Abstract
Characterization of components 17 and 23 of the inner mitochondrial membrane translocase (TIM17:23) from Arabidopsis indicated that there were three genes present for TIM17 and TIM23 and two for TIM44. AtTIM17 differed from the yeast (Saccharomyces cerevisiae) and mammalian homologs in that two genes encoded proteins that were longer and one gene encoded a shorter protein. All Arabidopsis TIM23 predicted proteins appeared to lack the first 34 amino acids compared with yeast TIM23. All AtTIM17 and AtTIM23 genes were expressed but displayed different tissue and developmental profiles. Complementation of deletion mutants in yeast indicated that for AtTIM17, the extension at the C terminus not present in yeast had to be removed to achieve complementation, whereas for TIM23, a preprotein and amino acid transporter domain had to be present for complementation. Import assays with AtTIM17 and AtTIM23 indicated that they both contained internal signals for integration into the inner mitochondrial membrane in a membrane potential-dependent manner. The C terminus of imported AtTIM17-2 was susceptible to degradation by externally added protease with intact mitochondria. Removal of the 85 C-terminal amino acids resulted in import and full protection of the truncated protein. This suggests that the novel extension at the C terminus of AtTIM17-2 links the outer and inner membrane in a manner analogous to yeast TIM23.
Mitochondria import several hundred nuclear encoded cytosolically synthesized proteins via the combined action of multisubunit protein complexes present in the outer and inner mitochondrial membranes (Bauer et al., 2000; Pfanner and Geissler, 2001). The translocase of the outer mitochondrial membrane (TOM) contains the receptor(s) for recognizing mitochondrial proteins and forms a pore in the outer membrane to pass the imported protein to one of the two translocase complexes of the inner membrane (TIM), which form pores in the inner mitochondrial membrane (Moro et al., 1999; Donzeau et al., 2000; Stan et al., 2000; Truscott et al., 2001; Kovermann et al., 2002; Model et al., 2002). The proposed uniform nomenclature will be used for components of the mitochondrial import apparatus (Pfanner et al., 1996). A number of chaperone proteins in the cytosol, small TIM proteins in the inter membrane space, chaperone proteins, and peptidases in the matrix act with the membrane bound translocases to import the hundreds of different proteins necessary to maintain mitochondrial function (Hartl et al., 1989; Koehler, 2000; Matouschek et al., 2000; George et al., 2002).
The TOM and TIM complexes were first characterized in yeast (Saccharomyces cerevisiae) and subsequently identified to different extents in Neurospora crassa, mammalian, and plant systems. The primary components of the TOM complex, the receptor subunits of 20 and 70 kD, the central organizer subunit of 22 kD, and pore-forming subunit of 40 kD appear well conserved in structural organization between yeast and mammalian systems (Hill et al., 1998; Rapaport et al., 1998; Brix et al., 1999; van Wilpe et al., 1999; Kanaji et al., 2000; Saeki et al., 2000; Stan et al., 2000; Suzuki et al., 2000, 2002; Yano et al., 2000; Model et al., 2002). The plant TOM complex differs from the characterized yeast and mammalian complexes in that it migrates as a smaller complex on Blue-Native-PAGE, 250 kD compared with 400 kD for yeast and mammalian complexes (Pfanner and Geissler, 2001; Werhahn et al., 2001; Suzuki et al., 2002). This plant TOM complex on BN-PAGE contains the TOM20 receptor in contrast to the 400-kD core complex of yeast, which does not contain TOM20 or TOM70 (Pfanner and Geissler, 2001; Werhahn et al., 2001). A noteworthy difference with the plant Tom complex is the absence of a 22-kD subunit. A subunit with a molecular mass of 9 kD, which is structurally similar to TOM22 but lacks the cytosolic receptor domain, is the likely replacement for TOM22 (Mascasev et al., 2000). Furthermore plant TOM20 may be anchored in the outer membrane by the C terminus in contrast to the N terminus evident in yeast and mammalian systems (Werhahn et al., 2001).
Genetic rather than biochemical approaches have been used to characterize the two TIM complexes in yeast (Rehling et al., 2001). The TIM17:23 complex was the first identified and is responsible for the import of proteins that generally contain cleavable targeting amino acid extensions. The TIM22 translocase is responsible for the import of proteins with internal targeting sequences (Rehling et al., 2001). TIM23 and TIM22 form voltage sensitive channels in the inner membrane that are activated by mitochondrial targeting signals (Truscott et al., 2001; Kovermann et al., 2002). TIM17, TIM22, and TIM23 all contain four transmembrane regions with the N and C termini on the intermembrane space side of the mitochondrial inner membrane (Rassow et al., 1999). Structural and genetic characterization of human TIM17:23 led to the conclusion that this translocase is highly conserved in all eukaryotes (Rassow et al., 1999). Gene sequences from a variety of organisms indicate that TIM22 and associated small TIM proteins 8, 9, 10, and 13 are well conserved (Bauer et al., 1999). Only one report exists concerning functional characterization of TIM components in plants (Lister et al., 2002), where it was shown that TIM9 and TIM10 are necessary for import of carrier proteins into the inner membrane using a biochemical reconstitution assay with potato (Solanum tuberosum) mitochondria.
Although the protein import process has been studied in plants for more than 10 years, progress on the identification and role of various import components is still at an early stage. The availability of the Arabidopsis and yeast genome sequences with the detailed characterization of the components of the import apparatus in yeast opens the way for a comparative analysis of the plant import apparatus (Goffeau, 1987; Arabidopsis Genome Initiative, 2000). It is well established that plant mitochondria differ from their yeast and mammalian counterparts in several respects, including biochemistry, genome structure, expression, use of genetic code, coding capacity, and the process of programmed cell death (Douce and Neuberger, 1989; Schuster et al., 1991; Schuster and Brennicke, 1994; Neuburger et al., 1996; Lam et al., 2001). The limited analysis of plant mitochondrial components to date indicates major differences in targeting requirements and specificity (Tanudji et al., 1999; Mascasev et al., 2000) and in the location of processing peptidases (Glaser and Dessi, 1999). Plant mitochondria import tRNA (Marechal-Drouard et al., 1993), and dual targeting of several proteins to mitochondria and plastids takes place (Small et al., 1998; Peeters et al., 2000). Thus homologous components in plants may carry out different or additional functions compared with other organisms.
Biochemical methods can be used to identify and characterize the TOM complex of plant mitochondria because it is the only major complex visible on BN-PAGE analysis (Werhahn and Braun, 2002). This direct approach cannot be used to characterize TIMs due to the presence of several multisubunit respiratory chain complexes in excess to the TIM complexes. We used genome information available from Arabidopsis to characterize homologs of TIM17 and TIM23 because they appeared to differ substantially in nature from their well-characterized yeast counterparts. We characterized the expression, function, import signals, and topology of the Arabidopsis homologs to TIM17 and TIM23 to gain a better understanding of these components compared with other eukaryotes.
