Abstract
In contrast to 16:3 plants like rapeseed (Brassica napus), which contain α-linolenic acid (18:3Δ9,12,15) and hexadecatrienoic acid (16:3Δ7,10,13) as major polyunsaturated fatty acids in leaves, the silica-less diatom Phaeodactylum tricornutum contains eicosapentaenoic acid (EPA; 20:5Δ5,8,11,14,17) and a different isomer of hexadecatrienoic acid (16:3Δ6,9,12). In this report, we describe the characterization of two cDNAs having sequence homology to Δ12-fatty acid desaturases from higher plants. These cDNAs were shown to code for a microsomal and a plastidial Δ12-desaturase (PtFAD2 and PtFAD6, respectively) by heterologous expression in yeast (Saccharomyces cerevisiae) and Synechococcus, respectively. Using these systems in the presence of exogenously supplied fatty acids, the substrate specificities of the two desaturases were determined and compared with those of the corresponding rapeseed enzymes (BnFAD2 and BnFAD6). The microsomal desaturases were similarly specific for oleic acid (18:1Δ9), suggesting that PtFAD2 is involved in the biosynthesis of EPA. In contrast, the plastidial desaturase from the higher plant and the diatom clearly differed. Although the rapeseed plastidial desaturase showed high activity toward the ω9-fatty acids 18:1Δ9 and 16:1Δ7, in line with the fatty acid composition of rapeseed leaves, the enzyme of P. tricornutum was highly specific for 16:1Δ9. Our results indicate that in contrast to EPA, which is synthesized in the microsomes, the hexadecatrienoic acid isomer found in P. tricornutum (16:3Δ6,9,12) is of plastidial origin.
Diatoms (Bacillariophyceae) represent a significant group of eukaryotic microalgae found in marine and freshwater habitats and in terrestrial environments. In ocean ecosystems, they are thought to be responsible for as much as 25% of the global primary productivity (Scala and Bowler, 2001). Furthermore, they play a key role in the biogeochemical cycling of silica because most of them are surrounded by a highly structured silica cell wall (Tréguer et al., 1995). The plastids of diatoms contain xanthophylls like fucoxanthin as the major accessory pigments for photosynthesis, which give these organisms their brownish color and their denomination as chromophytes (Bhaya and Grossman, 1991). Phylogenetically, diatoms are thought to have originated from the engulfment of a photoautotrophic eukaryotic cell, most probably an ancestor of the modern red algae, by a heterotrophic heterokont flagellate (McFadden, 2001). Because of this secondary endocytobiosis, the chromophytic plastids of diatoms are surrounded by four membranes and referred to as “complex plastids.” The two inner membranes of the plastid are thought to represent the original envelope of the plastid, whereas the two outer ones most probably evolved from the plasma membrane of the first endosymbiont and the phagotrophic membrane of the host cell (Kroth and Strotmann, 1999). The transport of nuclear-encoded proteins into such plastids was shown to depend on multisignal presequences and to be a two-step process (Lang et al., 1998).
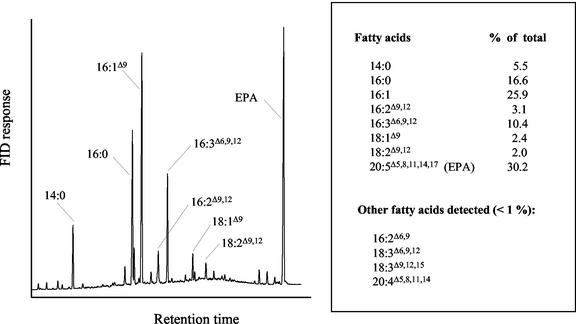
The object of this study, Phaeodactylum tricornutum, is a silica-less diatom mainly known as a potential source for the industrial production of eicosapentaenoic acid (EPA; 20:5Δ5,8,11,14,17; Molina Grima et al., 1996). In the fatty acid profile of P. tricornutum shown in Figure 1, EPA (30%), palmitoleic acid (16:1Δ9; 26%), palmitic acid (16:0; 17%), hexadecatrienoic acid (16:3Δ6,9,12; 10%), and myristic acid (14:0; 5%) are the major fatty acids. The first data concerning the fatty acid metabolism of P. tricornutum were obtained from labeling experiments. Incubation of the diatom with [14C]acetate suggested that palmitic and oleic acid are the main products of the de novo synthesis (Moreno et al., 1979). Incubation with C18 or C20-[14C]fatty acids showed that EPA was synthesized by desaturation and elongation of oleic acid (Arao and Yamada, 1994). In Figure 1, oleic acid and all the intermediates of the EPA biosynthetic pathway (18:2Δ9,12, 18:3Δ6,9,12, 20:3Δ8,11,14, and 20:4Δ8,11,14,17) are only present in trace amounts. This may indicate that this organism has developed highly efficient mechanisms to accumulate specifically EPA. Recently, we reported the cloning and characterization of the Δ5- and the Δ6-desaturases involved in EPA biosynthesis in P. tricornutum (Domergue et al., 2002). Both desaturases are microsomal enzymes, which indicates that several steps involved in EPA biosynthesis are taking place in the endoplasmic reticulum (ER). On the other hand, most of the EPA found in P. tricornutum is present in the plastidial glycolipids (Arao et al., 1987; Yongmanitchai and Ward, 1993), suggesting an import of EPA into the plastid after its synthesis in the ER. Although the subcellular origin of EPA appears to be microsomal, that of the hexadecatrienoic acid isomer found in P. tricornutum, 16:3Δ6,9,12, remains unclear.
Figure 1.
Fatty acid composition of P. tricornutum. P. tricornutum UTEX 646 was grown in brackish water medium with moderate shaking (80 rpm) under long-day light conditions (15 h at 30 μE m−2 s−1) at 23°C. Cells were harvested in the stationary phase and fatty acid methyl esters (FAMEs) from whole cells were prepared as indicated in “Materials and Methods.”
In contrast to the fatty acid metabolism of diatoms, that of higher plants is well documented. In higher plants, the de novo fatty acid synthesis is catalyzed in the plastid by a type II (dissociable) fatty acid synthase, leading primarily to the synthesis of 16:0-ACP. Most of this 16:0-ACP is then elongated to 18:0-ACP and desaturated to 18:1Δ9-ACP by a soluble Δ9-acyl-ACP desaturase. Genetic and biochemical analyses of mutants of the model plant Arabidopsis have shown that two interconnected pathways are then responsible for the synthesis of 16:3Δ7,10,13 and 18:3Δ9,12,15, the two major polyunsaturated fatty acids (PUFAs) found in the leaves of 16:3 plants like Arabidopsis and rapeseed (Brassica napus; Browse and Somerville, 1991; Wallis and Browse, 2002). The so-called “eukaryotic” pathway is located in the ER and involved in the synthesis of 18:2Δ9,12 and α-linolenic acid, 18:3Δ9,12,15. In the chloroplast, the “prokaryotic” pathway catalyzes similar reactions, but is also responsible for the entire synthesis of the hexadecatrienoic acid isomer, 16:3Δ7,10,13. Each pathway possesses its own set of Δ12- and Δ15-fatty acid desaturases (FADs), but they differ in both lipid substrates and electron donors. Although the plastidial desaturases of the prokaryotic pathway use primarily glycolipids as acyl-carriers and ferredoxin/ferredoxin oxidoreductase as electron donors, the microsomal desaturases involved in the eukaryotic pathway use phospholipids and cytochrome b5/cytochrome b5 oxidoreductase (Los and Murata, 1998). Such parallel sets of FADs may also exist in the plastidial and ER compartments of diatoms, and the cloning and functional characterization of these activities should help in understanding the origin of the predominating fatty acids found in P. tricornutum.
