Abstract
Expression of KNOX (KNOTTED1-like homeobox) genes in the shoot apical meristem of Arabidopsis is required for maintenance of a functional meristem, whereas exclusion of KNOX gene expression from leaf primordia is required for the elaboration of normal leaf morphology. We have constructed a steroid-inducible system to regulate both the amount and timing of KN1 (KNOTTED1) misexpression in Arabidopsis leaves. We demonstrate that lobed leaf morphology is produced in a dose-dependent manner, indicating that the amount of KN1 quantitatively affects the severity of lobing. The KN1-glucocorticoid receptor fusion protein is not detected in leaves in the absence of steroid induction, suggesting that it is only stable when associated with steroid in an active state. By using a second inducible fusion protein to mark exposure of leaf primordia to the steroid, we determined the stage of leaf development that produces lobed leaves in response to KN1. Primordia as old as plastochron 7 and as young as plastochron 2 were competent to respond to KN1.
Leaves arise from the shoot apical meristem (SAM) at regular intervals of time termed plastochrons. An Arabidopsis leaf consists of a distal lamina and proximal petiole with a pair of stipules flanking the leaf base. Like most plants, Arabidopsis exhibits heteroblasty such that juvenile rosette leaves have small, round lamina with smooth margins, and adult leaves have larger, elongate lamina with serrated margins. Cauline leaves differ from rosette leaves by lacking a petiole, and the lamina tends to be narrower.
Molecular evidence of the transition from meristem to leaf identity comes from a change in the expression of KNOX (KNOTTED1-like homeobox) genes. In Arabidopsis, the class I KNOX genes STM (SHOOTMERISTEMLESS), KNAT1 (KNOTTED1-like in Arabidopsis1), and KNAT2 are expressed in the SAM but excluded from cells destined to become leaf primordia (Lincoln et al., 1994; Long et al., 1996; Pautot et al., 2001). This expression pattern suggests that KNOX gene products are required for meristem function. Genetic evidence supports such a hypothesis: Loss-of-function mutations in STM result in a failure to initiate or maintain a SAM (Long et al., 1996). In addition, double-mutant analysis between weak stm alleles and the KNAT1 loss-of-function mutation bp (brevipedicellus) demonstrate that KNAT1 assumes a redundant role with STM in the SAM (Byrne et al., 2002).
Conversely, ectopic expression of KNOX genes alters normal leaf morphology by conferring less determinate characters to the leaf. Expression of KNAT1 driven by the constitutive CaMV 35S promoter transforms simple Arabidopsis leaves into lobed leaves with ectopic meristems and stipules (Lincoln et al., 1994; Chuck et al., 1996). Lobed leaf margins with ectopic stipules are produced in both 35S:KNAT2 and DEX-induced 35S:KNAT2-GR plants (G. Chuck and S. Hake, unpublished data; Pautot et al., 2001). Ectopic expression of STM also results in alterations of leaf shape, meristem formation on the adaxial surface of the cotyledons, and growth arrest (K. Barton, personal communication; Williams, 1998; Gallois et al., 2002).
Mutations at two independent loci, AS1 (ASYMMETRIC LEAVES1) and AS2, condition misexpression of KNAT1, KNAT2,and KNAT6 in Arabidopsis leaves, correlating with lobed leaf morphology (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001). AS1 encodes a myeloblastosis domain protein and is likely to act as a transcriptional regulator of KNOX expression (Byrne et al., 2000). AS2 is a member of the LATERAL ORGAN BOUNDARIES gene family and contains a Leu zipper motif and Cys repeats, suggesting that AS2 may associate with DNA or protein partners to negatively regulate KNOX gene expression (Iwakawa et al., 2002; Shuai et al., 2002). Less is known regarding the components that act downstream of KNOX function. In Arabidopsis, KNOX transcription factors repress the GA biosynthetic gene AtGA20ox1, thus promoting low GA conditions favorable for meristematic activity (Hay et al., 2002).
In contrast to Arabidopsis, the dissected leaves of tomato (Lycopersicon esculentum) plants express KNOX genes. Overexpression of maize (Zea mays) KN1 or tomato KNOX genes leads to a dramatic increase in leaf dissection (Hareven et al., 1996; Janssen et al., 1998). These results suggest that dissected leaves have a greater capacity for indeterminate growth than do simple leaves and that differential regulation of KNOX genes confers this indeterminacy to the development of dissected leaves. A recent study further supports this idea by demonstrating that KNOX expression early in leaf development correlates with formation of complex leaf primordia across a broad spectrum of vascular plants (Bharathan et al., 2002).
To regulate KN1 misexpression in vivo, a steroid-inducible fusion between the maize KN1 open reading frame (Vollbrecht et al., 1991) and the steroid-binding domain of the glucocorticoid receptor (GR), driven by the CaMV 35S promoter (Lloyd et al., 1995), was constructed and transformed into Arabidopsis. The GR steroid-binding domain maintains the constitutively expressed transcription factor in an inactive state by tethering it in the cytoplasm. This inactivation is reversed by application of the steroid dexamethasone (DEX), which triggers translocation of the fusion protein into the nucleus. As a homeodomain-containing protein, KN1 is expected to function as a transcriptional regulator in the nucleus. The subcellular localization of KN1 in the nucleus (Smith et al., 1992) and its ability to specifically bind DNA support this expectation (Smith et al., 2002).
