Abstract
Methionine (Met) S-methyltransferase (MMT) catalyzes the synthesis of S-methyl-Met (SMM) from Met and S-adenosyl-Met (Ado-Met). SMM can be reconverted to Met by donating a methyl group to homocysteine (homo-Cys), and concurrent operation of this reaction and that mediated by MMT sets up the SMM cycle. SMM has been hypothesized to be essential as a methyl donor or as a transport form of sulfur, and the SMM cycle has been hypothesized to guard against depletion of the free Met pool by excess Ado-Met synthesis or to regulate Ado-Met level and hence the Ado-Met to S-adenosylhomo-Cys ratio (the methylation ratio). To test these hypotheses, we isolated insertional mmt mutants of Arabidopsis and maize (Zea mays). Both mutants lacked the capacity to produce SMM and thus had no SMM cycle. They nevertheless grew and reproduced normally, and the seeds of the Arabidopsis mutant had normal sulfur contents. These findings rule out an indispensable role for SMM as a methyl donor or in sulfur transport. The Arabidopsis mutant had significantly higher Ado-Met and lower S-adenosylhomo-Cys levels than the wild type and consequently had a higher methylation ratio (13.8 versus 9.5). Free Met and thiol pools were unaltered in this mutant, although there were moderate decreases (of 30%–60%) in free serine, threonine, proline, and other amino acids. These data indicate that the SMM cycle contributes to regulation of Ado-Met levels rather than preventing depletion of free Met.
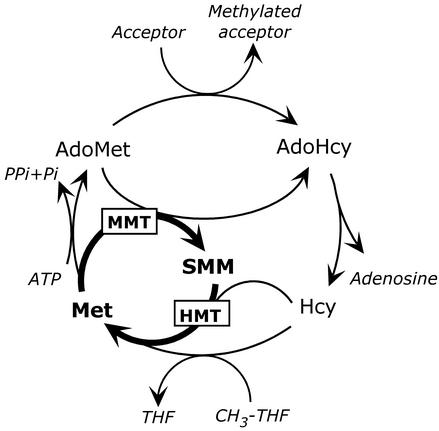
S-Methyl-Met (SMM) synthesis is a unique feature of plant sulfur and one-carbon metabolism (Pokorny et al., 1970; Mudd and Datko, 1990; Ranocha et al., 2001). SMM is formed by the S-adenosyl-Met (Ado-Met)-dependent methylation of Met, catalyzed by Met S-methyltransferase (MMT; Bourgis et al., 1999). SMM can be reconverted to Met by transferring a methyl group to homo-Cys in a reaction mediated by homo-Cys S-methyltransferase (HMT; Ranocha et al., 2000). The tandem action of MMT and HMT, together with that of Ado-Met synthetase and S-adenoyslhomo-Cys (AdoHcy) hydrolase, sets up a futile cycle (the SMM cycle) in which Met is converted to SMM, and SMM is reconverted to Met (Mudd and Datko, 1990). This cycle in effect short-circuits the activated methyl cycle (Fig. 1), and each of its turns hydrolyzes a molecule of ATP to adenosine, pyrophosphate, and phosphate. The SMM cycle operates throughout the plant, and consumes one-half the Ado-Met produced in Arabidopsis leaves (Ranocha et al., 2001).
Figure 1.
The SMM cycle and its relationship to the activated methyl cycle. The reactions catalyzed by MMT and HMT are bolded. CH3-THF, 5-Methyltetrahydrofolate; THF, tetrahydrofolate.
The functions of SMM and its seemingly wasteful cycle are for the most part unknown. The only established role of SMM is in transporting reduced sulfur in the phloem, for which there is qualitative evidence in a range of plants including Arabidopsis (Bourgis et al., 1999). The importance of SMM relative to other translocated forms of sulfur has been quantified only in wheat (Triticum aestivum), where it accounts for one-half the sulfur moving to developing grains (Bourgis et al., 1999). However, the contribution of SMM to sulfur transport may be less in other species (Bourgis et al., 1999) and may depend on developmental stage and sulfur nutrition (Fitzgerald et al., 2001). A hypothetical role for SMM is as methyl donor for a plant-specific reaction (Giovanelli et al., 1980). This role has not been tested but is attractive because it would obviously explain why plants alone have SMM.
