pollen, so additional studies will be needed to ascertain whether a signaling role for xATP in pollen germination, and a role for apyrase in quenching this signal, is likely.
Regarding a role for ectoapyrase activity in the export of inhibitors from pollen, Thomas et al. (2000) reported that the steepness of the ATP gradient across the plasma membrane, which is increased by ectoapyrase activity, can influence the transport of toxins out of cells across an MDR-like transporter. On the basis of this model, one could propose that a naturally occurring inhibitor of pollen germination must be exported from pollen grains to release them from dormancy and that ectoapyrase activity promotes this export. To our knowledge, there are no reports of any transport out of pollen being essential for pollen germination, although auxin levels in pollen drop after germination (Liu and Lee, 1995). Evidence for the necessity of some kind of export event would be required to make plausible this explanation of why apyrase activity is critical for pollen germination.
The addition of potato (Solanum tuberosum) apyrase to mutant pollen did not restore pollen germination (data not shown), a result that could be interpreted to contradict an extracellular role of apyrase. However, the potato apyrase used was soluble (Kettlun et al., 1992; Handa and Guidotti, 1996), and membrane proximity, which an externally added apyrase might not be able to achieve, may be important for the function of Arabidopsis apyrase in pollen. In addition, the required roles of AtAPY1 and AtAPY2 may involve activities, such as protein-protein interactions, beyond just enzymatic.
An extensive effort has been undertaken to identify key players in pollination (for reviews, see Franklin-Tong, 2002; Johnson and Preuss, 2002). Here, a crucial role for apyrases in pollen germination has been identified, and potential mechanisms to explain this role have been discussed. The importance of apyrases has most likely escaped forward genetics approaches so far, because both apyrase genes need to be mutated to unveil the phenotype. Learning the pollen-specific function of apyrases will contribute to a better understanding of plant apyrases in general and to plant reproduction specifically.
MATERIALS AND METHODS
Growth of Plants
Arabidopsis of ecotype Wassilewskija was either grown under continuous light directly on soil or on 0.8% (w/v) agar plates with Murashige and Skoog basal salt mixture for 7 d before being transferred to soil.
Screen for T-DNA Insertion Lines
A collection of T-DNA insertion mutants, ecotype Wassilewskija, generated by the Wisconsin Arabidopsis KO facility was screened simultaneously for insertions in the genes AtAPY1 and AtAPY2 according to the facility's guidelines (http://www.biotech.wisc.edu/NewServicesAndResearch/Arabidopsis; Sussman et al., 2000). The gene-specific primers ApyF (5′-GGACGGCACGAATCCCTTGCTGACAAG-3′) and ApyR (5′-TCTCCGTATTTCACCTTCTTCACTAACGT-3′) near the 5′ and 3′ end, respectively, of the highly conserved open reading frame of both genes (Steinebrunner et al., 2000) and the facility's recommended left border primer JL-202 were used. Positive PCR products were identified by Southern-blot analysis using the AtAPY1 cDNA as a probe and confirmed by sequencing. Homo- and heterozygosity of plants were determined by PCR. The number of T-DNA loci per line was assayed by analyzing the segregation pattern of kan resistance in the progeny of self-pollinated heterozygous lines.
RT-PCR
Total RNA (2 μg) from whole plants was reverse transcribed using oligo(dT) primers and Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA). One-tenth of the reaction volume was used as template in the subsequent PCR reactions with the following cycling conditions: 96°C for 15 s, 36 cycles of 94°C for 15 s, annealing temperature as specified for 30 s, 72°C for 90 s concluded by 72°C for 10 min. The primers AraF172 (5′-GCAGCCGTAACTTGCAATC-3′) and AraR2024 (5′-TGCGGTAAGCAGTTTAGATTAAC-3′) amplified the AtAPY1-specific product. The primers Arapy2F (5′-GCTTTCCCAAATTCACCGT-3′) and ApyR (see above) produced the AtAPY2-specific band. Both primer combinations were used at an annealing temperature of 56°C. AraF172, AraR2024, and Arapy2F are located in the untranslated regions of the respective genes. Taq PCR mastermix (Qiagen USA, Valencia, CA) was used at a final MgCl2 concentration of 2.5 mm.
Crosses
The plants used as females were emasculated by hand and pollinated by touching the anthers from donor plants onto stigmas. The pollinated flowers were labeled and all remaining flowers, opened or unopened, from the same plant were removed to avoid any confusion at harvest.