RESULTS
Identification of Arabidopsis Homologs of TIM17:23
We searched the Arabidopsis genome for homologs of TIM17:23. Three homologs for TIM17 on chromosomes 1, 2 and 5 were found and called AtTIM17-1, AtTIM17-2, and AtTIM17-3, respectively (At1g20350, At2g37410, and At5g11690). The predicted proteins display 52% similarity and 44%, 47%, and 42% identity, respectively, with ScTIM17. Three homologs for TIM23 were found, two on chromosome 1 (AtTIM23-1 [At1g17530] and AtTIM23-2 [At1g72750]) and one on chromosome 3 (AtTIM23-3 [At3g04800]), predicted proteins displaying 35% similarity and 27%, 26%, and 22% identity, respectively, to ScTIM23. Two homologs to TIM44 (AtTIM44-1 [At2g20510] and AtTIM44-2 [At2g36070]) were detected on chromosome 2, predicted proteins both displayed 23% identity and 34% similarity to ScTIM44. Analysis of the predicted proteins for these homologs indicated that there were significant differences in the structures of the predicted TIM17 and TIM23 proteins (Fig. 1; Supplementary Fig. 1; supplementary data can be viewed at www.plantphysiol.org). In comparison with ScTIM17 with 158 amino acids, the plant homologs AtTIM17-1 and AtTIM17-2 contain a 60 and 85 amino acid extension at the C terminus of the protein, respectively (Supplementary Fig. 1). In contrast, AtTIM17-3 was 25 amino acids shorter at the C terminus (Supplementary Fig. 1). All three were predicted to have four transmembrane regions. However, with AtTIM17-3, the fourth predicted transmembrane region was at the very end of the protein (Supplementary Fig. 1). All three of the predicted TIM23 proteins were very similar, containing 187 or 188 amino acids, 34 and 35 amino acids shorter than ScTIM23. Four transmembrane regions were predicted, similar to the yeast protein. The Arabidopsis predicted TIM44 proteins were 42 and 69 amino acids longer than ScTIM44 at the N terminus of the protein and were not predicted to contain any transmembrane regions. The N-terminal extension of the Arabidopsis proteins are predicted by TargetP to represent a mitochondrial targeting sequence and contain a potential −3R processing signal of RRF*S between amino acids 113 and 114 (Emanuelsson et al., 2000; Zhang et al., 2001). There is no evidence of N-terminal cleavable targeting signals for the TIM17 and TIM23 proteins. None of the Arabidopsis TIM17 or TIM23 genes contains any introns. The two genes encoding TOM9 in Arabidopsis are the only other import components that do not appear to contain introns (Lister et al., 2003).
Figure 1.
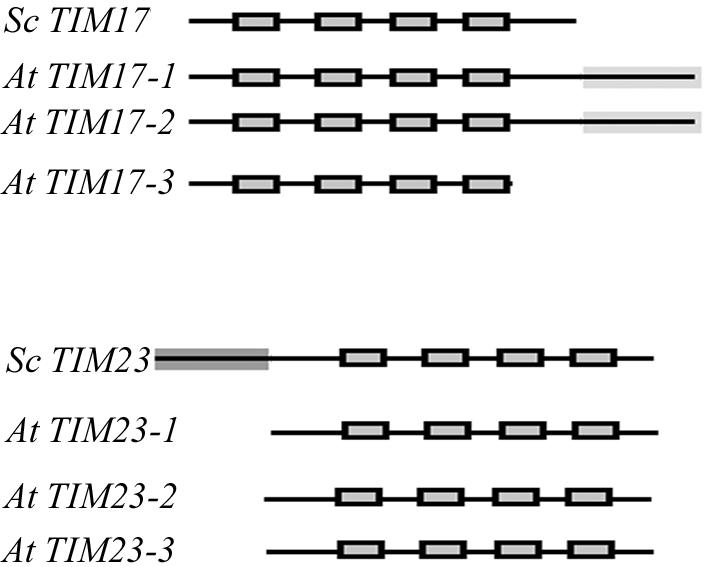
Diagrammatic representation of the TIM17 and TIM23 genes in Arabidopsis. The predicted proteins encoded by Arabidopsis homologs to TIM17 and TIM23 are indicated with the yeast proteins as comparison. Predicted transmembrane regions are indicated by a black box.
Expression of Arabidopsis AtTIM17-1, AtTIM17-2, AtTIM17-3, AtTIM23-1, AtTIM23-2, and AtTIM23-3
We analyzed the expression of all of the genes for AtTIM17 and AtTIM23 and compared them with three AtTIM22, which represents the other TIM (Pfanner and Geissler, 2001). The expression of three nuclear-encoded ribosomal genes were analyzed, with AtRPS15a located in the cytosol, AtRPS1 located in the plastid, and AtRPS13 located in the mitochondrion (Adams et al., 2002; Fig. 2). All TIM17, TIM22, and TIM23 genes were expressed. However changes in expression were evident between tissues and with development (Supplementary Figs. 2 and 3). Notable differences were that AtTIM17-1was expressed at very low levels in roots compared with all other TIM genes analyzed. In fact, it was similar to AtRPS1, which encoded a chloroplast-located ribosomal protein (Supplementary Fig. 2). AtTIM17-2 and AtTIM23-1 and AtTIM23-2 were the highest expressed forms for these genes and peaked at the earliest and latest stages of cotyledon development. The peak at the later stages of cotyledon development was not observed for AtTIM22-2, the highest expressed form for this translocase (Supplementary Fig. 3).
Figure 2.
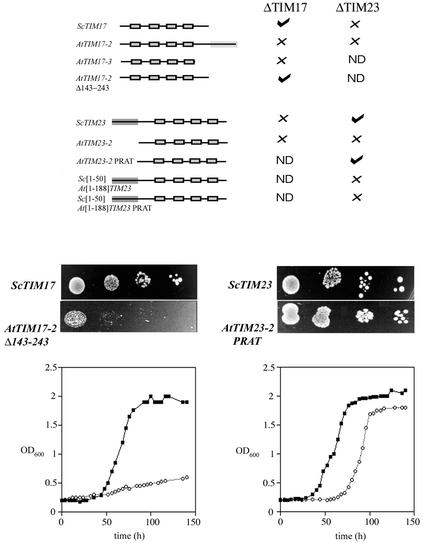
Ability of Arabidopsis TIM17 and TIM23 homologs to complement yeast deletion strains. The ability of AtTIM17 and AtTIM23 and various chimerics and mutants to complement yeast deletion strains was tested by their ability to support growth. A, The constructs tested and ability to complement are indicated. ScTIM17 and ScTIM23 on plasmids were used as positive controls. AtTIM17-2 and AtTIM17-3 failed to complement, but a deletion of AtTIM17-2 with 100 amino acids at the C terminus removed supported growth. AtTIM23-2 failed to complement growth, but insertion of a PRAT domain as is evident in AtTIM23-3 supported growth. The presence of the first 50 amino acids of ScTIM23 did not support growth with AtTIM23-2 or AtTIM23-2(PRAT). B, Growth of the complemented strains at 22°C. The top panels represent the growth of yeast supported by the corresponding gene on a plasmid, 10-fold serial dilutions of the inoculum are shown after growth for 7 d. C, Growth rates for the complemented strains compared with that supported by the yeast genes on a plasmid over a period of 7 d.
Complementation of Yeast Deletion Strains for ScTIM17 and ScTIM23 with Arabidopsis Homologs
Yeast TIM17 and TIM23 are essential proteins for viability, so we tested the ability of the Arabidopsis homologs to complement these genes using a gene replacement strategy (Winzeler et al., 1999; Rehling et al., 2001). This strategy also had the advantage of being less harsh in that the ability of the Arabidopsis homologs to complement under normal growth conditions and was not dependent on the ability of the Arabidopsis homologs to support sporulation. The constructs tested and the ability to complement are shown in Figure 2. AtTIM17-3 and AtTIM17-2 were not able to complement a defect in the corresponding yeast gene. When the region encoding the 85-amino acid extension of AtTIM17-2 was deleted, it supported growth of the deleted ScTIM17 strain. AtTIM23-2 was unable to complement a yeast deletion in ScTIM23. Examination of the predicted AtTIM23 proteins indicated that although AtTIM23-3 has a preprotein and amino acid transporters (PRAT) domain, AtTIM23-1 and AtTIM23-2 lack this domain (Rassow et al., 1999). This domain, G/AX2F/YX10RX3DX6[G/A/S]GX3G, is present in all predicted TIM23 proteins from yeast and mammalian systems (Rassow et al., 1999). In AtTIM23-1 and AtTIM23-2, the Arg is replaced by a Thr. When this Thr codon at position 132 was changed to an Arg codon in AtTIM23-2, complementation of a yeast deletion in TIM23 was observed. Therefore complementation was dependent on the presence of the PRAT domain.
We made a chimeric construct consisting of the region encoding first 50 amino acids of ScTIM23 fused to AtTIM23-2 (y[1-50]At[1-188]TIM23), because it had been previously reported that this portion of ScTIM23 connects the inner and outer membranes and is necessary for efficient protein import and cell growth (Donzeau et al., 2000). This construct failed to complement the deleted ScTIM23 strain, even when it was modified to contain a PRAT domain. Therefore although an Arabidopsis TIM23 gene with a PRAT domain can complement a deletion of ScTIM23, it cannot do this if the region encoding first 50 amino acids of ScTIM23 are placed in front. This suggests that while functionally similar, the plant and yeast proteins are structurally different.