Roles in transport or methylation might explain why plants produce SMM, but not why there is futile cycling of SMM throughout the plant. Two hypotheses have been advanced to justify this cycling. The first is that the SMM cycle prevents overshoots in Ado-Met synthesis from depleting the free Met pool required for protein synthesis (Mudd and Datko, 1990) by providing a way to convert Met moieties locked up in Ado-Met back to free Met. The second hypothesis is that the SMM cycle is a means whereby plants control Ado-Met level in the absence of the feedback loops between Ado-Met and the enzymes involved in its synthesis that occur in other eukaryotes (Ranocha et al., 2001; Roje et al., 2002). Controlling the levels of Ado-Met and AdoHcy is considered crucial to the many methyl transfer reactions that take place in cells: AdoHcy is a potent competitive inhibitor of methyltransferases (Cantoni et al., 1979) so that the Ado-Met:AdoHcy ratio (the methylation ratio) determines the activity of these enzymes (Cantoni, 1977). Computer modeling of the SMM cycle in Arabidopsis leaves, based on data for wild-type plants, favored the hypothesis that the SMM cycle contributes to the control of Ado-Met level. Thus, when the SMM cycle was eliminated in silico, the Ado-Met level increased by up to 160%, but steady-state free Met levels did not change. Moreover, the free Met pool recovered from a simulated overshoot in Ado-Met synthesis almost as fast in the absence of the SMM cycle as in its presence (Ranocha et al., 2001).
In the present study, we investigated the function of SMM and its cycle by isolating and characterizing insertional knockout mutants of MMT in Arabidopsis and maize (Zea mays), which both have single MMT genes (Bourgis et al., 1999). We found that SMM is dispensable but that eliminating it caused an increase in Ado-Met level and in the methylation ratio.
RESULTS
Isolation of Arabidopsis and Maize mmt Insertional Mutants
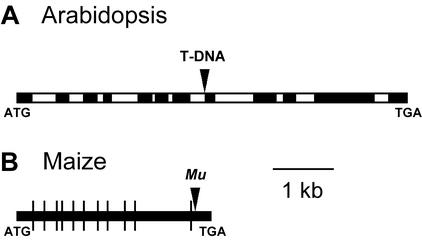
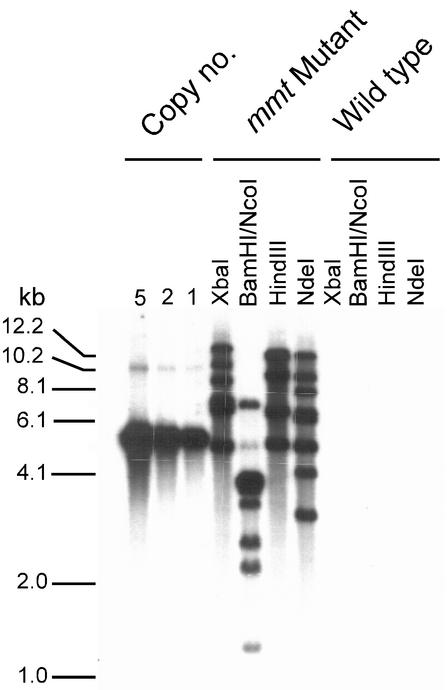
An Arabidopsis mmt mutant was isolated by PCR screening a T-DNA insertion population (Krysan et al., 1999), and a homozygous line was selected for study. Sequencing of genomic DNA showed that the insertion was located close to the 3′ end of intron 7 (Fig. 2A). An insertion at this point is expected to stop formation of a competent enzyme, given the large size of the T-DNA (Campisi et al., 1999). Southern-blot analysis of plants homozygous for the T-DNA-tagged mmt allele and their wild-type siblings demonstrated that only the former harbored inserts, indicating that the T-DNA is inserted only at the MMT locus (Fig. 3). All further experiments with Arabidopsis were carried out with these two sibling populations or their self-pollinated progeny.