In the present paper, we report the cloning of two Δ12-FADs from P. tricornutum and their functional characterization as microsomal and plastidial desaturase by expression in yeast (Saccharomyces cerevisiae) and a cyanobacterium, respectively. The substrate specificity of each desaturase was determined in these heterologous expression systems and compared with those of the corresponding homologs from rapeseed. The microsomal desaturase of P. tricornutum was shown to be most active with oleic acid, whereas the plastidial desaturase was highly specific for 16:1Δ9. It could be concluded from these experiments that the microsomal desaturase is most probably involved in the biosynthesis of EPA, whereas the plastidial enzyme contributes to the synthesis of the hexadecatrienoic acid isomer characteristic to P. tricornutum, 16:3Δ6,9,12, which in contrast to EPA, is most probably of prokaryotic origin.
RESULTS
Isolation of Two Δ12-Desaturase cDNA Clones
Two full-length clones coding for putative Δ12-desaturases were isolated from a P. tricornutum cDNA library by mass sequencing. The first clone was 1,651 bp long and contained an open reading frame (ORF) of 1,488 bp, which encoded a polypeptide of 495 amino acids. The second clone was 1,526 bp long, with an ORF of 1,311 bp coding for a polypeptide of 436 amino acids. The two proteins encoded by these ORFs were 28% identical (42% similar), and both showed high sequence similarities to Δ12-desaturases from various organisms, including the rapeseed ω6-desaturase (BnFAD2; GenBank accession no. AF243045). This latter was 25% identical (38% similar) to the first P. tricornutum clone and 35% identical (49% similar) to the second one. When the N-terminal amino acid sequences were analyzed for the presence of targeting signals (Emanuelsson et al., 2000), the protein encoded by the first clone was predicted to be of plastidial origin. With respect to the nomenclature developed for such enzymes in Arabidopsis (Falcone et al., 1994; Okuley et al., 1994), the first and second ORFs identified in P. tricornutum were annotated as PtFAD6 and PtFAD2, respectively.
Figure 2 shows the amino acid sequences of the proteins encoded by PtFAD2 and PtFAD6 together with the plastidial and microsomal homologs from rapeseed (BnFAD6 and BnFAD2, respectively). All these proteins contain the three conserved His clusters most likely involved in the coordination of the diiron center of the active site (Shanklin and Cahoon, 1998) and the four potential transmembrane helices of the topological model developed for membrane-bound desaturases (Shanklin et al., 1994). Both FAD6 proteins contain an N-terminal extension, the one of PtFAD6 being longer, as expected for the bipartite structure of the presequence encountered in diatoms (Lang et al., 1998).
Figure 2.
Amino acid sequences of PtFAD6 and PtFAD2 in comparison with the plastidial and microsomal homologs from rapeseed. For alignment, the ClustalX program (Thompson et al., 1997) was used (gap opening 12, gap extension 0.05). The conserved amino acids are on gray background. The three His boxes are indicated by subscripts, and the potential transmembrane domains are framed. The broken line, dots, and the arrow at the N terminus of PtFAD6 indicate an hydrophobic domain, hydroxylated fatty acids (Thr and Ser), and the N-terminal sequence of PtFAD6 fused to EGPF, respectively. The sequence data from P. tricornutum have been submitted to the DNA Data Bank of Japan/EMBL/GenBank sequence data bank (accession nos. AY165023 and AY165024 for PtFAD2 and PtFAD6, respectively).
Presequence Analysis and Expression of Enhanced Green Florescent Protein (EGFP) Fusion Proteins
The presequence of PtFAD6 contains the two domains that are typical for the import of nuclear-encoded proteins into the complex plastids of diatoms. The N-terminal domain of PtFAD6 is basic with an Arg in the third position and contains a hydrophobic portion (Fig. 2, broken line) similar to a classical ER-targeting signal. This sequence is followed by a domain rich in hydroxylated amino acids (Ser and Thr; Fig. 2, dots), which is characteristic for transit peptides involved in the transport into the plastid.
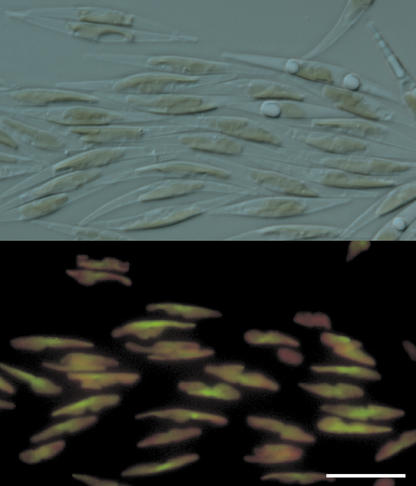
To confirm the cellular localization of PtFAD6, the DNA sequence encoding the first 113 amino acids (Fig. 2, arrow) was fused with the EGFP gene and cloned in pPha-T1. The resulting vector was used to transform P. tricornutum (see “Materials and Methods”). As shown in Figure 3, the EGFP fluorescence was colocalized with the red fluorescence of chlorophyll in the plastid. Although the EGFP fluorescence was not evenly distributed within the plastid, but most probably accumulated within the pyrenoid, these results strongly support that PtFAD6 is a plastidial desaturase. In contrast, when the first 55 amino acids of PtFAD2 were fused to the N terminus of EGFP, the fluorescence appeared to be cytoplasmic (data not shown).
Figure 3.
Expression of the PtFAD6-EGFP fusion protein in P. tricornutum. The DNA sequence encoding the first 113 amino acids of PtFAD6 was fused to the 5′ end of the EGFP gene and cloned in pPha-T1. The resulting vector was used to transform P. tricornutum UTEX 646 as indicated in “Materials and Methods.” Top, Light microscopical image of P. tricornutum with the plastids visible in brown. Bottom, Red (chlorophyll) and green (EGFP) fluorescence. Scale bar = 10 μm.
Functional Expression of the Desaturases in Yeast
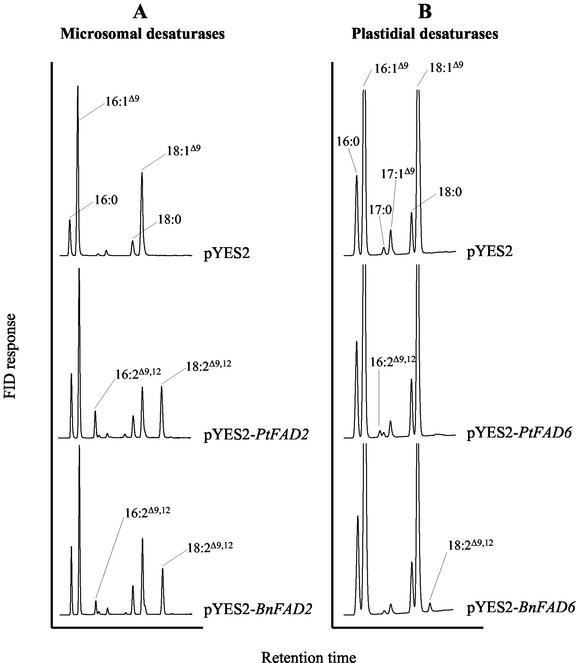
The ORF of the putative microsomal desaturase PtFAD2 was cloned in the yeast expression vector pYES2 (Invitrogen, Leek, Netherlands) and expressed in yeast to confirm its enzymatic activity. Using the empty vector pYES2 as control and pYES2-BnFAD2 for comparison, the different constructs were transformed into the yeast strain C13ABYS86 (Bröker et al., 1991) and expressed for 48 h at 20°C. The yeast cells transformed with the empty vector pYES2 showed the typical yeast fatty acids (16:0, 16:1Δ9, 18:0, and 18:1Δ9) and traces of 17:0 and 17:1Δ9 (Fig. 4A, top). The expression of PtFAD2 and BnFAD2 resulted in two additional peaks corresponding to 16:2Δ9,12 and 18:2Δ9,12 (Fig. 4A, middle and bottom, respectively). The high proportions of 18:2Δ9,12 were correlated with a significant decrease of 18:1Δ9 as expected for an educt-product relationship. PtFAD2 desaturated as much as 51% of oleic acid and about 14% of palmitoleic acid, whereas BnFAD2 was slightly less active, converting 40% and 8% of oleic and palmitoleic acid, respectively.