The ΔScTIM17 and ΔScTIM23 deletion strains complemented with AtTIM17-2Δ143-243 and AtTIM23-2(PRAT) displayed better growth at 22°C compared with 30°C. In both cases, this was slower than growth supported by the yeast gene on a plasmid, most dramatically for TIM17.
Import of AtTIM17 and AtTIM23 into Mitochondria
We investigated the import of AtTIM17 and AtTIM23 into isolated soybean (Glycine max) cotyledon mitochondria. Additionally, we used yeast mitochondria and ScTIM17 and ScTIM23 to compare the import and topology of the plant TIMs to this well-characterized system. We then characterized the topology and import characteristics of AtTIM17-2 and AtTIM23-2 in detail because the genes encoding these proteins were highly expressed. To verify the location and intactness of mitochondria used in the in vitro import assays, we carried out western-blot analysis against proteins present in different mitochondrial locations (Fig. 3). The outer membrane protein TOM20 was susceptible to protease digestion in both mitochondria and mitoplasts. The inter membrane space protein cytochrome c was only accessible to protease digestion in mitoplasts, whereas the inner membrane uncoupling protein and matrix-located HSP60 were resistant to protease digestion. These results verify the intactness of mitochondria, the rupture of the outer membrane by osmotic shock, and that the concentration of protease used did not digest known integral membrane proteins.
Figure 3.
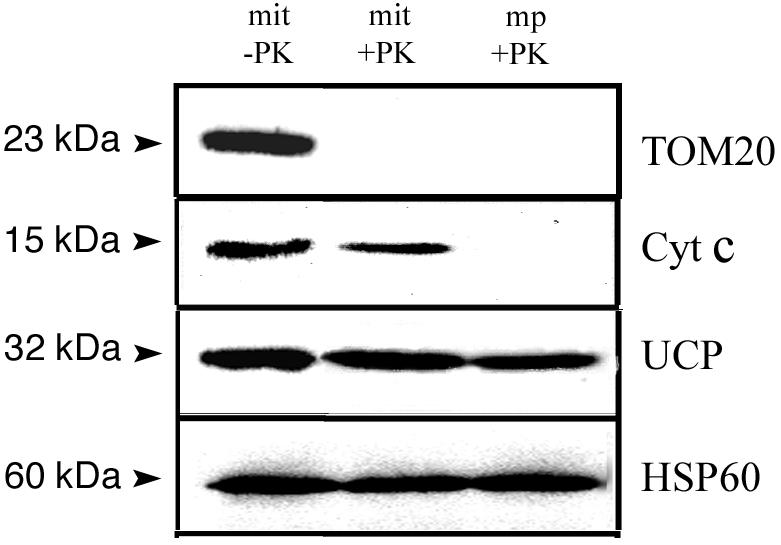
Western-blot analysis of mitochondria, mitochondria treated with PK, and mitoplasts treated with PK. The apparent molecular mass of the protein detected upon probing with various antibodies is indicated in kilodaltons. Cyt c, The inter membrane space protein cytochrome c; UCP, the inner membrane uncoupler protein; HSP60, matrix heat shock protein 60; TOM20, TOM of the outer membrane 20.
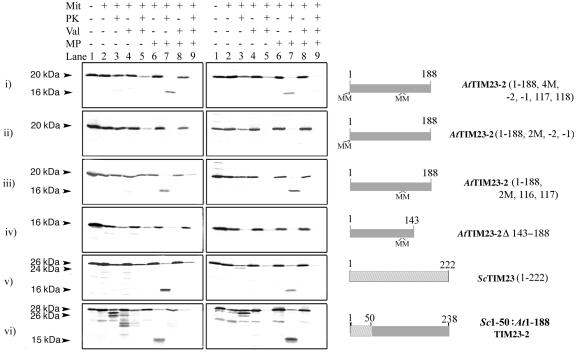
Because ScTIM23 links the inner and outer membrane, we carried out import studies on AtTim23-2 to investigate if it was also accessible to externally added protease with intact mitochondria. AtTIM23-2 contains a single Met at residue 1, which provides a convenient point to investigate the location of the N terminus of this protein. We either placed two additional Met residues in front of the start codon to produce AtTIM23-2 (2M, −2, −1) or changed two Ile residues at positions 116 and 117 to Met residues to produce AtTIM23-2 (2M, 116, 117), and a combination of both, AtTIM23-2 (4M, −2, −1, 116, 117). AtTIM23-2 (4M, −2, −1, 116, 117) was imported into isolated mitochondria and produced a membrane-protected fragment of 16 kD when the outer membrane was ruptured by osmotic swelling to allow access of added protease to the inter membrane space (Fig. 4, i). Import studies with AtTIM23-2 (2M, −2, −1) yielded the same results except that no protected fragment was evident when the outer membrane was ruptured (Fig. 4, ii). This was due to the position of the radiolabeled Met residues at the N terminus of the protein, making them accessible to added protease when the outer membrane was ruptured. However an inner membrane-protected fragment was detected with AtTIM23-2 (2M, 116, 117), because the added Met labels in this case were located in the predicted transmembrane region 2 and thus would be protected from digestion by added protease (Supplementary Fig. 1; Fig. 4, iii). This indicated that under normal import conditions with intact mitochondria, the N terminus of AtTIM23-2 was not accessible to externally added protease, in contrast to what has been observed with ScTIM23 (Donzeau et al., 2000; Fig. 4, v). A chimeric protein between the first 50 amino acids of ScTIM23 and AtTIM23-2 (y[1–50]At[1–188]TIM23) was imported into both soybean and yeast mitochondria and produced an inner membrane-protected fragment upon rupture of the outer membrane. It also generated a small breakdown fragment with protease-treated mitochondria, indicating that the first 50 amino acids of ScTIM23 also appear to have some sequence at the N-terminal exposed on the outer membrane upon import into plants (Fig. 4, vi). However, this smaller breakdown product was not observed in soybean mitochondria when ScTIM23 alone was used, only when the four membrane-spanning segments of TIM23 were from AtTIM23-2 (Fig. 4, soybean v and vi). The import signal for insertion into the inner membrane of AtTIM23-2 is located at the C terminus of the protein as deletion of residues 143 to 188 abolishes import (Fig. 4, iv). In summary, AtTIM23-2 contains an internal targeting sequence between residues 143 and 188 similar to ScTIM23. However, in contrast to the situation in yeast, the N terminus of the plant protein does not appear to be accessible to protease in intact mitochondria.
Figure 4.
Import of AtTIM23-2 into soybean and yeast mitochondria. AtTIM23-2 precursor protein synthesized in a rabbit reticulocyte translation system was used with in vitro import assays with purified soybean or yeast mitochondria. The left set of panels represent import assays into soybean mitochondria, and right panels represent import assays into yeast mitochondria. The apparent molecular mass of the precursor and products generated upon import and/or protease treatment are indicated in kilodaltons. The constructs used to produce the precursor protein are indicated on the right of the figure, inserted Met residues are indicated by M, and positions are indicated relative to the start Met residue of AtTIM23-2. The additions or treatment to each import assay is indicated on the top. MP, Mitochondria ruptured after import assay by osmotic shock; Val, valinomycin added before commencement of import assay; PK, addition of Proteinase K after import assay or mitoplast preparation.