Figure 2.
Insertion sites in MMT sequences. A, The Arabidopsis MMT locus showing the T-DNA insertion in intron 7; black boxes represent exons and white boxes introns. B, The maize MMT cDNA showing the site of the Mu insertion near the end of the coding sequence. Intron positions predicted from alignments to Arabidopsis and rice (Oryza sativa) genomic sequences are shown by vertical lines.
Figure 3.
Southern-blot analysis of T-DNA-containing sequences in mmt mutant and wild-type Arabidopsis plants. Genomic DNA was extracted from plants shown by PCR to be homozygous for the mutant mmt allele or the wild-type MMT allele. The DNA was digested with the enzymes indicated. Blots were probed with the whole T-DNA region from the pD991 vector, containing the GUS and kanamycin resistance genes (Campisi et al., 1999). Genomic reconstructions were made with linearized pD991 DNA equivalent to one, two, and five copies per haploid genome; the results are consistent with insertion of a small number of copies of the T-DNA at a single locus in the mmt mutant. The positions of DNA size markers are indicated on the left.
An analogous strategy was used to screen a population of maize plants mutagenized by Robertson's Mutator (Mu) element (Bensen et al., 1995; Meeley and Briggs, 1995). This procedure identified mutants harboring a Mu element inserted after the codon specifying amino acid 989 of the 1,091-residue protein (Fig. 2B). An MMT protein truncated at this point is unlikely to be functional because it would lack a putative ligand-binding region (Bourgis et al., 1999). Because plants from the Mu-mutagenized population that we screened typically harbor many Mu elements and have low vigor, mutants were outcrossed once or twice to vigorous genotypes. Progeny heterozygous for the mutant mmt allele were identified by PCR and selfed; the progeny were screened by PCR to identify homozygous mutant and wild-type individuals, and these were used for experiments.
SMM Levels and MMT Activities
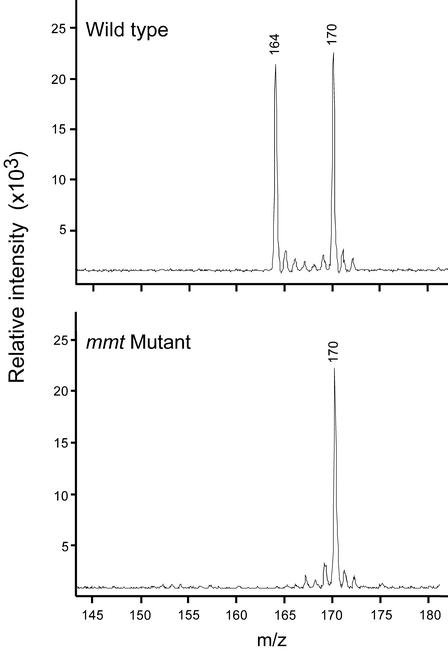
As a first test of the metabolic phenotype of the mmt mutants, leaf SMM levels were determined by matrix-assisted laser desorption/ionization (MALDI)-MS analysis of a cationic fraction from which inorganic salts had been removed. An internal standard of [methyl-2H6]SMM was used for quantification. This procedure gave spectra with strong signals for endogenous SMM and the standard, and no other significant peaks (Fig. 4). In both Arabidopsis and maize, the level of SMM in mmt mutant homozygotes was below the detection limit, which was 1% to 2% of the level in the corresponding wild type (Table I). To corroborate this result, MMT activities were measured in vivo by supplying leaves with tracer doses of [35S]Met and measuring 35S incorporation into SMM (Table II). No activity was detected in Arabidopsis mutants, and very little activity was detected in maize (Table II). The trace of [35S]SMM synthesis seen in maize mutants (0.8% of the wild type) could be due to vestigial activity in the truncated MMT protein resulting from Mu element insertion (Fig. 2B) or to a secondary activity of another methyltransferase (Katz and Gerhardt, 1990). In any case, the data of Table II confirm that our insertional mutants of Arabidopsis and maize are in effect MMT knockouts. Table II also shows that [35S]Met incorporation into protein was normal in mmt mutants, which suggests that the endogenous metabolic pool of free Met is unaltered. A larger free Met pool would reduce labeled protein synthesis via isotope dilution, and a smaller pool would increase it.