Figure 4.
Fatty acid profiles of transgenic yeast expressing microsomal (A) and plastidial (B) desaturases. The C13ABYS86 yeast strain was transformed with the indicated constructs. The transformants were grown for 48 h at 20°C, and FAMEs from whole cells were prepared and analyzed by gas-liquid chromatography (GLC) as indicated in “Materials and Methods.” The traces in B were magnified to show the small peaks representing the products of the plastidial desaturases, with the consequence that the peaks corresponding to 16:1Δ9 and 18:1Δ9 go off scale.
The yeast expression system was then used to determine the substrate specificity of PtFAD2 in more detail. The most efficiently desaturated substrate was oleic acid (50% conversion), but 16:1Δ9 and 17:1Δ9 were also accepted as substrates (15% and 22% conversion to 16:2Δ9,12 and 17:2Δ9,12, respectively; Table I). No activity was measured with 18:1Δ11 or 22:1Δ13, but about 4% of 20:1Δ11 was converted to 20:2Δ11,14. Similar results were obtained with BnFAD2 (data not shown), indicating that PtFAD2 has the substrate specificity typical for FAD2 enzymes. Although such enzymes can be denominated as ω6- or Δ12-desaturases, it should be noted that they convert 16:1Δ9 to 16:2Δ9,12, 17:1Δ9 to 17:2Δ9,12, and 20:1Δ11 to 20:2Δ11,14 (Table I), inserting the new double bond in the ω4-, ω5-, and ω6/Δ14-position, respectively. Therefore, a correct assignment of the regioselectivity for FAD2 enzymes would be μ + 3 because the position of the incipient double bond is determined relative to a preexisting double bond by placing it three carbons closer to the methyl end (Meesapyodsuk et al., 2000).
Table I.
Substrate specificity of PtFAD2 expressed in Saccharomyces cerevisiae
| Substrate | Total Fatty Acids (Educt) | Product | Total Fatty Acids | Desaturation |
|---|---|---|---|---|
| % | % | % | ||
| a16:1Δ9 | 38.4 ± 1.6 | 16:2Δ9,12 | 6.6 ± 0.5 | 14.7 |
| b17:1Δ9 | 2.9 ± 0.2 | 17:2Δ9,12 | 0.8 ± 0.1 | 22.3 |
| a18:1Δ9 | 15.9 ± 1.1 | 18:2Δ9,12 | 16.1 ± 0.9 | 50.3 |
| 18:1Δ11 | 36.6 ± 2.2 | 18:2Δ11,14 | 0 | 0 |
| c20:1Δ11 | 9.5 ± 0.9 | 20:2Δ11,14 | 0.4 ± 0.1 | 4.1 |
| c22:1Δ13 | 2.0 ± 0.3 | 22:2Δ13,16 | 0 | 0 |
Yeast strain C13ABYS86 transformed with pYES2-PtFAD2 was grown for 48 h at 20°C in the presence or absence of different fatty acid substrates (500 μm or as indicated) in separate experiments. FAMEs from the whole cells were prepared and analyzed by GLC as indicated in “Materials and Methods.” Each value represents the percent of total fatty acids and is the mean ± sd from three independent experiments. Desaturation (percentage) was calculated as (product × 100)/(educt + product).
In the absence of exogenously fed fatty acid.
In the presence of 2 mm exogenously fed 17:0.
In the presence of 2 mm exogenously fed fatty acid.
Interestingly, when the FAD6 desaturases from P. tricornutum and rapeseed were expressed in yeast, small but significant proportions of 16:2Δ9,12 and 18:2Δ9,12 were detected, respectively (Fig. 4B). Repeatedly, 16:2Δ9,12 accounted for 0.5% to 1.5% of the total fatty acids when PtFAD6 was expressed, whereas 18:2Δ9,12 represented up to 2% of the total fatty acids in yeast expressing BnFAD6. Yeast is known to be the model of choice for the functional characterization of microsomal FADs because it contains the short electron transport system required by such desaturases (i.e. cytochrome b5 and NADH-cytochrome b5 reductase). Nevertheless, the low desaturation levels evident from Figure 4B suggest that desaturases of plastidial origin, which usually require ferredoxin and NADPH-ferredoxin reductase, are supplied to some extent with reducing equivalents in yeast cells.
Functional Expression of the Desaturases in Synechococcus PCC 7942
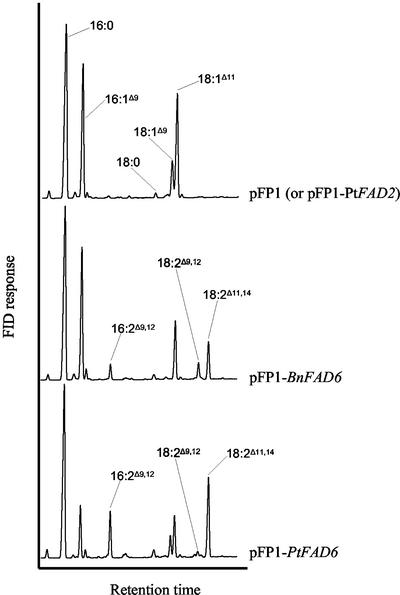
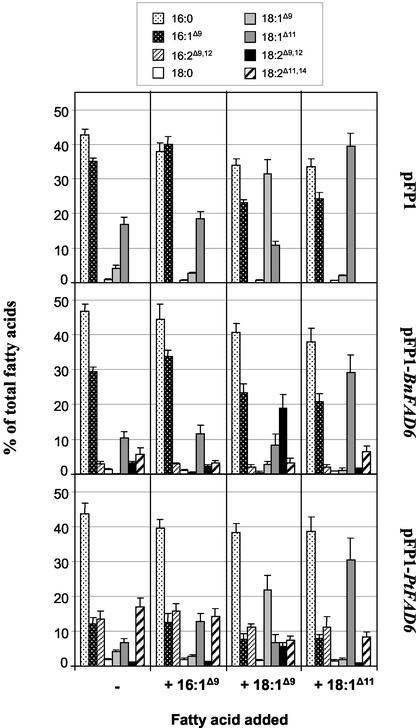
For the functional characterization of the putative plastidial FADs, the Synechococcus R2-PIM8 strain was used as a heterologous expression system (Fig. 5). Synechococcus transformed with the empty vector pFP1 presented a rather simple fatty acid profile with 16:0, 16:1Δ9, 18:1Δ9, and 18:1Δ11 as the major fatty acids, whereas 18:0 was present in trace amounts (Fig. 5, top). When Synechococcus had been transformed with pFP1-PtFAD2, the fatty acid profile did not change, indicating that the microsomal desaturase was not active in the cyanobacterium. In contrast, transformation with pFP1-BnFAD6 resulted in the expression of an active desaturase as indicated by the presence of three new fatty acids (Fig. 5, middle) corresponding to 16:2Δ9,12, 18:2Δ9,12, and 18:2Δ11,14 as characterized by GLC-MS analysis of the 4,4-dimethyloxazoline derivatives. The expression of BnFAD6 resulted in the conversion of nearly all of the 18:1Δ9 to 18:2Δ9,12, whereas only a small proportion of 16:1Δ9 was desaturated to 16:2Δ9,12. The presence of 18:2Δ11,14 could result either from the desaturation of 18:1Δ11 or from the elongation of 16:2Δ9,12, similar to the formation of 18:1Δ11 from 16:1Δ9. When PtFAD6 was expressed in Synechococcus, the same new fatty acids were detected but in different proportions (Fig. 5, bottom). 18:2Δ9,12 was present in a very low percentage, whereas 16:2Δ9,12 and 18:2Δ11,14 were highly accumulated at the expense of 16:1Δ9 and 18:1Δ11.