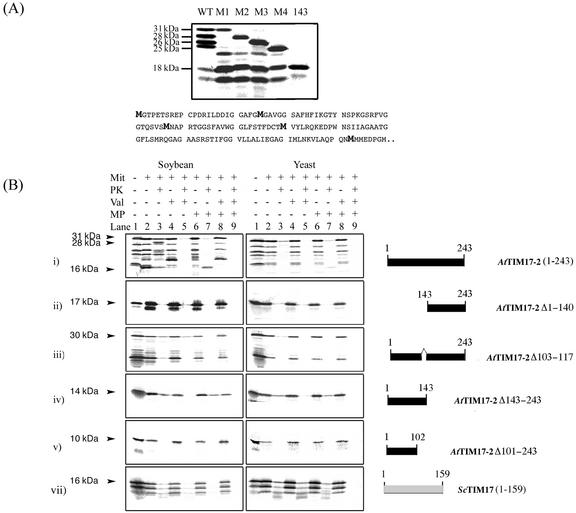
Import studies were carried out with AtTIM17-2 to investigate its topology and to elucidate the signals that define mitochondrial localization. In contrast to AtTIM23-2, AtTIM17-2 contains numerous Met residues and a Met-rich region beginning at residue 143. This causes the production of several products upon in vitro translation. Using site-directed mutagenesis to change Met residues 1, 25, 57, and 90 to Ile residues, it was concluded that the first four bands with apparent molecular masses of 31, 28, 26, and 25 kD arose from precocious translation initiation at each of these sites, respectively (Fig. 5A). Additionally, the major band at 18 kD likely corresponds to translation initiated at the triple Met residues (position 143–146) based on a similar size comparison when this fragment was expressed alone (Fig. 5A).
Figure 5.
Characterization of the translation products of AtTIM17-2 (A) and import of AtTIM17-2 into soybean and yeast mitochondria (B). A, To elucidate the nature of the several products observed with translation of AtTIM17-2, Met residues at various positions were changed to Ile residues. Each Met in the sequence was simply designated by which Met it represented. M1, Met residues at position 1; M2, Met residues at position 25; M3, Met residues at position 57; and M4, Met residue at position 90. Lane WT, Wild-type AtTIM17-2 with the apparent molecular mass of the main translation products indicated. Lane M1, Met residues M2, M3, and M4 have been converted to Ile. Lane M2, Met residues M1, M3, and M4 have been converted to Ile residues. Lane 3, Met residues M1, M2, and M4 have been converted to Ile. Lane M4, Met residues M1, M2, and M3 have been converted to Ile. Lane 143, A gene fragment where the coding sequence for the first 142 amino acids have been deleted so that translation may commence at Met residue 143. B, As in Fig 4 except that AtTIM17-2 was used in import assays.
Translation of AtTIM17-2 produced a precursor protein with an apparent molecular mass of 31 kD, which upon import into mitochondria and protease treatment produced a breakdown fragment of 28 kD in a membrane potential-dependent manner (Fig. 5B, i). Rupture of the outer membrane before protease treatment yielded an inner membrane-protected fragment with an apparent molecular mass of 16 kD. With yeast mitochondria import of AtTIM17-2 was reduced and thus only a faint breakdown fragment was apparent with intact mitochondria, but an inner membrane-protected fragment was produced upon protease treatment of mitoplasts (Fig. 5B, i). The C-terminal extension of AtTIM17-2 apparently does not contain mitochondrial targeting information because it was not imported into either soybean or yeast mitochondria when expressed (Fig. 5B, ii). The first 143 amino acids contained the mitochondrial targeting signal for insertion into the inner membrane between residues 103 and 117 (Fig. 5B, iii–v). AtTIM17-2Δ143-243, which complemented a yeast deletion in ScTIM17, upon translation was imported into both soybean and yeast mitochondria with equal efficiency and protected in mitoplasts (Fig. 5B, iv). This indicates that the first 143 amino acids contained the targeting signal for insertion into the inner membrane between residues 103 and 117 (Fig. 5B, iii–v). There was no change in apparent molecular mass of this product in mitoplasts treated with protease because it was indistinguishable in size from the inner membrane-protected product seen with AtTIM17-2.
In summary, under the import conditions used, AtTIM17-2 is imported into mitochondria under the direction of an internal targeting signal in a membrane potential-dependent manner. A portion of the protein appears to be accessible to externally added protease. A similar pattern of import for TIM17 and TIM23 was observed with purified Arabidopsis mitochondria, however, we routinely used purified soybean cotyledon mitochondria due to the availability of antibodies to verify mitochondrial purity and intactness and to probe intramitochondrial location. All three TIM17 and TIM23 proteins were tested for uptake into mitochondria. All proteins except AtTIM17-3 were imported into mitochondria, but none were imported into chloroplasts (data not shown).
DISCUSSION
The complete genome sequence from Arabidopsis allows the characterization of components using homology-based approaches from well-characterized organisms such as yeast. Using this approach, it was apparent that although the central components of both TIM complexes were present, major differences existed in the predicted proteins for AtTIM17 and AtTIM23. AtTIM17 displays high homology with ScTIM17, especially in the region encompassing the four inner membrane transmembrane regions (Supplementary Fig. 1A). One gene for AtTIM17 present on chromosome 2 (AtTIM17-2) contains an 85-amino acid extension at the C terminus, it is 243 amino acids long compared with the 158 amino acids of ScTIM17 (Fig. 1A). In contrast, AtTIM17-3 is only 133 amino acids long. The C-terminal extension of AtTIM17-2 does not display homology with any other proteins in GenBank but was characterized by a tripeptide repeat of GMQ/P from amino acids 149 to 178. The remainder of the protein of 65 amino acids was enriched with 16 hydroxylated amino acids and 14 charged residues. The only noticeable hit on searching GenBank with this region was that the tripeptide repeat displayed high homology with the M region of the signal recognition particle protein of 54 kD (SRP54). This protein, and in particular this region, is involved in binding mRNA in the signal recognition particle translation arrest cycle and in protein-protein interactions between the chloroplast homolog of SRP54 and chloroplast signal recognition particle protein of 43 kD (Jonas-Straube et al., 2001; Keenan et al., 2001). A similar extension is present on TIM17 homologs from several other plant species, including Medicago spp. (GenBank accession no. Aw559460) and soybean (GenBank accession no. Aw309106). TIM17 from these species display 67% and 60% identity with AtTIM17-2, although the C-terminal extension is less conserved displaying 49% and 47% identity. Structural predictions of the C-terminal extension indicate an α-helix, β-sheet, and α-helix structure, with maximum hydrophobicity in the β-sheet-forming region (Chou and Fasman, 1979; Deleage and Roux, 1987; Cserzo et al., 1997).
In contrast to AtTIM17-2, AtTIM23 predicted proteins are 34 and 35 amino acids shorter than their yeast counterpart. The AtTIM23 proteins do not appear to contain the N-terminal region corresponding to the domain shown to be present in the outer membrane in yeast (Fig. 1B; Supplementary Fig. 1; Donzeau et al., 2000).
To characterize the function of these various isoforms in plants, we investigated the expression, function, and topology of AtTIM17 and AtTIM23 of the plant inner mitochondrial membrane. Expression analysis indicated that all homologs of AtTIM17 and AtTIM23 were transcribed, but all were not expressed in the same pattern or amount. Notably, gene expression for the TIM17:23 translocase did not show the same developmental profile to that of TIM22. This may indicate differential regulation of the general and carrier import pathways.
Functional analysis of the Arabidopsis homologs was carried out by testing their ability to complement yeast deletion mutants. The abundantly expressed genes of AtTIM17 and AtTIM23 could not complement deletions of these genes in yeast. However, a truncated version of AtTIM17-2 and AtTIM23-2 encoding a PRAT domain could complement yeast. The latter did not require the yeast N-terminal region for complementation. Insertion of the coding region for the first 50 amino acids of ScTIM23 in front of the AtTIM23-2 gene prevented complementation.
TIM17 and TIM23 are imported via the carrier import pathway in yeast. This pathway uses the TIM22 translocase on the inner membrane and differs from the general import pathway that uses TIM17:23. Import via the carrier pathway is defined via internal import signals as we have demonstrated here for Arabidopsis TIM17 and TIM23. The carrier import pathway can be conveniently divided into a number of stages, from synthesis in the cytosol (stage I), interaction with receptor on the mitochondrial surface (stage II), partial membrane potential-independent translocation across the outer membrane (stage IIIa), and membrane potential interaction with the small TIMs of the inter membrane space (stage IIIb). Membrane potential-dependent insertion into the inner membrane (stage IV) and assembly (stage V) together result in a functional protein complex (Rehling et al., 2001). However, for different carrier proteins, variations on this theme occur at stage IIIa where membrane potential-independent translocation to a soluble intermediate in the inter-membrane space can be observed. This is seen with the dicarboxylate carrier in yeast (Zara et al., 2001). Consequently, in import assays of AtTIM17-2 and AtTIM23-2 into soybean mitochondria, we deemed import had occurred only when products were protease protected after rupture of the outer membrane and when import was membrane potential dependent. This indicated insertion into the inner membrane. We observed that AtTIM23-2 and AtTIM17-2 did display some protease insensitivity even in the absence of a membrane potential with soybean but not yeast mitochondria (Figs. 4 and 5). Notably, when AtTIM23 was only radiolabeled at the N terminus, this signal was much reduced. Thus, these products protected from protease in the absence of a membrane potential may represent import intermediates. The differences observed in this study are between the sources of mitochondria. The differences in the plant TOM complex, together with the apparent absence of TIM54, TIM18, and TIM12 from plants, may contribute to this difference between organisms (Jansch et al., 1998; Werhahn et al., 2001; Lister et al., 2002).