Figure 4.
MALDI-MS analysis of the cation fractions from leaves of wild-type and mmt mutant maize. The peaks at m/z 164 and 170 correspond to endogenous SMM and the [methyl-2H6]SMM internal standard, respectively. The spectra shown are for representative samples.
Table I.
SMM levels in leaves of wild-type and mmt mutant Arabidopsis and maize
| Arabidopsis
|
Maize
|
||
|---|---|---|---|
| Wild Type | mmt Mutant | Wild Type | mmt Mutant |
| nmol g−1 fresh wt | |||
| 41 ± 5 | <1 | 105 ± 27 | <1 |
SMM was determined by MALDI-MS in samples (approximately 0.5 g fresh weight) comprising pooled rosette leaves from five Arabidopsis plants harvested just before bolting or leaves from individual maize plants at the 5- to 6-leaf stage. Data are means of three (Arabidopsis) or four (maize) replicates ± se.
Table II.
In vivo conversion of tracer amounts of [35S]Met to [35S]SMM by leaf tissues of wild-type and mmt mutant Arabidopsis and maize
| Genotype | [35S]SMM | 35S-Labeled Protein | [35S]Met Remaining |
|---|---|---|---|
| nCi | |||
| Arabidopsis wild type | 50 ± 8 | 132 ± 13 | 142 ± 4 |
| Arabidopsis mmt mutant | <0.4a | 112 ± 17 | 154 ± 7 |
| Maize wild type | 84 ± 26 | 147 ± 27 | 340 ± 26 |
| Maize mmt mutant | 0.8 ± 0.2b | 150 ± 10 | 437 ± 15 |
Sets of three Arabidopsis leaves or single 25-mm maize leaf segments were supplied with 1.0 μCi (120–130 pmol) of [35S]Met for 1.5 or 2 h in the light and then washed in 0.1 mm Met for 0.5 h to remove unabsorbed label. This [35S]Met dose is very small compared with endogenous free Met content (Ranocha et al., 2001). At least 80% of the [35S]Met was absorbed. Data are expressed as nCi per three leaves (Arabidopsis) or per segment (maize) and are means of three replicates ± se.
The detection limit was 0.4 nCi.
The identity of the trace of [35S]SMM in mmt mutant maize was confirmed by comigration with authentic SMM in three thin-layer separatory systems and by decomposition in hot base.
Growth Characteristics and Seed Sulfur Contents
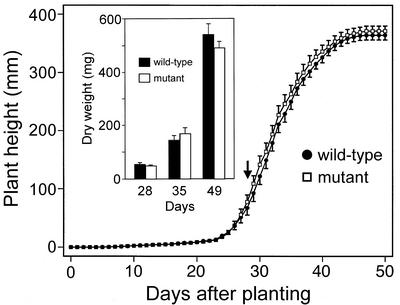
Despite the loss of SMM, neither Arabidopsis nor maize mutants appeared to differ from wild-type plants in morphology or fertility. This was verified for Arabidopsis by measuring the growth rates, flowering dates, and seed yields of mutant and wild-type homozygotes. Throughout growth, there were no significant differences in height, in dry weight (Fig. 5), or in fresh weight (not shown). The flowering dates of mutant and wild type coincided (Fig. 5). Nor did the total weight of seeds per plant or seed size differ significantly: The wild type and mutant produced 144 ± 15 and 138 ± 15 mg seeds plant−1 (mean ± se, n = 20), with average seed weights of 16.2 and 17.1 μg, respectively.
Figure 5.
Growth of wild-type and mmt mutant Arabidopsis plants. Heights were measured daily on 10 individuals in each of two experiments; dry weights (inset) were measured weekly for 10 individuals. Data are means ± se. The flowering date (arrow) did not differ significantly between wild-type and mutant plants (28.9 ± 0.4 and 28.5 ± 0.4 d after planting, respectively).