Here, we demonstrate a dose-dependent response of Arabidopsis leaves to KN1 induction in the production of lobed margins, broad petioles, and arrested growth. We show that the KN1-GR fusion protein is only detected in leaves after DEX treatment. We also define a window of competence in which primordia as old as plastochron 7 (P7) and as young as P2 are competent to respond to KN1 by producing lobed margins.
RESULTS
35S:KN1-GR Confers a Lobed Morphology to Arabidopsis Leaves upon DEX Induction
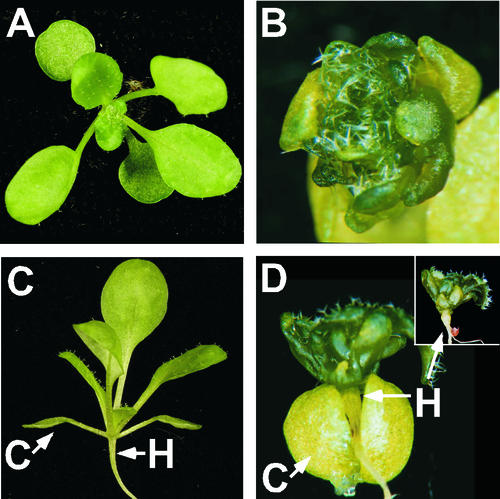
35S:KN1-GR seeds germinated on media containing DEX have perturbed leaf development (Fig. 1, B and D) similar to severe 35S:KNAT1 transformants and DEX-induced 35S:KNAT2-GR and 35S:STM-GR plants (Chuck et al., 1996; Pautot et al., 2001; Gallois et al., 2002). In the absence of DEX, 35S:KN1-GR plants show no phenotypic effects (Fig. 2A). Leaf growth is reduced in DEX-treated 35S:KN1-GR plants, resulting in a tight rosette of small, lobed leaves lacking petioles (Fig. 1B). Leaf primordia show very delayed lateral expansion of the lamina, such that primordia appear radial for some time before expansion occurs to produce a lobed morphology. Cotyledons are normal in size and shape but they demonstrate epinastic growth in response to KN1-GR activity. Elongation of the hypocotyl is inhibited, resulting in a short, thick hypocotyl (Fig. 1D). No ectopic shoot meristems were seen at the concentration of DEX used. Wild-type seeds germinated on DEX showed no phenotypic effects (Fig. 1, A and C).
Figure 1.
35S:KN1-GR plants show severe perturbations in leaf development when germinated on DEX-containing media. A, Wild-type top view; B, 35S:KN1-GR top view; C, wild-type side view, arrows indicate hypocotyl (H) and cotyledon (C); D, 35S:KN1-GR side view, arrows point to short, thick hypocotyl and epinastic cotyledon. Insert, Hypocotyl with cotyledons removed.
Figure 2.
Dose response of 35S:KN1-GR phenotype. Whole-plant and heteroblastic leaf series from cotyledons to leaf number 8 of 35S:KN1-GR plants in response to a single application of: A, no DEX; B, 10−9 m DEX, arrows point to serration on leaf 4; C, 10−8 m DEX, arrows point to serrations on leaves 3 and 4; D, 10−7 m DEX, arrows point to lobe on leaf 2; E, 10−6 m DEX, arrows point to deep lobes on leaves 3 and 4; and F, 10−5 m DEX, arrows point to small, deeply lobed leaves 3 to 6. Only one cotyledon is shown in F. Each panel is representative of 36 plants per treatment.
To further investigate the effects of KN1 misexpression on leaf development, we analyzed the response of 35S:KN1-GR plants to increasing doses of DEX hormone. The effect of the hormone application overlays a heteroblastic leaf series that is identical in 35S:KN1-GR and wild-type plants grown under long-day conditions. We applied a single dose of increasing concentration of DEX at the time when the first true leaves were visible. This allowed us to assess the effects of KN1 misexpression on the development of juvenile leaves, which have a more entire margin than adult leaves. Lobed leaf morphology was produced in a hormone-dependent manner with a dose-dependent increase in both the number and depth of lobes and the number of leaves affected (Fig. 2, A–F).