In vitro import assays suggested that the C terminus of AtTIM17-2 was accessible to protease digestion in intact mitochondria, whereas the remainder of the protein was present in the inner membrane. Kinetic and chase experiments with radiolabeled precursor protein did not result in all of the precursor being converted to this cleaved product (data not shown). Notably this degradation of AtTIM17-2 was only observed in the presence of a membrane potential, indicating that insertion into the inner membrane was a prerequisite for the generation of this product. Removal of 85 amino acids from the C-terminal of AtTIM17-2 resulted in no fragment being generated. Thus, this region appears to be inhibitory to the function of AtTIM17-2 in yeast and appears to have an inhibitory effect on import of AtTIM17-2 in yeast compared with soybean mitochondria. Because Arabidopsis TIM23 isoforms do not appear to be inserted into the outer membrane as yeast TIM23, it is possible that AtTIM17-2 exerts this function in Arabidopsis and links the inner and outer membrane, which has been reported to be necessary for efficient import in yeast (Donzeau et al., 2000). To date, several attempts to raise antibodies to AtTIM17-2 using either antibodies or overexpressed protein to verify the topology of the protein in vivo have been unsuccessful.
The TIM17:23 translocase of plant mitochondria appears to display significant differences to that in yeast and mammalian systems. Structurally, TIM17 appears to be different with a C-terminal extension that is accessible to external protease when the four predicted transmembrane regions are in the inner membrane. In yeast, the outer membrane-exposed domain is on ScTIM23. A chimeric construct that placed the yeast N-terminal 50 amino acids in front of AtTIM23-2 indicated that the plant TIM23 did not appear to function when the N terminus of the protein was located in the outer membrane. A C-terminal-deleted AtTIM17-2, on the other hand could complement a yeast deletion mutant. Overall, this may indicate that anchoring in the outer membrane may occur via different proteins of the translocase in yeast and plants. The role of the C-terminal extension in plants is unclear, but the homologous Met-rich region in SRP54 binds to RNA. Plant mitochondria import tRNA (Dietrich et al., 1996b), in a process that may require the components of the protein import apparatus (Dietrich et al., 1996a,b; Kolesnikova et al., 2000). The C-terminal extension of TIM17, which appears to be present in several plants, may serve the biological function of importing RNA. Because the type and extent of tRNA import differs dramatically between plant species, the C-terminal extension on plant TIM17 may not be highly conserved (Kumar et al., 1996).
MATERIALS AND METHODS
Identification of Arabidopsis Homologs of the Yeast (Saccharomyces cerevisiae) Mitochondrial Protein Import Apparatus
All bioinformatic programs were used with the default settings unless specified otherwise. BioNavigator (http://www.entigen.com) was used to facilitate the analysis, including the use of programs by the Genetics Computer Group (University of Wisconsin, Madison). Sequence information for the yeast import components was obtained from the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/). The yeast gene and protein sequences were used to search GenBank (http://www. ncbi.nlm.nih.gov/) and The Institute for Genomic Research (http://www.tigr.org/) Arabidopsis sequence databases for homologs by BLASTN, BLASTP, and TBLASTN alignment (Altschul et al., 1997). Protein sequences were deduced from nucleic acid sequences using Translate (Genetics Computer Group, University of Wisconsin). Protein alignments were generated using PileUp (Genetics Computer Group, University of Wisconsin). Percentage identity and similarity between yeast and Arabidopsis proteins was calculated using Gap (Genetics Computer Group, University of Wisconsin). Prediction of transmembrane regions was performed using the DAS transmembrane prediction server (Cserzo et al., 1997; http://www.sbc.su.se/approximately miklos/DAS/).
Gene Expression of TIM Components
Cloning of TIM Components
Total RNA was isolated from Arabidopsis tissue using the RNeasy Plant mini protocol (Qiagen, Clifton Hill, Australia). Total RNA was reverse transcribed with the appropriate reverse primer (refer to PCR primers below) using Expand Reverse Transcriptase as per the manufacturer's recommendations (Roche Diagnostics, Sydney). Five microliters of the resulting cDNA was used in a PCR reaction with the appropriate forward and reverse primers (30 pmol each) using the Expand High Fidelity PCR system (Roche Diagnostics) according to the manufacturer's instructions. The amplification profile consisted of 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min, and a final extension cycle of 72°C for 5 min and then 22°C for 6 min. Fragments amplified by PCR were purified by either the QIAquick Gel Extraction Kit or QIAquick PCR purification kit (Qiagen) and cloned into the pCR2.1 vector (Invitrogen, Sydney) according to the manufacturers' instructions.
The following PCR primers were used to clone the genes from plants and yeast: AT TIM17-1 fwd, CCCTAAAAAGTTACTTGTAG; AT TIM17-1 rev, TCAGACCTCAATAATTCCATC; AT TIM17-2 fwd, ATGGGAACACCAGAGACATC; AT TIM17-2 rev, TTACTTGAACTCAAATGATGGCACCG; AT TIM17-3 fwd, ATGGACACTAAGAAGAAATC; AT TIM17-3 rev, TTACTTGCTCCCGAACGGAGGG; AT TIM22-1 fwd, ATGGCTGATTCGAGTGCTGC; AT TIM22-1 rev, CTACTCAGGGATGCTAGTTG; AT TIM22-2 fwd, ATGGCGGCGAACGATTCTTC; AT TIM22-2 rev, TTAGAACTTCCTTGGTTTAGCTAAGG; AT TIM22-3 fwd, ATGGCGGCCGAGAATTCTTC; AT TIM22-3 rev, TCAACGAGCATGAGGAAATTTGAGC; AT TIM23-1 fwd, CGTCTCCCGTCTTCTTCTAATG; AT TIM23-1 rev, GGGCTCTAAAGATCACTCCGG; AT TIM23-2 fwd, ATGGCGGCTAATAACAGATC; AT TIM23-2 rev, TCAAATGGGCACATACCGC; AT TIM23-3 fwd, ATGGCGGATCCGATGAACCATAG; AT TIM23-3 rev, TTAGATTGGAACGAATCGC; Yeast TIM17 fwd, AATAGAGTACACGGGAGCG; Yeast TIM17 rev, CTAAGCTTGCAGAGGTTGAGAG; Yeast TIM23 fwd, ATGTCGTGGCTTTTTGGAGATAAG; and Yeast TIM23 rev, TCATTTTTCAAGTAGTCTTTTC.
Isolation of Total RNA and cDNA Synthesis
Total RNA was isolated three separate times from Arabidopsis tissue using the RNeasy Plant mini protocol (Qiagen). Each batch of total RNA was treated with DNaseI (Roche Diagnostics) to remove contaminating DNA and reverse transcribed using Expand Reverse Transcriptase as per the manufacturer's recommendations (Roche Diagnostics). Primers used in the reverse transcription were random primers (Roche Diagnostics) for the analysis of transcripts in different tissues and poly-A primer (Roche Diagnostics) for the cotyledon transcript analysis. This yielded three separate lots of cDNA. The QIAquick PCR purification kit (Qiagen) was used to purify the cDNA before real-time PCR analysis. Three ribosomal protein genes, one for a cytosolic ribosomal protein RPS15A, one for a nuclear encoded plastid ribosomal protein RPS1, and one for a nuclear encoded mitochondrial ribosomal protein RPS13, were used for comparison against the gene expression profiles of the TIM components (Adams et al., 2002).