In view of the potential role of SMM in long-distance sulfur transport (Bourgis et al., 1999), we measured the sulfur contents of wild-type and mutant Arabidopsis seeds. The values obtained were not significantly different: 8.08 ± 0.17 mg g−1 for the wild type and 8.02 ± 0.12 mg g−1 for the mutant (means ±se for six replicates). This suggests that loss of SMM reduced neither protein nor glucosinolate accumulation in seeds, the sulfur in Arabidopsis seeds being about equally divided between these categories (Haughn et al., 1991). Consistent with there being no difference in seed sulfur reserves, wild-type and mutant seed germination rates and seedling vigor were the same.
Ado-Met and AdoHcy Contents
Because SMM synthesis is a major fate of Ado-Met in mature Arabidopsis leaves (Ranocha et al., 2001), we measured levels of Ado-Met and AdoHcy in wild-type and mutant leaves by HPLC-fluorescence analysis of isoindole derivatives (Capdevila and Wagner, 1998). Ado-Met level was significantly higher in the mutant, and the AdoHcy level was significantly lower, so that the methylation ratio increased from 9.5 to 13.8 (Table III). The Ado-Met and AdoHcy levels that we observed in wild-type leaves are comparable with those recently reported for Arabidopsis by Moffatt et al. (2002).
Table III.
AdoMet and AdoHcy levels in leaves of wild-type and mmt mutant Arabidopsis
| Genotype | AdoMet | AdoHcy | Methylation Ratio (AdoMet:AdoHcy) |
|---|---|---|---|
| nmol g−1 fresh wt | |||
| Wild type | 13.0 ± 0.4 | 1.37 ± 0.06 | 9.5 ± 0.4 |
| mmt Mutant | 14.9 ± 0.8* | 1.09 ± 0.06** | 13.8 ± 0.5*** |
AdoMet and AdoHcy were determined as their isoindole derivatives by fluorescence-HPLC using samples (0.15–0.2 g fresh wt) drawn from pooled rosette leaves harvested from five plants just before bolting. Data are means of four replicates ± se and have been corrected for recovery. Differences between wild type and mutant that are significant at P = 0.05, 0.01, or 0.001 are indicated by one, two, or three asterisks, respectively.
Amino Acid and Thiol Contents
The free amino acid contents of Arabidopsis rosette leaves were determined by GC and GC-MS to assess the impact of eliminating SMM on amino acid metabolism as a whole. The contents of all 12 amino acids that were detected were either unaltered or modestly reduced, so that the total amino acid content of mutant leaves was 38% lower than wild type (Table IV). Two amino acids, Met and Thr, were of special interest because (a) SMM is synthesized from Met, and (b) Thr synthesis competes with Met (and SMM) synthesis for a common precursor and may be regulated by Ado-Met (Galili and Höfgen, 2002). There was no change in Met content, in agreement with the prediction based on radiolabeling data (see above). The Thr content was 50% lower in the mutant, but this was not a specific effect because other amino acids showed similar decreases (Table IV).
Table IV.
Free amino acid levels in leaves of wild-type and mmt mutant Arabidopsis plants
| Amino Acid | Amino Acid Level
|
|
|---|---|---|
| Wild Type | mmt Mutant | |
| nmol g−1 fresh wt | ||
| Met | 28 ± 1 | 31 ± 5 |
| Thr | 361 ± 17 | 179 ± 21 |
| Ala | 518 ± 46 | 522 ± 178 |
| Gly | 84 ± 16 | 61 ± 14 |
| Val | 71 ± 3 | 49 ± 6 |
| Ser | 974 ± 89 | 415 ± 20 |
| Leu | 27 ± 1 | 23 ± 4 |
| Ile | 26 ± 1 | 21 ± 3 |
| Pro | 236 ± 66 | 124 ± 14 |
| Asxa | 709 ± 14 | 463 ± 36 |
| Phe | 21 ± 1 | 14 ± 2 |
| Glxb | 1,680 ± 36 | 1,021 ± 87 |
| Total | 4,735 ± 290 | 2,923 ± 389 |
Amino acids were determined using pooled rosette leaves (0.4–0.5 g) from five plants harvested just before bolting. Data are means of three replicates ± se.