To describe the severity of the leaf margin phenotypes, we focused on the depth of the sinuses. We defined serrations as having sinuses less than one-fourth of the distance to the midvein, whereas lobes were defined as having sinuses extending one-fourth or more of the distance to the midvein (Groot and Meicenheimer, 2000). Application of low concentrations of DEX (10−9 and 10−8 m) results in single, conspicuous serrations on juvenile 35S:KN1-GR leaves (arrows, Fig. 2, B and C). At 10−7 m DEX, a number of more deeply indented lobes and serrations form on the juvenile leaves and a broader petiole forms in lobed leaves (arrow, Fig. 2D). At 10−6 m DEX, deep lobes form resembling leaflets with round lamina and narrow petiolules (arrow, Fig. 2E). The petiole is broader and shorter in these lobed leaves. At 10−5 m DEX, reduced growth results in small, highly lobed leaves that lack petioles (arrows, Fig. 2F), similar to those produced when seeds are germinated on 10−6 m DEX (Fig. 1, B and D). Four consecutive leaves are fully affected by the induction of KN1 activity with 10−5 m DEX, compared with one to two leaves with lower concentrations. These DEX-dependent changes in leaf morphology seen in 35S:KN1-GR plants suggest that KN1 is indeed regulating target gene expression upon steroid induction.
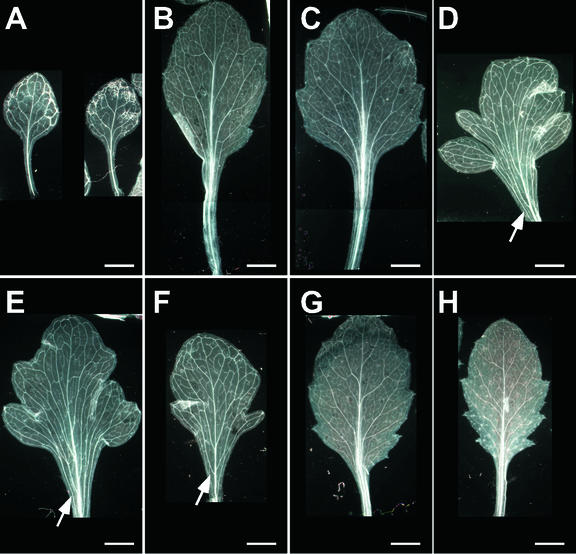
Variation in vascular patterning is associated with different leaf morphologies (Nelson and Dengler, 1997). Lobed 35S:KN1-GR leaves, induced using a single application of 10−6 m DEX, show prominent, widely spaced veins on either side of the midvein, correlated with a broad petiole (Fig. 3, D–F). No significant alterations in vascular pattern are apparent in the leaves immediately preceeding or after those that are lobed (Fig. 3, C, G, and H; data not shown), suggesting that changes in vasculature are tightly correlated with the lobed morphology.
Figure 3.
Vascular pattern of leaves 1 to 9 of a mature 35S:KN1-GR plant after DEX induction. A to C, Leaves 1 to 4 showing a normal vascular pattern in the petiole in which the veins are appressed to the midvein; D to F, leaves 5 to 7 with short, broad petioles; arrows indicate widely spaced veins, not appressed to the midvein; G and H, leaves 8 and 9 with normal vascular pattern in the petiole. Scale bars = 5 mm.
KN1-GR Is Stably Expressed Only in the Leaves of DEX-Treated Plants
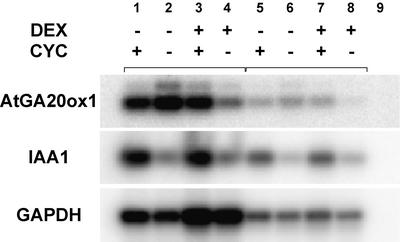
The induction of activity of a transcription factor, such as KN1, is predicted to alter expression levels of downstream genes. We previously demonstrated that the GA biosynthetic pathway is a downstream component of KNOX function in Arabidopsis (Hay et al., 2002). Transcript levels for the GA biosynthetic gene AtGA20ox1 are reduced in response to KNOX misexpression. By inducing KN1 activity for different time intervals in 35S:KN1-GR seedlings, we established that AtGA20ox1 expression is reduced within 30 min of DEX application (Fig. 4, lanes 4 and 8; Hay et al., 2002). This rapid repression of AtGA20ox1 expression suggests that it may be directly regulated by KN1, similar to the direct interaction between the tobacco (Nicotiana tabacum) KNOX protein, NTH15, and Ntc12 (Sakamoto et al., 2001). We tested this hypothesis by analyzing AtGA20ox1 expression after DEX induction in the presence of the protein synthesis inhibitor CYC. The effect of CYC was monitored by expression of the CYC-inducible gene, IAA1 (Fig. 4; Abel et al., 1995). The presence of CYC inhibits the repression of AtGA20ox1 transcription by KN1-GR (Fig. 4, lanes 3 and 7).
Figure 4.
Reverse transcriptase-PCR gel-blot analysis of AtGA20ox1 expression in 35S:KN1-GR seedlings 30 min after DEX and/or cyclohexamide (CYC) induction. Lane 1, CYC application only; lane 2, surfactant application only; lane 3, both DEX and CYC application; lane 4, DEX application only. Lanes 5 to 8, as for lanes1 to 4 with PCR performed using one-tenth dilution of cDNA. Lane 9, PCR performed with no cDNA. IAA1 expression is induced in response to CYC, and GAPDH expression indicates a similar abundance of cDNA in each sample.