Real-Time PCR Primers
The primers used in the real-time PCR reactions for analysis of gene expression are listed below: AT TIM17-1 fwd, CGTTCAAGCTTTGAGAATG; AT TIM17-1 rev, GCTCGTTATGCGCAGTAC; AT TIM17-2 fwd, GTGAGCATGAACGCACCTCG; AT TIM17-2 rev, CAGGCATTCCTTGCATTCCAGG; AT TIM17-3 fwd, CTAAGGAACATGGCCTATACC; AT TIM17-3 rev, CAGAACTCCACCTGTAGC; AT TIM22-1 fwd, GATAGGCATCTCTTGTATGG; AT TIM22-1 rev, CTCTTGGTGTATGGGAAACG; AT TIM22-2 fwd, CACTGACGGTGACGAAGC; AT TIM22-2 rev, CAGACTGCCCTGCATCTG; AT TIM22-3 fwd, ACAAACCAGAAGTCCCCAAC; AT TIM22-3 rev, TCAACGAGCATGAGGAAATTTG; AT TIM23-1 fwd, CGTAGCTCCGATCATGAATCC; AT TIM23-1 rev, GGCTAGATAACCCGTCCCTG; AT TIM23-2 fwd, ATCCGATCATGGGTCAGACG; AT TIM23-2 rev, TGACCAGAAGAGTTCAAGATCC; AT TIM23-3 fwd, ATGGCGGATCCGATGAACC; AT TIM23-3 rev, CCGCAATTGTACCCTTGAAG; AT RPS1 fwd, TATCGCAACTGTTCTTCAGCC; AT RPS1 rev, TCAGAACTCAGCGTCAGTCC; AT RPS15A fwd, CACTGGAGGCAAGCAGAAGC; AT RPS15A rev, AAGTGTCTTAGTACGTACAAGC; AT RPS13 fwd, ATCAGCCAGTCTCTGCTTCG; and AT RPS13 rev, TCAGTTCATCACCATGCTGG.
Real-Time PCR Analysis of Transcript Levels
Transcript levels were assayed using the LightCycler and FastStart DNA Master SYBR Green I kit (Roche Diagnostics). Reactions were carried out in a total volume of 10 μL with a final concentration of 0.008% (w/v) bovine serum albumin under conditions optimized to minimize primer-dimer formation and to maximize amplification efficiency. The LightCycler protocol consisted of four programs: denaturation, 95°C for 10 min; amplification, 40 cycles at 95°C for 15 s, touchdown annealing from 85°C to 45°C decreasing 2°C per cycle for 5 s, 72°C for 15 s with single data acquisition; melting curve analysis, 95°C for 0 s, 70°C for 60 s, 95°C for 0 s with a transition rate of 0.1°C s−1 and continuous data acquisition; cooling, 40°C for 30 s.
DNA was amplified from the cloned genes by PCR using the cloning primers and purified with the QIAquick PCR purification kit (Qiagen) for use as standard template DNA in real-time PCR reactions. The concentration of standard template DNA was quantitated using the PicoGreen dsDNA quantitation kit (Molecular Probes, Eugene, OR) according to the manufacturer's instructions. A standard curve was generated by real-time PCR analysis of 10-fold serial dilutions of the standard template DNA. Transcript levels were assessed using 10−1 dilutions of the cDNA prepared from Arabidopsis tissue of different ages. Absence of primer-dimer and/or nonspecific product accumulation was checked by melting curve analysis and confirmed by agarose gel electrophoresis. Template abundance was quantified using the second derivative maximum method of the LightCycler v3.5 software to determine the cycle at which each PCR reaction reached exponential amplification (Roche Diagnostics). From the three independent cDNA preparations, each transcript was analyzed twice giving a minimum of six replicates for each data point. The se was calculated for every data point. Absolute transcript levels were calculated from the standard curve of known DNA concentration. The data for each gene was normalized by setting the data point with the most abundant transcript to 100 and adjusting the other data points of that gene relative to it.
Complementation of Yeast Deletion Mutants Using Arabidopsis TIM17 and TIM23 Homologs
Diploid deletion strains for ScTIM17 (YJL143w) and ScTIM23 (YNR017w) were obtained from EUROSCARF (http://www.uni-frankfurt.de/fb15/mikro/euroscarf; Winzeler et al., 1999).
BY4743; Mat a/α'3b his3Δ1/his3Δ1′3b leu2Δ0/leu2Δ0; lys2Δ0/LYS2Δ'3b MET15/met15Δ0′3b ura3Δ0/ura3Δ0′3b YJL143w::kanMX4/YJL143w.
BY4743; Mat a/α'3b his3Δ1/his3Δ1′3b leu2Δ0/leu2Δ0; lys2Δ0/LYS2Δ'3b MET15/met15Δ0′3b ura3Δ0/ura3Δ0′3b YNR017w::kanMX4/YNR017w.
The KANMX4 insertion in the appropriate gene was confirmed by PCR. Confirmation of the strains was carried out using the primer sets: Sc-TIM17-fwd, GGGTAACCCGGCATTCTTGCG; Sc-TIM17-rev, CATGGTTGAGAGAAATGGTTGG; Sc-TIM23-fwd, GGTTATTGCATTCGCCCTCC; Sc-TIM23-rev, CCGGGTTTCTACTTTCATTGG; and kanMX4-rev, CAATCATAGATTGTCGCACC.
The ability of Arabidopsis homologs to complement the deletion in the yeast genes was tested by a gene replacement approach. The yeast genes for TIM17 and TIM23 were cloned into Yep352 plasmid under the control of the alcohol dehydrogenease promoter (Hill et al., 1986). The Arabidopsis homologs and various deletions and chimerics to be tested for their ability to complement were cloned into p425 Gal1 (Mumberg et al., 1994). The yeast genes were transformed into the appropriate deletion strains (ScTIM17 into YJL143w and ScTIM23 into YNR017w), and transformants were selected on Western Australia-Glc minus uracil plates. These diploid cells were induced to sporulate, and tetrad dissection was carried out for each strain (Sherman and Hicks, 1991). Tetrads were grown on WA minus uracil and Met or WA minus uracil and Lys to ensure the accuracy of the tetrad dissection. Tetrads that showed a 2:2 segregation for above were grown on WA minus uracil with Geneticin G418 (400 μg mL−1), and resistant colonies were used to transform with p425 Gal1 containing the Arabidopsis gene construct. Transformants were selected on WA minus uracil and Leu plates and replica plates onto WA-Gal minus Leu plus uracil (50 mg L−1), plus 5-fluoro-orotic acid (1 g L−1). The ability to grow on plates containing 5-fluoro-orotic acid was monitored at 22°C and 30°C, and colonies were replica plated at least three subsequent times over a period of 1 month so that the ability to grow was accurately determined. At least 10 individual transformants were monitored for each gene construct.
Colonies that displayed growth were cultured in liquid WA-Gal minus Leu medium and were used to determine the growth rate from an A600 of 0.2 over a period of a week. This growth rate was compared with the parental haploid that was Geneticin resistant so the only difference between the two strains was the plasmid that carried either the ScTIM gene (Yep 352) or the plasmid that carried the Arabidopsis gene (p425 Gal1) that supported growth.
Synthesis of Precursor Proteins
cDNA for AtTIM17-2, AtTIM23-2, ScTIM17, and ScTIM23 in pCR 2.1 (Invitrogen) where subcloned into pGEM-3Zf(+) (Promega, Melbourne). The various constructs and deletions of TIM17 and TIM23 were produced using site-directed mutagenesis (Stratagene, La Jolla, CA) and standard molecular biology techniques. Precursor proteins where translated using the TNT-coupled transcription-translation in reticulocyte lysate in the presence of [35S]Met under either the SP6 or T7 promoter (Promega).
Isolation of Mitochondria
Mitochondria and mitoplasts from 7-d-old soybean (Glycine max) cotyledons and yeast were isolated as outlined previously (Moro et al., 1999; Donzeau et al., 2000; Lister et al., 2002).