Asp + Asn.
Glu + Gln.
Perturbations of sulfur metabolism were sought by determining Cys and glutathione levels in Arabidopsis leaves and roots. The leaves and roots of mutant plants did not differ in their levels of Cys or glutathione from the corresponding wild-type organs when sulfate supply was adequate (1.5 mm) or when the sulfate concentration in the medium was reduced to 30 μm (Table V).
Table V.
Thiol levels in leaves and roots of wild-type and mmt mutant Arabidopsis plants
| Sulfate Level | Thiol | Leaves
|
Roots
|
||
|---|---|---|---|---|---|
| Wild Type | mmt Mutant | Wild Type | mmt Mutant | ||
| nmol g−1 fresh wt | |||||
| 1.5 mm | Cys | 34 ± 6 | 33 ± 3 | 41 ± 4 | 39 ± 4 |
| Glutathione | 492 ± 52 | 483 ± 46 | 179 ± 34 | 180 ± 19 | |
| 30 μm | Cys | 9 ± 1 | 10 ± 1 | 31 ± 12 | 21 ± 3 |
| Glutathione | 120 ± 19 | 101 ± 46 | 100 ± 19 | 75 ± 15 | |
Leaves and roots were harvested from 3-week-old homozygous wild-type and mutant plants cultured on medium containing 1.5 mm or 30 μm sulfate. Data are means of five replicates ± se. None of the differences between wild-type and mutant plants is significant at P = 0.05.
DISCUSSION
Our results show that eliminating MMT, and hence SMM, has no appreciable effect on growth or development in normal culture conditions. Normal growth has also been reported for an independent Arabidopsis mmt knockout line that was isolated to study selenium volatilization (Tagmount et al., 2002), although SMM levels were not quantified. It is therefore unlikely that SMM is a methyl donor for any essential reaction. This long-standing hypothetical role for SMM (Giovanelli et al., 1980) can thus be struck off the list of possibilities, at least for plants growing in favorable environments. Likewise, the normal pattern of growth, thiol levels, and seed sulfur contents in mmt mutants argue against an essential role for SMM in long-distance sulfur transport. Because SMM occurs in Arabidopsis and maize phloem (Bourgis et al., 1999), the lack of impact of eliminating SMM suggests that other sulfur sources such as glutathione or sulfate can readily replace it, or perhaps that SMM is normally only a minor form of phloem sulfur in these plants.
Our data also permit discrimination between the alternative hypothetical functions of the SMM cycle, namely that it forestalls depletion of the free Met pool due to overshoots in Ado-Met synthesis (Mudd and Datko, 1990), or that it controls Ado-Met levels (Ranocha et al., 2001). The first hypothesis predicts that the metabolic pool of free Met will fluctuate in MMT knockouts, to the detriment of protein synthesis. Although fluctuations would presumably be damped by the relatively constant growth conditions we used, it is improbable that they would disappear—above all in meristematic regions where the metabolic Met pool turns over very fast and there is little or no vacuolar storage pool of Met to act as a reserve (Mudd and Datko, 1990; Ranocha et al., 2001). The lack of growth defects in mmt mutants therefore argues against this hypothesis. That the free Met content in mutant Arabidopsis leaves is normal likewise argues against it.
In contrast, the second hypothesis about the function of the SMM cycle is supported by our data. This hypothesis predicts a rise in Ado-Met level but not Met level in mutant leaves—which is what we find. It should be noted that the Ado-Met accumulation observed in the leaves of mmt knock-outs is much smaller (15% versus 160%) than predicted by in silico modeling of the effect of ablating the SMM cycle (Ranocha et al., 2001). However, the model used was designed to predict short-term metabolic responses and did not provide for long-term changes in gene expression, and hence enzyme level, in response to loss of SMM. Such long-term changes are likely a priori, and we have in fact seen many alterations in transcript levels in pilot DNA array hybridization experiments with wild-type and mmt mutant Arabidopsis (P. Ranocha and A.D. Hanson, unpublished data). Compensatory changes in the expression of genes involved in Ado-Met production or consumption may therefore mitigate the impact of the mmt mutation. In any event, coupled with the modest opposing change in AdoHcy, the rise in Ado-Met level increases the methylation ratio in mmt mutant leaves by 45%. Proportional changes of this scale in animal tissues are associated with altered DNA methyltransferase activity and gene expression (Caudill et al., 2001; Halsted et al., 2002; Pascale et al., 2002). It remains to be established whether this is the case in plants.