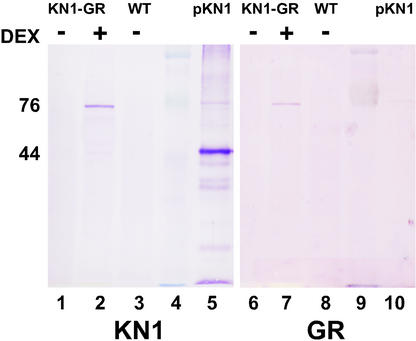
The inability of KN1-GR to repress AtGA20ox1 transcription in the absence of protein synthesis suggests that the regulation of AtGA20ox1 is indirect, or that the KN1-GR fusion protein is rapidly degraded in the absence of steroid binding. The stability of the KN1-GR fusion protein was investigated by western-blot analysis. Protein was extracted from leaves of wild-type plants and 35S:KN1-GR plants that had been treated with either 10−6 m DEX or a control treatment. Forty micrograms of each protein sample was blotted and immunoreacted with antibodies raised against either KN1 or GR (Fig. 5, lanes 1–10). The KN1 antibody recognizes a 44-kD protein in an extract from bacteria that express a recombinant KN1 protein (Fig. 5, lane 5). A 76-kD protein that corresponds to the full-length KN1-GR fusion protein is detected by both anti-KN1 and anti-GR antibodies in the DEX-treated 35S:KN1-GR sample (Fig. 5, lanes 2 and 7). No protein is detected in either wild-type or untreated 35S:KN1-GR leaves (Fig. 5, lanes 1, 3, 6, and 8).
Figure 5.
KN1-GR fusion protein is detected only in the presence of DEX. Western-blot analysis detects a 76-kD KN1-GR fusion protein using antibodies raised to both KN1 and GR in 35S:KN1-GR leaves after DEX application (lanes 2 and 7) but not in the absence of DEX (lanes 1 and 6). No fusion protein is detected in wild-type leaves (lanes 3 and 8). Forty micrograms of total protein of each sample was loaded. A 44-kD recombinant KN1 protein (pKN1) was detected only by the KN1 antibody (lanes 5 and 10). Prestained marker was used to determine protein size (lanes 4 and 9).
The absence of KN1-GR protein in untreated 35S:KN1-GR leaves suggests that the fusion protein is only stable in association with DEX. Perhaps KN1-GR is only stable upon translocation to the nucleus after DEX treatment and is unstable in the cytoplasm. Instability in the cytoplasm, however, is not true of all KN1 fusion proteins. The biologically active fusion between KN1 and green fluorescent protein is stably expressed in both the cytoplasm and nucleus (Kim et al., 2002). The GR steroid-binding domain maintains a transcription factor in an inactive state in the absence of ligand via interactions with a protein complex involving the heat shock protein hsp90 (Picard et al., 1988, 1990; Scherrer et al., 1993). It is possible that KN1-GR is unstable in association with this complex. If KN1-GR is degraded in the cytoplasm, then inhibition of protein synthesis by CYC would limit the amount of fusion protein available for DEX-induced nuclear translocation. It is still possible, therefore, that KN1 directly regulates AtGA20ox1, but in the presence of CYC, too little fusion protein is present to have an effect after DEX induction.
Developmental Competence of Leaf Primordia to Form Lobes in Response to KN1
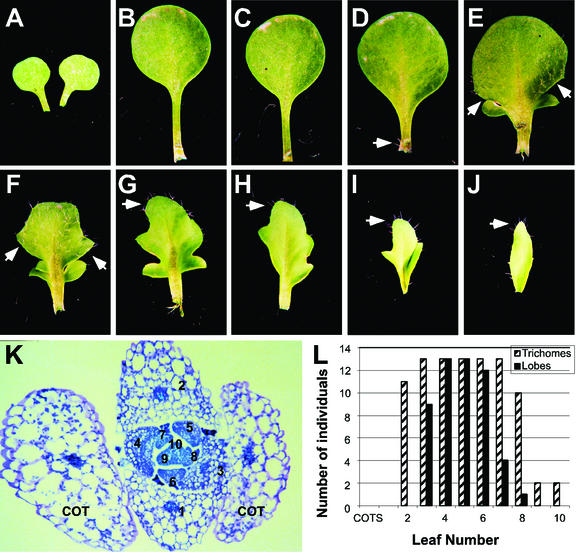
To analyze the competence of primordia to respond to KN1 activity, we marked primordia that had been exposed to DEX using a second DEX-inducible transgene, R-GR. R, a maize basic helix loop helix transcription factor, produces trichomes in a hormone-dependent background when the transgene is expressed in the trichomeless background of ttg (transparent testa glabrous; Lloyd et al., 1995). The range of plastochrons that respond to DEX treatment by producing trichomes in 35S:R-GR plants is wider than that which produces lobes in 35S:KN1-GR plants, indicating that R-GR is a suitable marker to assess the competence of primordia to respond to KN1. Plants were constructed that contained both 35S:KN1-GR and 35S:R-GR transgenes in a ttg background. These plants produced both lobed margins and trichomes in response to DEX induction (Fig. 6, A–J). Hence, each leaf of these plants could be scored for exposure to DEX by trichome production and response to DEX-induced KN1 activity by a lobed morphology.