Figure 5.
Fatty acid profiles of transgenic Synechococcus expressing plastidial desaturases. The Synechococcus R2-PIM8 strain was transformed with the indicated constructs. The transformants were grown at 23°C, and FAMEs from whole cells were prepared and analyzed by GLC as indicated in “Materials and Methods.”
Substrate Specificity of FAD6 Desaturases
The data presented in Figure 5 suggest that the favorite substrates of PtFAD6 and BnFAD6 differed and were 16:1Δ9 and 18:1Δ9, respectively, in agreement with the results obtained in yeast (Fig. 4B). Nevertheless, the origin of 18:2Δ11,14 remained unclear. To clarify the origin of 18:2Δ11,14 and to determine the substrate specificity of each desaturase in more detail, transgenic Synechococcus cultures were supplemented with 16:1Δ9, 18:1Δ9, or 18:1Δ11, and grown for 1 week before fatty acid analysis (Fig. 6). In Synechococcus that had been transformed with pFP1 as control (Fig. 6, top), exogenously supplied 16:1Δ9 increased only slightly the 16:1Δ9 and 18:1Δ11 content, confirming that 18:1Δ11 results from the elongation of 16:1Δ9. The fact that supplying 16:1Δ9 had only a slight impact on the fatty acid composition of Synechococcus wild type was probably due to the high proportion of the endogenous 16:1Δ9 (about 35%) present in the wild-type strain. In contrast, the proportion of 18:1Δ9, which represents only about 4% of the total fatty acids in the wild-type strain, was increased more than 7 times upon exogenous supply of 18:1Δ9. Finally, supplementing 18:1Δ11 doubled its proportion, reaching about 40% of the total fatty acids. In Synechococcus expressing BnFAD6 (Fig. 6, middle), exogenously supplied 16:1Δ9 barely changed the fatty acid composition, whereas supply of 18:1Δ9 resulted in a dramatic increase of 18:2Δ9,12, indicating that BnFAD6 had desaturated more than 87% of 18:1Δ9. In the presence of 18:1Δ11, the proportion of 18:1Δ11 was increased about 3 times but 18:2Δ11,14 remained nearly unchanged. When Synechococcus had been transformed with pFP1-PtFAD6 (Fig. 6, bottom), supplementing 16:1Δ9 doubled the proportions of 18:1Δ11 but did not change that of 16:1Δ9, 16:2Δ9,12, or 18:2Δ11,14. In the presence of 18:1Δ9, although the proportion of 18:2Δ9,12 was increased, 18:1Δ9 accumulated to high levels, indicating that PtFAD6 had desaturated only 20% of the 18:1Δ9. When 18:1Δ11 was supplemented, the 18:2Δ11,14 percentage was lowered by about 50%, although the 18:1Δ11 proportion had been increased by more than 4 times, suggesting that 18:2Δ11,14 was produced by elongation of 16:2Δ9,12 rather than by desaturation of 18:1Δ11. This was further supported by supplying exogenous 16:2Δ9,12 to Synechococcus transformed with pFP1. Although 16:2Δ9,12 was very poorly incorporated, 30% to 40% of it was converted to 18:2Δ11,14 in the absence of any Δ12-desaturase, confirming the presence in Synechococcus of an endogenous elongation activity converting 16:2Δ9,12 to 18:2Δ11,14. If we consider that none of the 18:2Δ11,14 was due to the action of PtFAD6 on 18:1Δ11, then more than 70% of 16:1Δ9 had been desaturated by PtFAD6.
Figure 6.
Fatty acid composition of transgenic Synechococcus expressing plastidial desaturases in the presence or absence of different exogenous fatty acids. The Synechococcus R2-PIM8 strain was transformed with the indicated constructs. The transformants were grown for a week at 23°C in the presence or absence of 75 μm 16:1Δ9, 18:1Δ9, or 18:1Δ11, and FAMEs from whole cells were prepared and analyzed by GLC as indicated in “Materials and Methods.” Each value is the mean ± sd from three to five independent experiments.
The specificity of both desaturases was further evaluated by feeding other potential substrates for ω6-desaturation to the transgenic Synechococcus cultures (Table II). Both desaturases did not accept 22:1Δ13 as substrate, in contrast to previous data obtained in vitro for the plastidial desaturase from spinach (Spinacia oleracea; Schmidt and Heinz, 1993). Nevertheless, because the authors used an ω6-desaturase solubilized by Triton X-100 from chloroplast envelopes, the high activity with 22:1Δ13 obtained in this study may be artifactual and due to the presence of detergent in the assays. Similar to the FAD2 desaturases (Table I), BnFAD6 converted about 19% of 20:1Δ11 to 20:2Δ11,14, but PtFAD6 did not display activity toward that fatty acid. Because the plastidial desaturases of the 16:3 higher plants like rapeseed are involved in the synthesis of 16:3Δ7,10,13, 16:1Δ7 was chemically synthesized and tested as substrate. In Synechococcus transformed with pFP1, 16:1Δ7 accumulated to about 10% of the total fatty acids and was most probably not elongated to 18:1Δ9 because the content of oleic acid was not increased (data not shown). In the transgenic cyanobacteria expressing PtFAD6, only about 5% of 16:1Δ7 was converted to 16:2Δ7,10, whereas upon expression of BnFAD6, more than 80% of 16:1Δ7 was desaturated to 16:2Δ7,10 (Table II). The results presented in Table II clearly show that PtFAD6 is highly specific for 16:1Δ9, whereas BnFAD6 is as active with 16:1Δ7 as with 18:1Δ9, in line with its involvement in the synthesis of the two majors PUFAs of rapeseed leaves, 16:1Δ7,10,13 and 18:3Δ9,12,15. Similar to the microsomal desaturases, both plastidial enzymes display ω6 as well as ω4 activity, as already suggested by Hitz et al. (1994) for BnFAD6. It can be added that 16:1Δ7 was not accepted as substrate by the microsomal desaturases (PtFAD2 and BnFAD2) expressed in yeast (data not shown).
Table II.
Substrate specificity of the plastidial desaturases BnFAD6 and PtFAD6 expressed in Synechococcus
| Substrate | Product(s) | Desaturation
|
|
|---|---|---|---|
| BnFAD6 | PtFAD6 | ||
| % | |||
| 16:1Δ7 | 16:2Δ7,10 | 79.6 ± 5.8 | 5.2 ± 0.8 |
| 16:1Δ9 | 16:2Δ9,12, 18:2Δ11,14a | 15.5 ± 1.9 | 70.3 ± 3.1 |
| 18:1Δ9 | 18:2Δ9,12 | 87.3 ± 4.9 | 20.6 ± 2.5 |
| 20:1Δ11 | 20:2Δ11,14 | 19.4 ± 2.2 | 0 |
| 22:1Δ13 | 22:2Δ13,15 | 0 | 0 |
Synechococcus was transformed with pFP1-BnFAD6 or pFP1-PtFAD6 and grown for 7 d at 23°C in the presence of different fatty acid substrates (75 μm) in separate experiments. FAMEs from the whole cells were prepared and analyzed by GLC as indicated in “Materials and Methods.” Desaturation (percentage) was calculated as (product(s) × 100)/(educt + product(s)) using values corresponding to percent of total fatty acids. Each value is the mean ± sd from three to five independent experiments.
Due to the endogenous elongation of the desaturase product 16:2Δ9,12.