Raamsdonk et al. (2001) have predicted that mutations that are silent when scored on the basis of growth rate should produce obvious effects on metabolite concentrations. This has a firm basis in metabolic control theory because concentration control coefficients (CSE), which relate changes in metabolite concentrations S to changes in enzyme levels E, can have large positive or negative values (Fell, 1997). Our results with mmt mutants conform to this pattern: The mutants look normal but have a definite metabolic phenotype. This is in principle encouraging for metabolic profiling efforts, which it is hoped will reveal the phenotype of many silent mutations (Raamsdonk et al., 2001; Trethewey, 2001). However, it is sobering to note that the GC-MS profile of free amino acids in the Arabidopsis mmt mutant was uninformative and that a clear metabolic phenotype was only evident by analyzing SMM. In high-throughput metabolic profiling projects, amino acids are typically analyzed by GC-MS (Trethewey, 2001), but SMM is not and cannot be because it is non-volatile. The phenotype of mmt mutants would thus be missed by GC-MS profiling technology.
The decrease in Thr level in Arabidopsis mmt mutants is noteworthy because a modest increase would in the simplest case be expected. The current view of regulation in the Asp amino acid family is that the level of Ado-Met controls flux partitioning at the branch point between Thr and Met synthesis by activating Thr synthase, a chloroplastic enzyme that competes with cystathionine γ-synthase for the common intermediate O-phosphohomo-Ser (Giovanelli et al., 1989; Galili and Höfgen, 2002). This view stems from the strong allosteric activation of Thr synthase by Ado-Met observed in vitro (Curien et al., 1998; Laber et al., 1999) but the role of Ado-Met in regulating flux to Thr in vivo is unclear and there are some contrary findings (Galili and Höfgen, 2002). For example, Arabidopsis mto3 mutants have lower Ado-Met levels but higher Thr levels, and mto1 mutants that overproduce Ado-Met and transgenics that presumably do so have normal Thr levels (Inaba et al., 1994; Kim et al., 2002; Shen et al., 2002). That mmt mutants have a higher level of Ado-Met but a lower level of Thr is thus another instance of the lack of an in vivo relationship between Ado-Met level and Thr synthase activation. One way to reconcile this is to suppose that Ado-Met is in part sequestered away from the site of Thr synthase in the chloroplast, and in this connection, it is interesting to note that yeast cells sequester Ado-Met in the vacuole (Schlenk, 1965).
MATERIALS AND METHODS
Plants and Growing Conditions
Arabidopsis was routinely grown in Super Fine germinating mix (Fafard, Agawam, MA) at 22°C for 12-h days (80–150 μmol photons m−2 s−1) and irrigated with water. Arabidopsis plants for thiol analysis were cultured at 22°C for 16-h days (30 μmol photons m−2 s−1) for 3 weeks on Murashige and Skoog medium containing 0.8% (w/v) agarose, 1% (w/v) Suc, and 1.5 mm or 30 μm sulfate. Maize (Zea mays) for seed production was grown in the field or in 34-cm pots of Fafard 3B mix in a naturally lit greenhouse with a minimum temperature of 18°C; greenhouse plants were fertilized with 20–20-20 soluble fertilizer (Masterblend International, Chicago). Maize plants for experiments were grown in the greenhouse or a growth chamber (12-h day, 200–300 μmol photons m−2 s−1, 25°C/21°C night).