In Vitro Import Assays
Import assays were performed as previously described for soybean (Whelan et al., 1996). Import assays into yeast mitochondria varied to contain 2 mm ATP, 2 mm GTP, and 2 mm NADH. Fifty milligrams of proteinase K was added to import assays where indicated to determine whether the precursor protein was imported into mitochondria, Protease treatments of mitochondria and mitoplasts were carried out as previously described (Murcha et al., 1999; Lister et al., 2002). Proteins were separated by 12% or 16% (w/v) SDS-PAGE, and gels were stained, dried, exposed to a BAS TR2040S plate for 24 h, and detected using a BAS 2500 (Fuji, Tokyo).
Preparation of Mitoplasts after Import
Mitoplasts were prepared after import via osmotic swelling. The mitochondria were pelleted after import and resuspended in 10 μL of SEH buffer (250 mm Suc, 1 mm EDTA, 10 mm HEPES, pH 7.4). One hundred and fifty-five microliters of 20 mm HEPES, pH 7.4, was added and incubated on ice for 20 min, and 25 μL of 2 m Suc and 10 μL of 3 m KCl was added and mixed. The sample was aliquoted into two where to one proteinase K was added and incubated on ice for 30 min. Phenylmethylsulfonyl fluoride was added to stop the reaction, and mitoplasts were pelleted.
Immunodetection
Mitochondrial proteins were transferred onto a nitrocellulose membrane (Bio-Rad, Sydney) using a semidry blotting apparatus (Millipore, Sydney) and were incubated with primary antibodies against matrix HSP60 (StressGen, Canada), intermembrane space cytochrome c (BD Biosciences, San Diego), inner membrane uncoupling protein (Considine et al., 2001) and TOM20 (Dr H.-P. Braun, University of Hannover, Germany). Chemiluminescence was used for detection using a LAS 1000 (Fuji).
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. E. Pratje for guidance with the yeast complementation protocols. We thank Harvey Millar and David Day for useful discussions.
Footnotes
This work was supported by the Australian Research Council (grant to J.W.).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.016808.
LITERATURE CITED
- Adams KL, Daley DO, Whelan J, Palmer JD. Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell. 2002;14:931–943. doi: 10.1105/tpc.010483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bauer MF, Hofmann S, Neupert W, Brunner M. Protein translocation into mitochondria: the role of the complexes. Trends Cell Biol. 2000;10:25–31. doi: 10.1016/s0962-8924(99)01684-0. [DOI] [PubMed] [Google Scholar]
- Bauer MF, Rothbauer U, Muhlenbein N, Smith RJ, Gerbitz K, Neupert W, Brunner M, Hofmann S. The mitochondrial TIM22 preprotein translocase is highly conserved throughout the eukaryotic kingdom. FEBS Lett. 1999;464:41–47. doi: 10.1016/s0014-5793(99)01665-8. [DOI] [PubMed] [Google Scholar]
- Brix J, Rudiger S, Bukau B, Schneider-Mergener J, Pfanner N. Distribution of binding sequences for mitochondrial import receptors Tom20, TOM22 and TOM70 in a presequence-carrying preprotein and a non-cleavable preprotein. J Biol Chem. 1999;274:16522–16530. doi: 10.1074/jbc.274.23.16522. [DOI] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. Prediction of beta turns. Biophys J. 1979;26:367–373. doi: 10.1016/S0006-3495(79)85259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Considine MJ, Daley DO, Whelan J. The expression of alternative oxidase and uncoupling protein during fruit ripening in mango. Plant Physiol. 2001;126:1619–1629. doi: 10.1104/pp.126.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- Deleage G, Roux B. An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- Dietrich A, Marechal-Drouard L, Carneiro V, Cosset A, Small I. A single base change prevents import of cytosolic tRNA(Ala) into mitochondria in transgenic plants. Plant J. 1996a;10:913–918. doi: 10.1046/j.1365-313x.1996.10050913.x. [DOI] [PubMed] [Google Scholar]
- Dietrich A, Small I, Cosset A, Weil JH, Marechal-Drouard L. Editing and import: strategies for providing plant mitochondria with a complete set of functional transfer RNAs. Biochimie. 1996b;78:518–529. doi: 10.1016/0300-9084(96)84758-4. [DOI] [PubMed] [Google Scholar]
- Donzeau M, Kaldi K, Adam A, Paschen S, Wanner G, Guiard B, Bauer MF, Neupert W, Brunner M. TIM23 links the inner and outer mitochondrial membranes. Cell. 2000;101:401–412. doi: 10.1016/s0092-8674(00)80850-8. [DOI] [PubMed] [Google Scholar]
- Douce R, Neuberger M. The uniqueness of plant mitochondria. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:371–414. [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminus amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- George R, Walsh P, Beddoe T, Lithgow T. The nascent polypeptide-associated complex (NAC) promotes interaction of ribosomes with the mitochondrial surface in vivo. FEBS Lett. 2002;516:213–216. doi: 10.1016/s0014-5793(02)02528-0. [DOI] [PubMed] [Google Scholar]
- Glaser E, Dessi P. Integration of the mitochondrial-processing peptidase into the cytochrome bc1 complex in plants. J Bioenerg Biomembr. 1999;31:259–274. doi: 10.1023/a:1005475930477. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Aert R, Agostini-Carbone ML, Aigle AM, Alberghina L, Albermann K, Albers M, Aldea M, Alexandraki D, Aljinoni G et al. The Yeast Genome Directory. Nature Suppl. 1997;387:1–105. [Google Scholar]
- Hartl FU, Pfanner N, Nicholson DW, Neupert W. Mitochondrial protein import. Biochim Biophys Acta. 1989;988:1–45. doi: 10.1016/0304-4157(89)90002-6. [DOI] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Jansch L, Kruft V, Schmitz UK, Braun HP. Unique composition of the preprotein translocase of the outer mitochondrial membrane from plants. J Biol Chem. 1998;273:17251–17257. doi: 10.1074/jbc.273.27.17251. [DOI] [PubMed] [Google Scholar]
- Jonas-Straube E, Hutin C, Hoffmann NE, Schunemann D. Functional analysis of the protein-interacting domains of chloroplast SRP43. J Biol Chem. 2001;276:24654–24660. doi: 10.1074/jbc.M100153200. [DOI] [PubMed] [Google Scholar]
- Kanaji S, Iwahashi J, Kida Y, Sakaguchi M, Mihara K. Characterisation of the signal that directs TOM 20 to the mitochondrial outer membrane. J Cell Biol. 2000;151:277–288. doi: 10.1083/jcb.151.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- Koehler CM. Protein translocation pathways of the mitochondrion. FEBS Lett. 2000;476:27–31. doi: 10.1016/s0014-5793(00)01664-1. [DOI] [PubMed] [Google Scholar]
- Kolesnikova OA, Entelis NS, Mireau H, Fox TD, Martin RP, Tarassov IA. Suppression of mutations in mitochondrial DNA by tRNAs imported from the cytoplasm. Science. 2000;289:1931–1933. doi: 10.1126/science.289.5486.1931. [DOI] [PubMed] [Google Scholar]
- Kovermann P, Truscott KN, Guiard B, Rehling P, Sepuri NB, Muller H, Jensen RE, Wagner R, Pfanner N. TIM22, the essential core of the mitochondrial protein insertion complex, forms a voltage-activated and signal-gated channel. Mol Cell. 2002;9:363–373. doi: 10.1016/s1097-2765(02)00446-x. [DOI] [PubMed] [Google Scholar]
- Kumar R, Marechal-Drouard L, Akama K, Small I. Striking differences in mitochondrial tRNA import between different plant species. Mol Gen Genet. 1996;252:404–411. doi: 10.1007/BF02173005. [DOI] [PubMed] [Google Scholar]
- Lam E, Kato N, Lawton M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature. 2001;411:848–853. doi: 10.1038/35081184. [DOI] [PubMed] [Google Scholar]
- Lister R, Mowday B, Whelan J, Millar AH. Zinc-dependent intermembrane space proteins stimulate import of carrier proteins into plant mitochondria. Plant J. 2002;30:555–566. doi: 10.1046/j.1365-313x.2002.01316.x. [DOI] [PubMed] [Google Scholar]