DISCUSSION
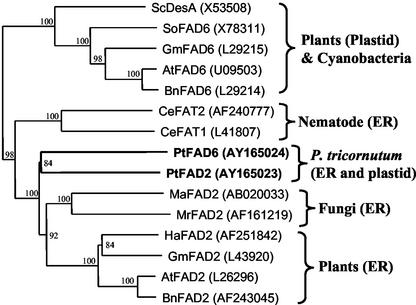
In this paper, we report the cloning and functional characterization of the microsomal and the plastidial Δ12-desaturase from the diatom P. tricornutum, PtFAD2 and PtFAD6, respectively. Similar to FADs from other species, both enzymes contain the three His clusters most likely coordinating the diiron center of the active site as well as long hydrophobic stretches involved in membrane binding (Fig. 2; Shanklin et al., 1994). Although PtFAD2 presents moderate sequence homology to microsomal desaturases from higher plants (about 35% identity/50% homology), PtFAD6 has no homology to the corresponding plastidial desaturases (less than 14% identity/27% homology). As shown in the phylogenetic tree of desaturases presented in Figure 7, PtFAD2 and PtFAD6 form a separate branch between the microsomal desaturases from fungi (Mucor rouxii and Mortierella alpina) and another group containing the ω3- and ω6-desaturases of the nematode Caenorhabditis elegans (FAT1 and FAT2, respectively). The cyanobacterial Δ12-desaturase (DesA) groups together with the plastidial Δ12-desaturases from higher plants (Fig. 7) in line with the phylogenetic origin of plastids. The large separation between DesA and PtFAD6 does not suggest that the latter one is derived from a cyanobacterial symbiont. On the other hand, it is accepted that a single primary endosymbiosis event represents the origin for the evolution of both the green (and the higher plants) and the red algae (Moreira et al., 2000). Because the evolution of diatoms is thought to have involved a secondary endocytobiosis, the plastidial desaturases from P. tricornutum may have a more complicated history. Because PtFAD6 is more similar to ER-localized desaturases, it could have evolved from the microsomal Δ12-desaturase of the host of either the first or the second endosymbiosis. In the latter case, the gene of the microsomal desaturase of the heterotrophic flagellate that engulfed the photoautotrophic eukaryote could have been duplicated and the protein encoded by one copy redirected to the newly acquired plastid. A similar gene duplication may have given rise to the ω3- and ω6-desaturase of C. elegans (FAT1 and FAT2, respectively; Fig. 7) and is supported by the presence of conserved intron-exon junctions in both genes (Napier and Michaelson, 2001). Although the phylogenetic relationship of PtFAD2 and PtFAD6 looks similar to that of CeFAT1 and CeFAT2 in Figure 7, the two Δ12-desaturases from P. tricornutum are much less similar and the absence of introns in the genomic sequences (data not shown) does not provide additional support for a similarly late gene duplication. It could also have happened that the gene of the microsomal Δ12-desaturase from the ancestral red algae, the host of the first endosymbiont, has been transferred to the nuclear genome of the second host and that, after appropriate modifications, the product of its expression was targeted back to the first host, which during evolution became the complex plastid of P. tricornutum. Whatever the true origin of PtFAD6, these results are in agreement with the recent analysis of about 1,000 expressed sequence tags (ESTs) from P. tricornutum, which has shown that many sequences were more similar to animals than to plant counterparts, reflecting the different phylogenetic histories of diatoms and higher plants (Scala et al., 2002).
Figure 7.
Phylogenetic tree of Δ12-desaturases from different organisms. The alignment was generated by the ClustalX program (Thompson et al., 1997), and the phylogenetic tree was constructed by the neighbor-joining method using Tree View (Page, 1996). Numbers indicate bootstrap values. The DNA Data Bank of Japan/EMBL/GenBank accession numbers of the different nucleic acid sequences coding for the desaturases used to construct this tree are indicated.
Because of their origin via secondary endocytobiosis, the plastids of the chromophytic diatoms are surrounded by four membranes. The transport of nuclear-encoded proteins into such complex plastids is a two-step process that relies on the bipartite structure of the targeting signal (Lang et al., 1998). The N-terminal sequence of the plastidial desaturase of P. tricornutum PtFAD6 (Fig. 2) contains a typical signal peptide for cotranslational transport through the ER membranes and a transit peptide for posttranslational protein targeting into the plastid. This bipartite presequence is sufficient to target EGFP into the plastid (Fig. 3). In contrast, fusing the N-terminal sequence of the microsomal desaturase PtFAD2 to EGFP led to cytoplasmic fluorescence (data not shown). In another study (Apt et al., 2002), the fusion of the N-terminal sequence of a lumenal protein to EGFP led to fluorescence within a network of membranes most probably representing the ER. In accordance, the N-terminal extremity of PtFAD2 preceding the first transmembrane domain (Fig. 2) seemed to contain no information for a localization within the microsomes. It should be added that the three microsomal desaturases from P. tricornutum characterized so far, PtFAD2 (this study), PtD5, and PtD6 (Domergue et al., 2002), do not contain the putative ER retention signal for diatoms, DDEL, at their C terminus. Immunocytological studies have shown that the FAD2 desaturases from higher plants are localized in the ER and face the cytosol (Dyer and Mullen, 2001). If PtFAD2 is similarly located in the ER, the information for its localization is most likely present within the transmembrane domains of the protein sequence.
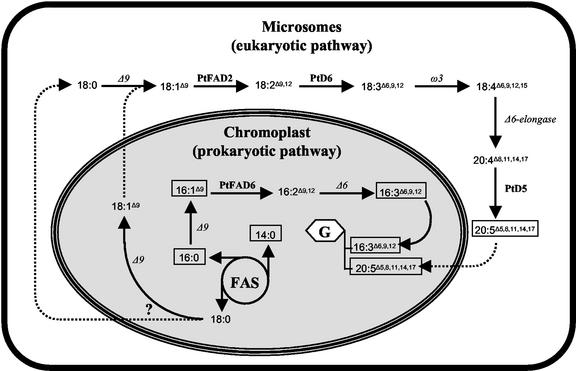
The functional characterization of PtFAD2 and PtFAD6 in yeast and Synechococcus, respectively, confirmed that the former enzyme is a microsomal desaturase that uses cytochrome b5 and cytochrome b5 oxidoreductase as electron donors, whereas the latter desaturase is a plastidial protein requiring ferredoxin and ferredoxin oxidoreductase for electron donation. In addition, these heterologous expression systems enabled a detailed study of substrate specificities (Table I and II, respectively), which in turn led to a better understanding of the fatty acid metabolism in P. tricornutum. The clear preference of PtFAD2 for 18:1Δ9 (50% conversion to 18:2Δ9,12; Table I) fits with previous labeling experiments that had shown that EPA (20:5Δ5,8,11,14,17) was synthesized by desaturation and elongation of oleic acid (Arao and Yamada, 1994). Together with the recently cloned Δ5- and Δ6-desaturases (Domergue et al., 2002), PtFAD2 is the third microsomal enzyme involved in the biosynthesis of EPA to be characterized. It should be added that the different intermediates of the EPA biosynthetic pathway were only present in trace amounts in the fatty acid profile obtained from P. tricornutum cells (Fig. 1) but found in much more abundant proportions in phospholipids, particularly in phosphatidylcholine (Arao et al., 1987). In contrast, the proportion of EPA was about the same in phosphatidylcholine, phosphatidylglycerol, and the total fatty acids (30%; Arao et al., 1987). From this, it may be concluded that in addition to the Δ12-, Δ6-, and Δ5-desaturation steps required for EPA biosynthesis, the Δ6-elongation and the ω3-desaturation steps are taking place in the microsomal fraction (Fig. 8). In contrast, the high specificity of PtFAD6 for 16:1Δ9 (Table II) suggests that 16:2Δ9,12 and most probably 16:3Δ6,9,12, the second major PUFA found in P. tricornutum, are synthesized within the plastid, similar to the hexadecatrienoic acid isomer found in 16:3 plants. Although 16:2Δ9,12 could leave the plastid, be converted to 16:3Δ6,9,12 by a microsomal Δ6-desaturase, and then re-incorporated into the plastid, this scenario seems unlikely. Because the microsomal Δ6-desaturase PtD6 is highly active with 18:2Δ9,12 but poorly active with both 16:1Δ9 (Domergue et al., 2002) and 16:2Δ9,12 (data not shown), we rather speculate that P. tricornutum contains a plastidial Δ6-desaturase that is involved in the synthesis of 16:3Δ6,9,12 (Fig. 8). It should be added that among the 3,860 ESTs that have been identified, no ESTs corresponding to an acyl-ACP desaturase, to a plastidial Δ6 desaturase, or to an ω3-desaturase were detected, and only one sequence having similarities to mammalian stearoyl-CoA desaturase was detected. Nevertheless, this clone was not full length and functionally characterized; therefore, its role in the fatty acid metabolism of P. tricornutum remains to be studied. In accordance, the number of Δ9-desaturases present in P. tricornutum remains unclear: Is there a microsomal Δ9-desaturase specific for 18:0 and a plastidial Δ9-desaturase specific for 16:0, or is a single desaturase in the plastid responsible for the synthesis of both 16:1Δ9 and 18:1Δ9 as in cyanobacteria? Alternatively, a soluble Δ9-acyl-ACP desaturase and a membrane-bound Δ9-acyl-lipid desaturase, responsible for the synthesis of 18:1Δ9 and 16:1Δ9, respectively, could co-exist in the plastid of diatoms, similar to the situation found in higher plants.