Phenotype Analysis
Growth assays involving cycloheximide and ATP were performed as described elsewhere (Thomas et al., 1999, 2000).
Screen for Double KOs
The genotypes were determined by PCR (for conditions see under RT-PCR). A leaf was cut from each plant and ground with a disposable pellet pestle (Kontes Glass, Vineland, NJ) in a 1.5-mL microcentrifuge tube in the presence of 150 μL of DNAzol (Invitrogen). After vortexing, the debris was pelleted, and the DNA in the supernatant was precipitated by adding 0.75 volume of 100% (v/v) ethanol. The DNA pellet was washed and resuspended in 30 μL of water. The DNA preparations were screened for intact copies of AtAPY1 and AtAPY2 first using the primer combinations AraF172/AraR693 (5′-GATATGCACTTAAACCCGG-3′) and then Apy2I6F (5′-GCCATTATCTTCGCCATCCTTTTC-3′)/ApyR, respectively. The former primer combination spanned the first intron and was used at an annealing temperature of 54°C. The annealing temperature for the latter primer pair was 65°C, and its forward primer was located in intron 6. If the intact versions of both apyrase genes could not be amplified, the presence of mutant copies was determined by using the primers ApyF/JL-202 for apy1-1 and ApyR/JL-202 for apy2-1 (see Fig. 1).
T-DNA Transmission Analysis
Plants were initially grown on kan plates (30 μg mL−1) to score for wild-type genotype. Resistant seedlings were transferred to soil and planted in a grid-like three by three arrangement per pot. The presence of the mutant alleles apy1-1 and apy2-1 was ascertained as described under screen for double KOs.
Complementation of the Apyrase Mutant
The almost full-length cDNAs of AtAPY1 and AtAPY2 were amplified using the primer pair AraF172/AraR2024 and Arapy2F/dT(19), respectively. The PCR products were cloned into pCR2.1 (Invitrogen) and sequenced. Constructs with correct orientation were cut with XhoI and SpeI located in the vector's multiple cloning site and inserted into the XhoI and SpeI site of the pTA7002 vector (Aoyama and Chua, 1997). Plants of the double heterozygous genotype were transformed, and progeny was selected on 50 μg mL−1 hygromycin. In addition, hygromycin resistant plants were screened for the presence of the DEX-construct by PCR using the primers GAL4–5′ and GAL4–3′ (Kang et al., 1999) at a MgCl2 concentration of 3 mm. For induction of the transgene, plants were watered with 30 nm DEX. Complemented genotypes were confirmed by two independent DNA isolations and PCRs.
Construction of Promoter-GUS Fusion and GUS Assay
The genomic sequences upstream of the AtAPY1 and AtAPY2 coding region were subcloned as a 3-kb HindIII/SalI fragment and a 2.8-kb SalI/XbaI fragment, respectively, into the binary vector pBI101 (BD Biosciences Clontech, Palo Alto, CA) which contained a promoterless GUS gene. Stable transformants were obtained by following the Green Lab protocol for vacuum infiltration. GUS staining was performed according to Lehman et al. (1996). Flower stages were adapted from Smyth et al. (1990).
Pollen Studies
For germination assays, pollen was used from flowers at stages 13 to 14 (Smyth et al., 1990), which were picked from plants no later than 1 week after bolting. Fifty to 100 μL of liquid germination medium (Torres et al., 1995) were used per convection of a hanging drop slide representing one experiment sample. Potato (Solanum tuberosum) apyrase (grade VI) was purchased from Sigma-Aldrich (St. Louis) and added at a concentration of 0.04 unit μL−1 to the germination medium. Apyrase inhibitor NGXT1913 (Windsor et al., 2002) was kindly donated by Texagen Inc. (Austin, TX). The pH of the germination medium was adjusted to 7.0 after the addition of ATP, ADP, AMP (Sigma-Aldrich), or Na2HPO4. Na2HPO4 was used as a source of phosphate ions. Free calcium was calculated according to http://www.stanford.edu/∼cpatton/maxc.html. Viability was assayed using fluorescein diacetate according to the protocol of Heslop-Harrison and Heslop-Harrison (1970). For DAPI staining, the procedure described by Howden et al. (1998) was followed.