- Lister R, Murcha MW, Whelan J. The Mitochondrial Protein Machinery of Plants database (MPIMP) Nucleic Acids Res. 2003;31:325–327. doi: 10.1093/nar/gkg055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal-Drouard L, Weil JH, Gietrich A. Transfer RNAs and transfer RNA in plants. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:13–32. [Google Scholar]
- Mascasev D, Newbigin E, Whelan J, Lithgow T. How do plant mitochondria avoid importing chloroplast proteins? Components of the import apparatus Tom20 and Tom22 from Arabidopsis differ from their fungal counterparts. Plant Physiol. 2000;123:811–816. doi: 10.1104/pp.123.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouschek A, Pfanner N, Voos W. Protein unfolding by mitochondria: the Hsp70 import motor. EMBO Rep. 2000;1:404–410. doi: 10.1093/embo-reports/kvd093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Model K, Prinz T, Ruiz T, Radermacher M, Krimmer T, Kuhlbrandt W, Pfanner N, Meisinger C. Protein translocase of the outer mitochondrial membrane: role of import receptors in the structural organization of the TOM complex. J Mol Biol. 2002;316:657–666. doi: 10.1006/jmbi.2001.5365. [DOI] [PubMed] [Google Scholar]
- Moro F, Sirrenberg C, Schneider HC, Neupert W, Brunner M. The TIM17.23 preprotein translocase of mitochondria: composition and function in protein transport into the matrix. EMBO J. 1999;18:3667–3675. doi: 10.1093/emboj/18.13.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha MW, Huang T, Whelan J. Import of precursor proteins into mitochondria from soybean tissues during development. FEBS Lett. 1999;464:53–59. doi: 10.1016/s0014-5793(99)01674-9. [DOI] [PubMed] [Google Scholar]
- Neuburger M, Rebeille F, Jourdain A, Nakamura S, Douce R. Mitochondria are a major site for folate and thymidylate synthesis in plants. J Biol Chem. 1996;271:9466–9472. doi: 10.1074/jbc.271.16.9466. [DOI] [PubMed] [Google Scholar]
- Peeters NM, Chapron A, Giritch A, Grandjean O, Lancelin D, Lhomme T, Vivrel A, Small I. Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J Mol Evol. 2000;50:413–423. doi: 10.1007/s002390010044. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Douglas MG, Endo T, Hoogenraad NJ, Jensen RE, Meijer M, Neupert W, Schatz G, Schmitz UK, Shore GC. Uniform nomenclature for the protein transport machinery of the mitochondrial membranes. Trends Biochem Sci. 1996;21:51–52. [PubMed] [Google Scholar]
- Pfanner N, Geissler A. Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol. 2001;2:339–349. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- Rapaport D, Kunkele KP, Dembowski M, Ahting U, Nargang FE, Neupert W, Lill R. Dynamics of the tom complex of mitochondria during binding and translocation of preproteins. Mol Cell Biol. 1998;18:5256–5262. doi: 10.1128/mcb.18.9.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J, Dekker PJ, van Wilpe S, Meijer M, Soll J. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J Mol Biol. 1999;286:105–120. doi: 10.1006/jmbi.1998.2455. [DOI] [PubMed] [Google Scholar]
- Rehling P, Wiedemann N, Pfanner N, Truscott KN. The mitochondrial import machinery for preproteins. Crit Rev Biochem Mol Biol. 2001;36:291–336. doi: 10.1080/20014091074200. [DOI] [PubMed] [Google Scholar]
- Saeki K, Suzuki H, Tsuneoka M, Maeda M, Iwamoto R, Hasuwa H, Shida S, Takahashi T, Sakaguchi M, Endo T et al. Identification of mammalian Tom22 as a subunit of the preprotein translocase of the mitochondrial outer membrane. J Biol Chem. 2000;272:31996–32002. doi: 10.1074/jbc.M004794200. [DOI] [PubMed] [Google Scholar]
- Schuster W, Brennicke A. The plant mitochondrial genome: physical structure, information content, RNA editing and gene migration to the nucleus. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:61–78. [Google Scholar]
- Schuster W, Wissinger B, Hiesel R, Unseld M, Gerold E, Knoop V, Marchfelder A, Binder S, Schobel W, Scheike R et al. Between DNA and protein: RNA editing in plant mitochondria. Physiol Plant. 1991;81:437–445. [Google Scholar]
- Sherman F, Hicks J. Micromanipulation and dissection of asci. Methods Enzymol. 1991;194:21–37. doi: 10.1016/0076-6879(91)94005-w. [DOI] [PubMed] [Google Scholar]
- Small I, Wintz H, Akashi K, Mireau H. Two birds with one stone: genes that encode products targeted to two or more compartments. Plant Mol Biol. 1998;38:265–277. [PubMed] [Google Scholar]
- Stan T, Ahting U, Dembowski M, Kunkele KP, Nussberger S, Neupert W, Rapaport D. Recognition of preproteins by the isolated TOM complex of mitochondria. EMBO J. 2000;19:4895–4902. doi: 10.1093/emboj/19.18.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Maeda M, Mihara K. Characterization of rat TOM70 as a receptor of the preprotein translocase of the mitochondrial outer membrane. J Cell Sci. 2002;115:1895–1905. doi: 10.1242/jcs.115.9.1895. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Okazawa Y, Komiya T, Saeki K, Mekada E, Kitada S, Ito A, Mihara K. Characterisation of rat TOM40, a central component of the preprotein translocase of the mitochondrial outer membrane. J Biol Chem. 2000;275:37930–37936. doi: 10.1074/jbc.M006558200. [DOI] [PubMed] [Google Scholar]
- Tanudji M, Sjoling S, Glaser E, Whelan J. Signals required for the import and processing of the alternative oxidase into mitochondria. J Biol Chem. 1999;274:1286–1293. doi: 10.1074/jbc.274.3.1286. [DOI] [PubMed] [Google Scholar]
- Truscott KN, Kovermann P, Geissler A, Merlin A, Meijer M, Driessen AJ, Rassow J, Pfanner N, Wagner R. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by TIM23. Nat Struct Biol. 2001;8:1074–1082. doi: 10.1038/nsb726. [DOI] [PubMed] [Google Scholar]
- van Wilpe S, Ryan MT, Hill K, Maarse AC, Meisinger C, Brix J, Dekker PJT, Moczko M, Wagner R, Meijer M et al. Tom22 is a multifunctional organiser of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- Werhahn W, Braun H-P. Biochemical dissection of the mitochondrial proteome from Arabidopsis thaliana by three-dimensional gel electrophoresis. Electrophoresis. 2002;23:640–646. doi: 10.1002/1522-2683(200202)23:4<640::AID-ELPS640>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Werhahn W, Niemeyer A, Jansch L, Kruft V, Schmitz UK, Braun H-P. Purification and characterisation of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis: identification of multiple forms of Tom 20. Plant Physiol. 2001;125:943–954. doi: 10.1104/pp.125.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan J, Tanudji MR, Smith MK, Day DA. Evidence for a link between translocation and processing during protein import into soybean mitochondria. Biochim Biophys Acta. 1996;1312:48–54. doi: 10.1016/0167-4889(96)00014-6. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Yano M, Hoogenraad N, Terada K, Mori M. Identification and functional analysis of human tom22 for protein import into mitochondria. Mol Cell Biol. 2000;20:7205–7213. doi: 10.1128/mcb.20.19.7205-7213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zara V, Palmisano I, Rassow J, Palmieri F. Biogenesis of the dicarboxylate carrier (DIC): translocation across the mitochondrial outer membrane and subsequent release from the TOM channel are membrane potential-independent. J Mol Biol. 2001;310:965–971. doi: 10.1006/jmbi.2001.4833. [DOI] [PubMed] [Google Scholar]
- Zhang XP, Sjoling S, Tanudji M, Somogyi L, Andreu D, Eriksson LE, Graslund A, Whelan J, Glaser E. Mutagenesis and computer modelling approach to study determinants for recognition of signal peptides by the mitochondrial processing peptidase. Plant J. 2001;27:427–438. doi: 10.1046/j.1365-313x.2001.01108.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.