Figure 6.
Developmental competence of leaf primordia to respond to KN1 activity. Heteroblastic leaf series of a representative 35S:R-GR;35S:KN1-GR;ttg plant after DEX induction. A to C, Cotyledons and leaves 1 and 2; D, leaf 3 with entire margin, arrow indicates trichomes at the petiole base; E to I, leaves 4 to 8 with lobed margins, arrows indicate trichomes; J, leaf 9 with entire margin, arrow indicates trichomes. K, Transverse section through the apex of a 35S:R-GR;35S:KN1-GR;ttg plant at the time of DEX induction, stained with Toluidine Blue O. Mature leaf number is indicated on the corresponding primordia where leaf 10 is at the P0 stage. L, Graphical representation of the number of 35S:R-GR;35S:KN1-GR;ttg individuals scored for the presence of trichomes (striped bar) and lobes (black bar) on each leaf number 1 to 10.
To correlate each mature leaf with its position at the apex at the time of DEX induction, a sample of plants was fixed at the time of induction (n = 10). Transverse sections were stained with Toludine Blue-O and the number of primordia initiated was determined. An average of 10.2 ± 0.3 leaf primordia had been initiated at the time of induction, with leaf 10 corresponding to P0 (Fig. 6K). The remainder of plants was allowed to grow until bolting, and each mature leaf was scored for both trichomes and lobed margins (n = 13). A heteroblastic leaf series for a representative plant is shown in Figure 6. Arrows point to trichomes on leaves 3 to 9 (Fig. 6, D–J), indicating that these leaves were exposed to DEX and competent to produce trichomes. A subset of these leaves (leaves 4–8) produced lobed margins (Fig. 6, E–I), indicating that a window of competence exists for leaf primordia to respond to KN1.
The number of lobes per leaf increases from two lobes in leaf 4 (Fig. 6E) to at least three lobes in leaves 5 and 6 (Fig. 6, F and G). The position of the lobes moves from the base of the lamina in leaf 4 to more distal positions in the lamina in leaves 5 and 6. This may reflect the age of each leaf in terms of the heteroblastic changes in margin shape, or it may reflect a temporal pattern in the competence to respond to KN1, such that the tip of the lamina differentiates and loses competence to respond to KN1 before the base.
Correlating each mature leaf number with its plastochron number at the time of induction reveals the developmental stages of leaf primordia that are competent to respond to KN1 activity. Leaf 3 is the oldest mature leaf that produces a lobed margin in the majority of individuals in this experiment (Fig. 6L). This leaf corresponds to P7 at the time of induction (Fig. 6K). P7 is a late stage of leaf development when vascular pattern has been established and basipetal tissue differentiation has occurred (Telfer and Poethig, 1994). Leaf 8 is the youngest leaf that produced lobes in one individual in this experiment (Fig. 6L) and corresponds to P2 at the time of induction (Fig. 6K). Although P0 and P1 are exposed to DEX, indicated by the presence of trichomes on leaves 9 and 10 of some individuals (Fig. 6L), these incipient primordia are not competent to respond to the presence of KN1.
DISCUSSION
We have constructed and characterized a steroid-inducible system to regulate KN1 misexpression in vivo. We show that lobed leaf morphology is produced in a hormone-dependent manner. The KN1-GR fusion protein appears to be unstable in the absence of hormone induction, suggesting that protein degradation may be one mechanism by which KNOX proteins are regulated. We also show that a window of competence exists during which leaf primordia can respond to KN1 activity by producing a lobed margin.
Hormone-Dependent Production of Lobed Leaf Morphology
Increasing concentrations of DEX produce a dose-dependent increase in the severity of the 35S:KN1-GR phenotype. The number of leaves affected also increases with DEX concentration, perhaps due to higher levels of residual hormone. The increased severity is manifest in the number and depth of lobes, length and width of the petiole, and overall growth of the leaf.
Serrations are produced at the margin in response to low concentrations of DEX and replaced by lobes as the concentration of DEX increases. These results suggest that serrations and lobes are both a consequence of KN1 misexpression and that the difference between them is due to the relative level of KN1 activity. The same range of phenotypes, mild serrations to deep lobes, was also seen in independent 35S:KNAT1 transformants, although no correlation with RNA levels could be detected (Chuck et al., 1996). The leaf lamina remains a constant size despite increases in the circumference of the leaf margin with increasing DEX concentrations. This finding supports the hypothesis that lobes result from a lack of growth in the sinus rather than an increase in growth of the lobe (Chuck et al., 1996).