In vivo hydration of pollen grains was performed as described by Hülskamp et al. (1995). Pollen grains in direct contact with papillar cells were counted under the microscope, and spherical grains were scored as hydrated.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or part of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Travis Graber, Collene Jeter, Alan Lloyd, Charlotte Song, Eldon Sutton, and Wenquiang Tang for critical reading of the manuscript.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN–0080363) and by The Texas Higher Education Coordinating Board.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.014308.
LITERATURE CITED
- Aoyama T, Chua N-H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- Bers D, Patton C, Nuccitelli R. A practical guide to the preparation of Ca buffers. In: Nuccitelli R, editor. A Practical Guide to the Study of Ca2+ in Living Cells. Vol. 40. San Diego: Academic Press; 1994. pp. 3–29. [DOI] [PubMed] [Google Scholar]
- Coe EH, McCormick SM, Modena SA. White pollen in maize. J Hered. 1981;72:318–320. [Google Scholar]
- Cohn J, Uhm T, Ramu S, Nam Y-W, Kim D-J, Penmetsa R, Wood T, Denny R, Young N, Cook D et al. Differential regulation of a family of apyrase genes from Medicago truncatula. Plant Physiol. 2001;125:2104–2119. doi: 10.1104/pp.125.4.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R, McAlvin C, Loh J, Denny R, Wood T, Young N, Stacey G. Differential expression of two soybean apyrases, one of which is an early nodulin. Mol Plant-Microbe Interact. 2000;13:1053–1070. doi: 10.1094/MPMI.2000.13.10.1053. [DOI] [PubMed] [Google Scholar]
- Franklin-Tong V. The difficult question of sex: the mating game. Curr Opin Plant Biol. 2002;5:14–18. doi: 10.1016/s1369-5266(01)00217-5. [DOI] [PubMed] [Google Scholar]
- Handa M, Guidotti G. Purification and cloning of a soluble ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum) Biochem Biophys Res Commun. 1996;218:916–923. doi: 10.1006/bbrc.1996.0162. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J, Heslop-Harrison Y. Evaluation of pollen viability by enzymatically induced fluorescence: intracellular hydrolysis of fluorescein diacetate. Stain Technol. 1970;45:115–120. doi: 10.3109/10520297009085351. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison Y, Heslop-Harrison J. Germination of monocolpate angiosperm pollen: evolution of the actin cytoskeleton and wall during hydration, activation and tube emergence. Ann Bot. 1992a;69:385–394. [Google Scholar]
- Heslop-Harrison J, Heslop-Harrison Y. Germination of monocolpate angiosperm pollen: effects of inhibitory factors and the Ca2+-channel blocker nifedipine. Ann Bot. 1992b;69:395–403. [Google Scholar]
- Howden R, Park S, Moore J, Orme J, Grossniklaus U, Twell D. Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics. 1998;149:621–631. doi: 10.1093/genetics/149.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh H-L, Tong C-G, Thomas C, Roux SJ. Light-modulated abundance of an mRNA encoding a calmodulin-regulated, chromatin-associated NTPase in pea. Plant Mol Biol. 1996;30:135–147. doi: 10.1007/BF00017808. [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Kopczak S, Horejsi T, Kihl B, Pruitt R. Identification of genes required for pollen-stigma recognition in Arabidopsis thaliana. Plant J. 1995;8:703–714. doi: 10.1046/j.1365-313x.1995.08050703.x. [DOI] [PubMed] [Google Scholar]
- Johnson M, Preuss D. Plotting a course: multiple signals guide pollen tubes to their targets. Dev Cell. 2002;2:273–281. doi: 10.1016/s1534-5807(02)00130-2. [DOI] [PubMed] [Google Scholar]
- Johnson SA, McCormick S. Pollen germinates precociously in the anthers of raring-to-go, an Arabidopsis gametophytic mutant. Plant Physiol. 2001;126:685–695. doi: 10.1104/pp.126.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsi G, Etzler M. Localization of a nod factor-binding protein in legume roots and factors influencing its distribution and expression. Plant Physiol. 2000;124:1039–1048. doi: 10.1104/pp.124.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H-G, Fang Y, Singh KB. A glucocorticoid-inducible transcription system causes severe growth defects in Arabidopsis and induces defense-related genes. Plant J. 1999;20:127–133. doi: 10.1046/j.1365-313x.1999.00575.x. [DOI] [PubMed] [Google Scholar]