Figure 8.
Hypothetical compartmentalization of fatty acid metabolism in P. tricornutum cells. The main fatty acids present in P. tricornutum (see Fig. 1) are framed. Desaturase genes that have been cloned are marked in bold, whereas putative desaturase (Δ6, ω3, and Δ9) and elongase are indicated with italics. Fatty acid modifications are indicated by arrows, and possible exchange reactions are indicated by dashed lines. The backbone with the framed G refers to the major glycolipids found in the plastid of P. tricornutum (monogalactosyldiacylglycerol, digalactosyldiacylglycerol, and suphoquinovosyldiacylglycerol). Thioester- or lipid-linked desaturation is not differentiated and actually not known in detail. FAS, Fatty acid synthase.
Although the expression of a rapeseed FAD6 cDNA in Synecchococcus has already been achieved (Hitz et al., 1994), this is the first time, to our knowledge, that the substrate specificity of a plastidial desaturase is characterized in detail. The use of the Synechococcus R2-PIM8 strain as a heterologous expression system was instrumental in this, as evidenced by the high desaturation activities reported in Figures 5 and 6 and in Table II. These results confirm the strong activity of the nptII promoter in Synechococcus and demonstrate the usefulness of the R2-PIM8 strain for the functional characterization of plastidial enzymes involved in fatty acid metabolism. The substrate specificity of BnFAD6 reported in Table II confirms that the plastidial Δ12-desaturases of 16:3 plants are involved in the synthesis of the two trienoic fatty acids found in the leaves of such plants, 16:3Δ7,10,13 and 18:3Δ9,12,15. In addition, the high activity of the microsomal desaturase BnFAD2 with 18:1Δ9 (Fig. 4A) is in agreement with the involvement of this enzyme in the synthesis of 18:2Δ9,12 in the ER. In 16:3 plants like rapeseed, two glycerolipid biosynthetic pathways co-exist: The prokaryotic pathway in the plastid leads to the synthesis of glycolipids with 18:3Δ9,12,15 and 16:3Δ7,10,13 at the sn-1 and sn-2 position, respectively, whereas the eukaryotic pathway in the ER is contributing to the synthesis of glycolipids with 18:3Δ9,12,15 at both positions (Browse and Somerville, 1991; Wallis and Browse, 2002). In P. tricornutum, the most abundant glycolipids contain EPA and unsaturated 16 carbon fatty acids at the sn-1 and sn-2 position, respectively (Fig. 8; Arao et al., 1987; Yongmanitchai and Ward, 1993), which is considered to be a typically prokaryotic diacylglycerol backbone. Such a denomination is nevertheless ambiguous in the case of P. tricornutum because the results presented in this study demonstrate that the 16 carbon fatty acids at the sn-2 position are most probably synthesized exclusively by a plastidial prokaryotic pathway, whereas EPA at the sn-1 position is of eukaryotic origin. Whether EPA is imported into the chromoplast of P. tricornutum as a free fatty acid in reversal of the fatty acid export from plastids, or whether it is linked to a glycerol backbone similar to the situation prevailing in higher plants, deserves further investigation. The lipid exchange between plastids and other compartments may differ to some extent regarding nature of intermediates and direction of transport when looking at organisms such as algae, which differ in fatty acid diversity from the monotonous situation encountered in higher plants.
MATERIALS AND METHODS
Materials
Restriction enzymes, polymerases, and DNA-modifying enzymes were obtained from New England BioLabs (Frankfurt) unless indicated otherwise. All other chemicals were from Sigma (St. Louis).
Culture of Phaeodactylum tricornutum
P. tricornutum UTEX 646 was grown in brackish water medium (Schlösser, 1993) at 23°C with moderate shaking under long-day light conditions (15 h at 30 μE m−2 s−1). Cells were harvested by centrifugation, washed with water, and used for fatty acid analysis.
Isolation of P. tricornutum cDNA Clones
A P. tricornutum cDNA library was constructed and subjected to random sequencing as previously described (Domergue et al., 2002). Among the 3,860 nonredundant clones obtained, three sequences presented high homologies to various Δ12-desaturases and were fully sequenced. Two clones originating from a single gene overlapped, but Pt001072031r and Pt001070010r differed and each contained a full-length ORF.
Isolation of Rapeseed Brassica napus cDNA Clones
Using an excised rapeseed cv Askari cDNA library as template and primers designed according to the sequences available in databases, clones corresponding to the rapeseed plastidial and microsomal desaturases (BnFAD6 and BnFAD2, respectively) were amplified by PCR and sequenced. The nucleotide sequences of the clones BnFAD6 and BnFAD2 presented in this study were 95% and 96% identical to that of the rapeseed plastidial ω6-desaturase (accession no. L29214) and that of the rapeseed Δ12-oleate desaturase (accession no. AF243045), respectively. The deduced proteins differed slightly from those already published (accession nos. AAF78778 and AAA50157). BnFAD2 presented three amino acid changes (T20N, A246V, and L266F) whereas BnFAD6 had eight substitutions at the N terminus (Q17H, C18S, P32Q, L53F, F66S, N76S, D79H, and E83D), respectively.
Functional Characterization in Yeast (Saccharomyces cerevisiae)
For functional characterization, the four desaturase sequences were cloned in the yeast expression vector pYES2 (Invitrogen). The ORFs of BnFAD6, PtFAD2, and PtFAD6 were modified by PCR to create BamHI and XhoI restriction sites adjacent to the start and stop codons, respectively, and to insert the yeast consensus sequence for enhanced translation (Donahue and Cigan, 1990) in front of the start codon. The ORF of BnFAD2 was similarly amplified but with KpnI and XbaI sites. All these PCR products were cloned into the pGEM-T vector (Promega, Madison, WI), and the ORFs were released by BamHI/XhoI (or KpnI/XbaI) digestion. Cloning of the PtFAD2, PtFAD6, BnFAD2, and BnFAD6 ORFs in pYES2 using the same sites yielded pYES2-PtFAD2, pYES2-PtFAD6, pYES2-BnFAD2, and pYES2-BnFAD6, respectively.