35S:KN1-GR petioles decrease in length and increase in width in response to increasing DEX concentrations. At high concentrations of DEX, leaves do not develop a petiole; instead, they assume a palmate leaf shape with lobes extending directly from the leaf base. A similar leaf shape is seen in 35S:KN1-GR plants germinated on DEX. Dominant KNOX mutants in maize also show a decrease in length and increase in width of the basal half of the leaf (Foster et al., 1999). Analysis of the changes in vascular pattern produced in these deeply lobed 35S:KN1-GR leaves suggests that the lack of petiole is correlated with precocious radiation of secondary veins out from the midrib. Rather than remaining appressed close to the midvein, secondary veins radiate out toward the lobes almost immediately at the base of the leaf. This change in vascular patterning may indicate that lobes have features of leaves, such that the vascular pattern of the leaf reiterates in each lobe.
A reduction in expansive leaf growth also correlates with increasing concentrations of DEX. The small mature leaves produced in response to high DEX concentrations undergo a period of growth arrest early in development from which they recover and expand to a highly dissected form. This stage of primordia arrest also occurs in 35S:KN1-GR plants germinated on media containing DEX. A reduction in localized growth, therefore, correlates with the level of KN1 activity.
A Window of Competence Exists in Which Primordia Respond to KN1
The steroid-inducible system enables regulation of not only the amount of KN1 misexpression but also the timing of misexpression. The ability to mark leaf primordia that have been exposed to DEX by expressing a second DEX-inducible fusion protein in 35S:KN1-GR plants allowed us to determine which leaf primordia were competent to respond to KN1 activity. Although it is technically possible that different leaf primordia received variable levels of DEX, and that R-GR was able to respond to lower levels of DEX than KN1-GR, we made the assumption that all leaf primordia could respond to DEX, given that the application was 10−6 m and we observed a leaf phenotype even at 10−9 m. By determining the number of leaf primordia that had been initiated at the time of DEX-induction, we could correlate mature leaves with their position from the apex at the time of induction.
These experiments revealed that a window of competence exists during which leaf primordia can respond to KN1. Primordia outside of this window, at both earlier and later plastochron stages, are not competent to respond to KN1 by producing a lobed morphology. The window defined in these transgenic lines is quite broad. Primordia as late as P7 and as early as P2 are competent to respond to KN1 activity. Thus, KN1 has a similar effect when misexpressed over a continuum of stages of leaf development.
Similarities in lobed morphology between 35S:KNAT1 leaves and the recessive mutants as1 and as2 led to the identification of the AS1 and AS2 gene products as regulators of KNAT1, 2, and 6 expression in the Arabidopsis leaf (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Iwakawa et al., 2002). Analysis of the pattern of KNOX expression in as1 and as2 by in situ hybridization demonstrates that KNAT1 is negatively regulated in P0 and P1 leaf primordia, as seen in normal plants, but is misexpressed in later primordia (Ori et al., 2000). This temporal pattern of KNOX misexpression fits within the window of competence defined by KN1-GR induction, suggesting that misexpression of any one KNOX gene within this window can result in lobed morphology. Loss of all three genes, KNAT1, 2, and 6, is, therefore, likely to be required to suppress the lobed leaf phenotype of as1 or as2.
The inability of P0 and P1 leaf primordia to respond to KN1 misexpression is reminiscent of earlier findings. Mutations in both Arabidopsis and maize that result in misexpression of KNOX genes all show correct down-regulation of KNOX proteins in P0, even when KNOX proteins accumulate as early as P1 (Schneeberger et al., 1998; Foster et al., 1999). Further, in a broad study of KNOX expression in various vascular plants, all species showed down-regulation of KNOX protein at P0, but differed in whether KNOX was expressed later in leaf development (Bharathan et al., 2002). This conservation in expression patterns suggests that mechanisms exist to exclude KNOX from the cells of incipient primordia that may be different from mechanisms that negatively regulate KNOX expression in older leaf primordia.
The KN1-GR fusion protein is only stable when associated with DEX in an active state in the nucleus. Therefore, one mechanism to exclude KNOX activity from incipient primordia may be to export KNOX proteins from the nucleus or block their import. KNOX proteins also contain PEST motifs (Vollbrecht et al., 1991; Nagasaki et al., 2001), which target proteins for degradation via proteolytic pathways. The context in which KNOX proteins are expressed is likely to influence whether these PEST domains are exposed or not. Interactions between KNOX proteins and/or other protein partners (Smith et al., 2002) may regulate exposure of protein degradation motifs.
It is also possible that incipient primordia fail to respond to KN1 for reasons unrelated to the stability or subcellular localization of the protein. The environment of these leaf founder cells is different from that of established leaf primordia. Lobed leaf morphology is thought to involve the juxtaposition of novel boundaries in the Arabidopsis leaf (Ori et al., 2000). Gene products required early in leaf development to specify the initiation of dorsiventral polarity could be involved in specifying such boundaries. Members of the YABBY gene family and both KANADI1 (KAN1) and KAN2 promote abaxial cell fate in Arabidopsis leaves, whereas dominant mutations in both PHABULOSA and PHAVOLUTA transform abaxial leaf fates to adaxial (Siegfried et al., 1999; Eshed et al., 2001; Kerstetter et al., 2001; McConnell et al., 2001). The expression patterns of these genes do not resolve into clear abaxial or adaxial domains until primordia become distinct from the SAM at the P2 stage (McConnell et al., 2001). Therefore, the domains of gene expression required to establish boundaries with ectopic KNOX expression may not exist in P0 and P1 primordia. This would suggest that young primordia are not competent to respond to the ectopic KNOX expression.