- Kettlun AM, Leyton M, Valenzuela MA, Mancilla M, Traverso-Co A. Identification and subcellular localization of two isoenzymes of apyrase from Solanum tuberosum. Phytochemistry. 1992;31:1889–1894. [Google Scholar]
- Komoszynski M, Wojtczak A. Apyrases (ATP diphosphohydrolases, EC 3.6.1.5): function and relationship to ATPases. Biochim Biophys Acta. 1996;1310:233–241. doi: 10.1016/0167-4889(95)00135-2. [DOI] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyls. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- Lew R, Dearnaley J. Extracellular nucleotide effects on the electrical properties of growing Arabidopsis thaliana root hairs. Plant Sci. 2000;153:1–6. [Google Scholar]
- Li H, Wu G, Ware D, Davis K, Yang Z. Arabidopsis Rho-related GTPases: differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 1998;118:407–417. doi: 10.1104/pp.118.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z-H, Lee B-H. Changes of endogenous indole-3-acetic acid, peroxidases, and auxin oxidases during pollen germination in maize. Bot Bull Acad Sin. 1995;36:53–58. [Google Scholar]
- Mo Y, Nagel C, Taylor L. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci USA. 1992;89:7213–7217. doi: 10.1073/pnas.89.15.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesner L. Ecto-ATPases: identities and functions. Int Rev Cytol. 1995;158:141–214. doi: 10.1016/s0074-7696(08)62487-0. [DOI] [PubMed] [Google Scholar]
- Procissi A, Laissardière de S, Férault M, Vezon D, Pelletier G, Bonhomme S. Five gametophytic mutations affecting pollen development and pollen tube growth in Arabidopsis thaliana. Genetics. 2001;158:1773–1783. doi: 10.1093/genetics/158.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt R, Hülskamp M. From pollination to fertilization. In: Meyerowitz E, Somerville C, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 467–483. [Google Scholar]
- Roberts NJ, Brigham J, Wu B, Murphy JB, Volpin H, Phillips DA, Etzler ME. A Nod factor-binding lectin is a member of a distinct class of apyrases that may be unique to the legumes. Mol Gen Genet. 1999;262:261–267. doi: 10.1007/s004380051082. [DOI] [PubMed] [Google Scholar]
- Ryan E, Grierson CS, Cavell A, Steer M, Dolan L. TIP1 is required for both tip growth and non-tip growth in Arabidopsis. New Phytol. 1998;138:49–58. [Google Scholar]
- Shibata K, Morita Y, Abe S, Stankovic B, Davies E. Apyrase from pea stems: isolation, purification, characterization and identification of a NTPase from the cytoskeleton fraction of pea stem tissue. Plant Physiol Biochem. 1999;37:881–888. [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinebrunner I, Jeter C, Song C, Roux S. Molecular and biochemical comparison of two different apyrases from Arabidopsis thaliana. Plant Physiol Biochem. 2000;38:913–922. [Google Scholar]
- Sussman M, Amasino R, Young J, Krysan P, Austin-Phillips S. The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 2000;124:1465–1467. doi: 10.1104/pp.124.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L, Hepler P. Pollen germination and tube growth. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:461–491. doi: 10.1146/annurev.arplant.48.1.461. [DOI] [PubMed] [Google Scholar]
- Taylor LP, Jorgensen R. Conditional male fertility in chalcone synthase-deficient petunia. J Hered. 1992;83:11–17. [Google Scholar]
- Thomas C, Rajagopal A, Windsor B, Dudler R, Lloyd A, Roux S. A role for ecto-phosphatase in xenobiotic resistance. Plant Cell. 2000;12:519–533. doi: 10.1105/tpc.12.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Sun Y, Naus K, Lloyd A, Roux S. Apyrase functions in plant phosphate nutrition and mobilizes phosphate from extracellular ATP. Plant Physiol. 1999;119:543–551. doi: 10.1104/pp.119.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M-A, Rigau J, Puigdomènech P, Stiefel V. Specific distribution of mRNAs in maize: growing pollen tubes observed by whole-mount in situ hybridization with non-radioactive probes. Plant J. 1995;8:317–321. [Google Scholar]
- Windsor JB, Thomas C, Hurley L, Roux SJ, Lloyd AM. An automated colorimetric screen for apyrase inhibitors. Biotechniques. 2002;33:1024–1030. doi: 10.2144/02335st02. [DOI] [PubMed] [Google Scholar]