Yeast strain C13ABYS86 (leu2, ura3, his, pra1, prb1, prc1, and cps; Bröker et al., 1991) was transformed with plasmid DNA by a modified polyethylene glycol/lithium acetate protocol (Ausubel et al., 1995). After selection on minimal medium agar plates without uracil, cells harboring the vector were cultivated in minimal medium lacking uracil but containing 2% (w/v) raffinose and 1% (v/v) Tergitol NP-40. Expression was induced by supplementing Gal to 2% (w/v) when the cultures had reached an A600 of 0.2 to 0.3. At that time, the appropriate fatty acids were added to a final concentration of 500 μm, unless indicated otherwise, and the cultures were further grown for 48 h at 20°C. Cells were harvested by centrifugation, and the pellet was washed once with 0.1 m NaHCO3 before being used for fatty acid analysis.
Functional characterization in Synechococcus
The Synechococcus PCC7972 strain R2-PIM8, which contains an integration platform in the metF gene for pBR322-derived plasmids (van der Plas et al., 1990), was used to express the PtFAD2, PtFAD6 and BnFAD6 desaturases. For this purpose, the plasmid pFP1–3 (Götz et al., 1999) was digested with SmaI, dephosphorylated with the calf intestinal alkaline phosphatase, purified, and self-ligated, yielding the plasmid pFP1. The PtFAD2, PtFAD6, and BnFAD6 ORFs were cut out of pYES2-PtFAD2, pYES2-PtFAD6, and pYES2-BnFAD6, respectively, by BamHI/XhoI digestion, blunt ended with Klenow enzyme, and phosphorylated with T4 polynucleotide kinase. Each ORF was then inserted in the correct orientation in pFP-1 opened with SmaI, yielding pFP1-PtFAD2, pFP1-PtFAD6, and pFP1-BnFAD6, respectively.
The Synechococcus R2-PIM8 strain was cultured in BG11 medium (Rippka et al., 1979) supplemented with 30 μg mL−1 Met and 10 μg mL−1 streptomycin at 23°C with moderate shaking and under long-day light conditions (15 h at 30 μE m−2 s−1). Transformation was conducted according to Windhövel et al. (1994), and transformants were selected on BG11 plates (1.5% [w/v] Bacto-agar) containing 30 μg mL−1 Met, 1 μg mL−1 ampicillin, and 10 μg mL−1 kanamycin. When fatty acids were exogenously supplied, growing cultures were sedimented by centrifugation and resuspended in 30 mL of BG11 containing Met, ampicillin, kanamycin, and 75 μm fatty acid. The cultures were further grown for a week at 23°C, harvested by centrifugation, and the pellet was washed once with 0.1 m NaHCO3 before being used for fatty acid analysis.
Fatty Acid Analysis
Cell sediments were directly transmethylated with 1 n sulfuric acid in methanol containing 2% (v/v) dimethoxypropane (1 h at 80°C) to prepare FAMEs. FAMEs were extracted in petroleum ether and analyzed by GLC as already described (Domergue et al., 2002). Fatty acids were identified by comparison with appropriate reference substances or by GLC-MS of 4,4-dimethyloxazoline derivatives as described earlier (Sperling et al., 2000).
GFP Expression in P. tricornutum
The N-terminal part of the plastidial or the microsomal desaturase from P. tricornutum was fused to the N-terminus of EGFP. For this purpose, the first 165 and 339 bp of the ORF of PtFAD2 and PtFAD6, respectively, were cloned in frame at the 5′ end of the nucleotidic sequence coding for EGFP. The resulting EGFP fusions were inserted into the P. tricornutum transformation vector pPha-T1, and transformation of the diatom was achieved by microparticle bombardment (Zaslavskaia et al., 2000). Selection and culture of the transformants and light and fluorescence microscopy were carried out according to Apt et al. (2002).
Chemical Synthesis of 16:1Δ7 and 16:2Δ9,12
The synthesis of (7Z)-hexadec-7-enoic acid (16:1Δ7) was achieved by a cis-selective Wittig reaction (Bestmann et al., 1976) of 7-phosphoranylheptanoic acid methyl ester and nonanal in 40% (w/v) yield. (9Z,12Z)-hexadeca-9,12-dienoic acid (16:2Δ9,12) was synthesized along a protocol described for the synthesis of homoconjugated trienoic acids (Pohnert and Boland, 2000; Zank et al., 2002). Successive treatment of the symmetrical bisphosphorane with butanal and methyl 9-oxononanoate, using a carefully optimized regime of temperatures, yielded the methyl ester of 16:2Δ9,12 in a single operation and 42% (w/v) yield. The free fatty acids were obtained by saponification of the respective methyl ester with LiOH in THF:water (3:1 [v/v]; Nicolaou et al., 1986).
Details of the syntheses and spectroscopic data are available as supplementary material (see www.plantphysiol.org for supplemental material).
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Dr. Gerhard Sandmann (Frankfurt, Germany) for providing the Synechococcus PIM8 strain and the pFP1–3 vector and BASF Plant Science GmbH (Ludwigshafen, Germany) for performing the random sequencing program.
Footnotes
This work was supported in part by the European Community program Human Potential (Marie Curie Fellowship under contract no. HPMF–CT–1999–00148 to F.D.), by the Fachagentur für Nachwachsende Rohstoffe (grant to P.S.), by the Deutsche Forschungsgemeinschaft (grant no. Transregio-SFB TR1, TP A1 to P.G.K. and grant no. SFB 436 to W.B.), by the Fonds der Chemischen Industrie, and by BASF Plant Science GmbH.
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.018317.
LITERATURE CITED
- Apt KE, Zaslavkaia L, Lippmeier JC, Lang M, Kilian O, Wetherbee R, Grossman AR, Kroth PG. In vivo characterization of diatom multipartite plastid targeting signals. J Cell Sci. 2002;115:4061–4069. doi: 10.1242/jcs.00092. [DOI] [PubMed] [Google Scholar]
- Arao T, Kawaguchi A, Yamada M. Positional distribution of fatty acids in lipids of the marine diatom, Phaeodactylum tricornutum. Phytochemistry. 1987;26:2573–2576. [Google Scholar]
- Arao T, Yamada M. Biosynthesis of polyunsaturated fatty acids in the marine diatom, Phaeodactylum tricornutum. Phytochemistry. 1994;35:1177–1181. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, Albright LM, Coen DM, Varki A. Current Protocols in Molecular Biology. New York: John Wiley and Sons; 1995. [Google Scholar]
- Bestmann HJ, Stransky W, Vostrowsky O. Reactions of Alkylidenetriphenylphosphoranes: 33. Preparation of lithium salt-free ylid solutions with sodium bis(trimethylsilyl) amide as base. Chem Ber-Recl. 1976;109:1694–1700. [Google Scholar]
- Bhaya D, Grossman A. Targeting proteins to diatom plastids involves transport through an endoplasmic reticulum. Mol Gen Genet. 1991;229:400–404. doi: 10.1007/BF00267462. [DOI] [PubMed] [Google Scholar]
- Bröker M, Bauml O, Gottig A, Ochs J, Bodenbenner M, Amann E. Expression of the human blood coagulation protein factor XIIIa in Saccharomyces cerevisiae: dependence of the expression levels from host-vector systems and medium conditions. Appl Microbiol Biotechnol. 1991;34:756–764. doi: 10.1007/BF00169346. [DOI] [PubMed] [Google Scholar]
- Browse J, Somerville C. Glycerolipid synthesis: biochemistry and regulation. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:467–506. [Google Scholar]
- Domergue F, Lerchl J, Zähringer U, Heinz E. Cloning and functional characterization of Phaeodactylum tricornutumfront-end desaturases involved in eicosapentaenoic acid biosynthesis. Eur J Biochem. 2002;269:4105–4113. doi: 10.1046/j.1432-1033.2002.03104.x. [DOI] [PubMed] [Google Scholar]