CONCLUSION
Lobed leaf morphology is produced in 35S:KN1-GR plants in a dose-dependent manner, indicating that the amount of KN1 quantitatively affects the severity of lobing. KN1-GR fusion protein is not detected in leaves in the absence of steroid induction, suggesting that it is only stable in an active state as a transcriptional regulator. P0 and P1 leaf primordia are not developmentally competent to produce lobed morphology in response to KN1. This lack of effect at early leaf stages could be due to the stability or subcellular localization of KN1-GR protein in these young primordia, or to a difference in the expression patterns of genes that are required for developmental competence.
MATERIALS AND METHODS
Plasmid construction: 35S:KN1-GR is a C-terminal fusion of the KN1 cDNA with the steroid-binding domain of GR from pBI-ΔGR (Lloyd et al., 1995). The cloning was performed by amplifying a PCR fragment using a 5′ primer inside the KN1 cDNA with the 3′ primer DJKBS2 (tcacggatccccgagccggtac), which replaces the KN1 stop codon with a BamHI site. This fragment was sequenced to confirm fidelity, then cut with KpnI and BamHI and cloned along with an XbaI/KpnI KN1 cDNA fragment cut from pKOC10 into XbaI/BamHI sites in the pBI-ΔGR plasmid.
Plant Transformation
The Columbia (Col) ecotype was transformed by the floral dip method using the Agrobacterium tumefaciens strain GV3101. A large number of transformants were selected on Murashige and Skoog media containing 50 μg mL−1 kanamycin, and at least 20 independent lines were screened on media containing 10−6 m dexamethazone. Three lines were pursued, which gave consistent inducible phenotypes. 35S:KN1-GR transgenic line 46-10 is used as a heterozygote in the Col background for all analyses presented here.
Plant Growth Conditions
Plants were grown to maturity in a greenhouse, and seedlings were grown on DEX plates in a growth chamber. Both greenhouse and growth chamber were set at long day conditions of 16 h of light and 8 h of dark, day temperature was 20°C, and night temperature was 18°.
Chemical Treatments
DEX (Sigma, St. Louis) was dissolved in water to 1 mm stock and applied by spraying at 10−6 m concentration, unless otherwise specified, with 0.2% (w/v) silwet surfactant. Control plants were treated with the same concentration of surfactant in water. Plants were treated when the first two leaves were visible and analyzed just before bolting unless otherwise specified. 35S:KN1-GR and Col plants were germinated on Murashige and Skoog media containing B5 vitamins and 10−6 m DEX.
Reverse Transcriptase-PCR Gel-Blot Analysis
One microgram of total RNA extracted using RNeasy (Qiagen USA, Valencia, CA) was used for cDNA synthesis with an oligo(dT) primer and SuperscriptII reverse transcriptase (Invitrogen, Carlsbad, CA). PCR primers specific for AtGA20ox1, IAA1, and GAPDH amplified products of 359, 508, and 542 bp, respectively, that were detected by Southern hybridization with gene-specific probes. Twenty-one PCR cycles were performed. Primers are as follows: AtGA20ox1-F, gccgtaagtttcgtaacaacatctcc; AtGA20ox1-R, gagagaggcatatcaaagaagcgg; IAA1-F, atggaagtcaccaatgggc; IAA1-R, tcataaggcagtaggagcttcggatcc; GAPDH-F, cacttgaagggtggtgccaag; and GAPDH-R, cctgttgtcgccaacgaagtc.
Histology
Tissue was fixed in 4% (w/v) paraformaldehyde and paraffin embedded as described (Jackson, 1992). Sections were stained with 0.1% (w/v) Toluidine Blue O. Tissue clearing was carried out as described (Aida et al., 1997) and viewed under dark-field microscopy.
Protein Extraction and Western-Blot Analysis
Total protein was extracted from leaves of five 16-d-old plants using the EZ extraction procedure (Martinez-Garcia et al., 1999), which allows protein quantification in the extract using the DC protein assay (Bio-Rad Laboratories, Hercules, CA). Forty micrograms of each protein sample was loaded in duplicate, separated by SDS-PAGE on a 10% (w/v) acrylamide gel, and transferred to a nitrocellulose membrane using standard techniques. Western-blot analysis was performed on the duplicate membranes using an antibody raised in rabbit to KN1 at 1:500 (w/v) dilution (Smith et al., 2002) and an antibody raised in rabbit to GR (Amersham, Buckinghamshire, UK) at 1:1,000 (w/v) dilution. Goat anti-rabbit alkaline phosphatase-conjugated secondary antibody was used at 1:5,000 (w/v) dilution, and signal was detected using nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate color detection.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Alan Lloyd for the gift of the pBI-ΔGR plasmid and 35S:R-GR seed. We also thank Miltos Tsiantis for helpful advice and discussion.
Footnotes
This work was supported by the National Science Foundation (grant no. 1BN–0131431).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.017434.
LITERATURE CITED
- Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR. Homologies in leaf form inferred from KNOXI gene expression during development. Science. 2002;296:1858–1860. doi: 10.1126/science.1070343. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408:967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development. 2002;129:1957–1965. doi: 10.1242/dev.129.8.1957. [DOI] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S. Knat1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. Establishment of polarity in lateral organs of plants. Curr Biol. 2001;11:1251–1260. doi: 10.1016/s0960-9822(01)00392-x. [DOI] [PubMed] [Google Scholar]
- Foster T, Veit B, Hake S. Gnarley is a dominant mutation in the knox4 homeobox gene affecting cell shape and identity. Plant Cell. 1999;11:1239–1252. doi: 10.1105/tpc.11.7.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois JL, Woodward C, Reddy GV, Sablowski R. Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development. 2002;129:3207–3217. doi: 10.1242/dev.129.13.3207. [DOI] [PubMed] [Google Scholar]
- Groot EP, Meicenheimer RD. Comparison of leaf plastochron index and allometric analyses of tooth development in Arabidopsis thaliana. J Plant Growth Regul. 2000;19:77–89. doi: 10.1007/s003440000008. [DOI] [PubMed] [Google Scholar]
- Hareven D, Gutfinger T, Parnis A, Eshed Y, Lifschitz E. The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell. 1996;84:735–744. doi: 10.1016/s0092-8674(00)81051-x. [DOI] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol. 2002;12:1557–1565. doi: 10.1016/s0960-9822(02)01125-9. [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002;43:467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- Jackson D. In SJ Gurr, MJ McPherson, DJ Bowles, eds, Plant Molecular Pathology: A Practical Approach. I. Oxford University Press; 1992. In situ hybridization in plants; pp. 163–174. [Google Scholar]
- Janssen BJ, Lund L, Sinha N. Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 1998;117:771–786. doi: 10.1104/pp.117.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;411:706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Cilia M, Khalfan-Jagani Z, Jackson D. Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc Natl Acad Sci USA. 2002;99:4103–4108. doi: 10.1073/pnas.052484099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A Knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis R. Epidermal-cell fate determination in Arabidopsis - patterns defined by a steroid-inducible regulator. Science. 1995;266:436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the SHOOTMERISTEMLESS gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Monte E, Quail PH. A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 1999;20:251–257. doi: 10.1046/j.1365-313x.1999.00579.x. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Nagasaki H, Sakamoto T, Sato Y, Matsuoka M. Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15. Plant Cell. 2001;13:2085–2098. doi: 10.1105/TPC.010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T, Dengler N. Leaf vascular pattern formation. Plant Cell. 1997;9:1121–1135. doi: 10.1105/tpc.9.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development. 2000;127:5523–5532. doi: 10.1242/dev.127.24.5523. [DOI] [PubMed] [Google Scholar]
- Pautot V, Dockx J, Hamant O, Kronenberger J, Grandjean O, Jublot D, Traas J. KNAT2: Evidence for a link between knotted-like genes and carpel development. Plant Cell. 2001;13:1719–1734. doi: 10.1105/TPC.010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Khursheed B, Garabedian MJ, Fortin MG, Lindquist S, Yamamoto KR. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- Picard D, Salser SJ, Yamamoto KR. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell. 1988;54:1073–1080. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Kamiya N, Ueguehi-Tanaka M, Iwahori S, Matsuoka M. KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 2001;15:581–590. doi: 10.1101/gad.867901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer LC, Picard D, Massa E, Harmon JM, Simons SS, Jr, Yamamoto KR, Pratt WB. Evidence that the hormone binding domain of steroid receptors confers hormonal control on chimeric proteins by determining their hormone-regulated binding to heat-shock protein 90. Biochemistry. 1993;32:5381–5386. doi: 10.1021/bi00071a013. [DOI] [PubMed] [Google Scholar]
- Schneeberger R, Tsiantis M, Freeling M, Langdale JA. The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development. 1998;125:2857–2865. doi: 10.1242/dev.125.15.2857. [DOI] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. The asymmetric leaves2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001;128:1771–1783. doi: 10.1242/dev.128.10.1771. [DOI] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Pena CG, Springer PS. The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 2002;129:747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- Smith HM, Boschke I, Hake S. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc Natl Acad Sci USA. 2002;99:9579–9584. doi: 10.1073/pnas.092271599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S. A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development. 1992;116:21–30. doi: 10.1242/dev.116.1.21. [DOI] [PubMed] [Google Scholar]
- Telfer A, Poethig RS. Leaf development in Arabidopsis. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1994. pp. 379–401. [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature. 1991;350:241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- Williams RW. Plant homeobox genes: many functions stem from a common motif. BioEssays. 1998;20:280–282. doi: 10.1002/(SICI)1521-1878(199804)20:4<280::AID-BIES2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]