- Donahue TF, Cigan AM. Sequence and structural requirements for efficient translation in yeast. Methods Enzymol. 1990;185:366–372. doi: 10.1016/0076-6879(90)85032-j. [DOI] [PubMed] [Google Scholar]
- Dyer JM, Mullen RT. Immunocytological localization of two plant fatty acid desaturases in the endoplasmic reticulum. FEBS Lett. 2001;494:44–47. doi: 10.1016/s0014-5793(01)02315-8. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Falcone DL, Gibson S, Lemieux B, Somerville C. Identification of a gene that complements an Arabidopsismutant deficient in chloroplast omega 6 desaturase activity. Plant Physiol. 1994;106:1453–1459. doi: 10.1104/pp.106.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz T, Windhövel U, Böger P, Sandmann G. Protection of photosynthesis against ultraviolet-B radiation by carotenoids in transformants of the cyanobacterium SynechococcusPCC7942. Plant Physiol. 1999;120:599–604. doi: 10.1104/pp.120.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitz WD, Carlson TJ, Booth JR, Kinney AJ, Stecca KL, Yadav NS. Cloning of a higher-plant plastid ω-6 fatty acid desaturase cDNA and its expression in a cyanobacterium. Physiol Plant. 1994;105:635–641. doi: 10.1104/pp.105.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroth PG, Strotmann H. Diatom plastids: secondary endocytobiosis, plastid genome and protein import. Physiol Plant. 1999;107:136–141. [Google Scholar]
- Lang M, Apt KE, Kroth PG. Protein transport into “complex” diatom plastids utilizes two different targeting signals. J Biol Chem. 1998;273:30973–30978. doi: 10.1074/jbc.273.47.30973. [DOI] [PubMed] [Google Scholar]
- Los DA, Murata N. Structure and expression of fatty acid desaturases. Biochem Biophys Acta. 1998;1394:3–15. doi: 10.1016/s0005-2760(98)00091-5. [DOI] [PubMed] [Google Scholar]
- McFadden GI. Primary and secondary endosymbiosis and the origin of plastids. J Phycol. 2001;37:951–959. [Google Scholar]
- Meesapyodsuk D, Reed DW, Savile CK, Buist PH, Ambrose SJ, Covello PS. Characterization of the regiochemistry and cryptoregiochemistry of a Caenorhabditis elegans fatty acid desaturase (FAT-1) expressed in Saccharomyces cerevisiae. Biochemistry. 2000;39:11948–11954. doi: 10.1021/bi000756a. [DOI] [PubMed] [Google Scholar]
- Molina Grima E, Robles Medina A, Giménez Giménez A, Ibáñez González MJ. Gram-scale purification of eicosapentaenoic acid (EPA, 20,5n-3) from wet Phaeodactylum tricornutumUTEX 640 biomass. J Appl Phycol. 1996;8:359–367. [Google Scholar]
- Moreira D, Le Guyader H, Philippe H. The origin of red algae and the evolution of chloroplasts. Nature. 2000;405:69–72. doi: 10.1038/35011054. [DOI] [PubMed] [Google Scholar]
- Moreno VJ, De Moreno JEA, Brenner RR. Biosynthesis of unsaturated fatty acids in the diatom Phaeodactylum tricornutum. Lipids. 1979;14:15–19. [Google Scholar]
- Napier JA, Michaelson LV. Genomic and functional characterization of polyunsaturated fatty acid biosynthesis in Caenorhabditis elegans. Lipids. 2001;36:761–766. doi: 10.1007/s11745-001-0782-9. [DOI] [PubMed] [Google Scholar]
- Nicolaou KC, Ladduwahetty T, Taffer IM, Zipkin RE (1986) A general strategy for the synthesis of monohydroxyeicosatetraenoic acids: total synthesis of 5(S)-hydroxy-6(E),8,11,14(Z)-eicosatetraenoic acid (5-Hete) and 12(S)-hydroxy-5,8,14(Z),10(E)-eicosatetraenoic acid (12-Hete). Synthesis 344–347
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. ArabidopsisFAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R. Tree View: an application to display phylogenic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pohnert G, Boland W. Highly efficient one-pot double-Wittig approach to unsymmetrical (1Z,4Z,7Z)-homoconjugated trienes. Eur J Org Chem. 2000;9:1821–1826. [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- Scala S, Bowler C. Molecular insights into the novel aspects of diatom biology. Cell Mol Life Sci. 2001;58:1666–1673. doi: 10.1007/PL00000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala S, Carels N, Falciatore A, Chiusano ML, Bowler C. Genome properties of the diatom Phaeodactylum tricornutum. Plant Physiol. 2002;129:993–1002. doi: 10.1104/pp.010713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlösser UG. SAG-Sammlung von Algenkulturen at the University of Göttingen: catalogue of strains 1994. Bot Acta. 1993;107:113–186. [Google Scholar]
- Schmidt H, Heinz E. Direct desaturation of intact galactolipids by a desaturase solubilized from spinach (Spinacia oleracea) chloroplast envelopes. Biochem J. 1993;289:777–782. doi: 10.1042/bj2890777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J, Cahoon EB. Desaturation and related modifications of fatty acids. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:611–641. doi: 10.1146/annurev.arplant.49.1.611. [DOI] [PubMed] [Google Scholar]
- Shanklin J, Whittle E, Fox BG. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- Sperling P, Lee M, Girke T, Zähringer U, Stymne S, Heinz E. A bifunctional delta 6-fatty acyl acetylenase/desaturase from the moss Ceratodon purpureus: a new member of the cytochrome b5 superfamily. Eur J Biochem. 2000;267:3801–3811. doi: 10.1046/j.1432-1327.2000.01418.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tréguer P, Nelson DM, Van Bennekom AJ, De Master D, Leynaert A, Quéguiner B. The silica balance in the world ocean: a reestimate. Science. 1995;268:375–379. doi: 10.1126/science.268.5209.375. [DOI] [PubMed] [Google Scholar]
- van der Plas J, Hegeman H, de Vrieze G, Tuyl M, Borrias M, Weisbeek P. Genomic integration system based on pBR322 sequences for the cyanobacterium Synechococcussp. PCC7942: transfer of genes encoding plastocyanin and ferredoxin. Gene. 1990;95:39–48. doi: 10.1016/0378-1119(90)90411-j. [DOI] [PubMed] [Google Scholar]
- Wallis JG, Browse J. Mutants of Arabidopsisreveal many roles for membrane lipids. Prog Lipid Res. 2002;41:254–278. doi: 10.1016/s0163-7827(01)00027-3. [DOI] [PubMed] [Google Scholar]
- Windhövel U, Geiges B, Sandmann G, Böger P. Expression of Erwinia uredovora phytoene desaturase in SynechococcusPCC7942 leading to resistance against a bleaching herbicide. Plant Physiol. 1994;104:119–125. doi: 10.1104/pp.104.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongmanitchai W, Ward OP. Positional distribution of fatty acids, and molecular species of polar lipids, in the diatom Phaeodactylum tricornutum. J Gen Microbiol. 1993;139:465–472. doi: 10.1099/00221287-139-3-465. [DOI] [PubMed] [Google Scholar]
- Zank TK, Zähringer U, Beckmann C, Pohnert G, Boland W, Holtorf H, Reski R, Lerchl J, Heinz E. Cloning and functional characterisation of an enzyme involved in the elongation of delta6-polyunsaturated fatty acids from the moss Physcomitrella patens. Plant J. 2002;31:255–268. doi: 10.1046/j.1365-313x.2002.01354.x. [DOI] [PubMed] [Google Scholar]
- Zaslavskaia LA, Lippmeier JC, Kroth PG, Grossman AR, Apt KE. Transformation of the diatom Phaeodactylum tricornutum(Bacillariophyceae) with a variety of selectable marker and reporter genes. J Phycol. 2000;36:379–986. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.