Arabidopsis Mutant Isolation
DNA pools of a collection of 60,480 Arabidopsis (ecotype Wassilewskija) lines mutagenized with the pD991 vector were screened for insertions in the MMT locus using the Arabidopsis Gene Knockout Service Facility (http://www.b.iotech.wisc.edu/Arabidopsis). Because the MMT locus is large (6.6 kb), we sought insertions into the 5′ and 3′ halves of the gene separately, using two pairs of MMT-specific forward and reverse primers: 5′-CGCTTTTTCTTCTCTATTACTGCAATCAC-3′ and 5′-TGATCATCTGATTTATCCATGCTAGTGTC-3′ for the 5′ half, and 5′-CTGATGAGAAGATTCCATTCCTAGCCTAT-3′ and 5′-GCGAGTTATTTAGAAACAACAGAACAAAG-3′ for the 3′ half. These primers yield fragments that overlap by 1.2 kb in the center of the gene. To identify and localize T-DNA insertions, these primers were used along with the T-DNA left-border primer, 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′. When an insertion was identified, the PCR-amplified product spanning the insertion site was sequenced to determine its position. A second, nested left-border primer (5′-TTTCTCCATATTGACCATCATACTCATTG-3′) was used to confirm the PCR result and the insert position. Genotypes of plants used for experiments were verified by PCR using cotyledon DNA as template plus either the second and third MMT-specific primers above (for wild-type plants) or the second MMT-specific primer and the first left-border primer (for mutants).
Maize Mutant Isolation
The Pioneer Hi-Bred collection of 42,000 F1 maize plants mutagenized with Robertson's Mu element was screened for Mu-containing MMT alleles by PCR as described (Bensen et al., 1995). Inserts in the 5′ and 3′ halves of the gene were sought separately as above; only the 3′ screening was successful, using the MMT-specific forward and reverse primers: 5′-CAGTACCTTCAGCAGTGAATGCGTCTGT-3′ and 5′-ATTGCTCCATCAGGCACCATTCATCTGAG-3′. Insertion site position was determined by cloning and sequencing MMT::Mu PCR products. Intron positions were predicted from multiple sequence alignments against Arabidopsis (GenBank accession no. AB025612) and rice (Oryza sativa; indica scaffold AAAA01000451) genomic DNA sequences. F2 plants heterozygous for the mmt mutation were crossed to inbred PHN46, followed by two generations of selfing with or without a subsequent outcross to hybrid 46,242 × 46,243. For use in experiments, homozygous mmt mutant and wild-type progeny from ears of selfed heterozygotes were identified by PCR using leaf DNA as template and the above primers together with a Mu-specific primer (5′-AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC-3′). The outcrossing procedure enhanced uniformity, and comparing progeny from the same ears reduced effects of background mutations.
Southern Analysis
Arabidopsis genomic DNA was isolated from 2-g batches of leaves pooled from 25 plants as described (Dellaporta, 1994). Isolated DNA (5 μg) was digested, separated by 0.7% (w/v) agarose gel, and transferred to NitroPure nitrocellulose membrane (Osmonics, Minnetonka, MN). Hybridization was at 65°C in 5× SSC, 5× Denhardt's solution, 1% (w/v) SDS, and 1 mm EDTA. The probe was the PCR-amplified T-DNA region of plasmid pD991 (Campisi et al., 1999) labeled with 32P by random hexamer priming. The final wash was at 65°C in 0.1× SSC containing 0.1% (w/v) SDS.
Arabidopsis Growth Measurements
Plants were grown (one per 3.5-cm pot) as above. Heights of 10 plants were measured daily. Ten shoots were harvested weekly and weighed both immediately and after lyophilizing. At the last harvest, shoots were dried (70°C, 12 h), weighed, and threshed. The separated seeds were then weighed.
In Vivo Measurement of MMT Activity
For Arabidopsis, sets of three mature rosette leaves from 3.5-week-old plants were given a pulse of [35S]Met (0.34 μCi, 40 pmol per leaf) for 90 min in the light at 24°C ± 1°C as described previously (Ranocha et al., 2001). For maize, 25-mm sections (with weight of about 50 mg) were cut from the mid-blade position of the youngest expanded leaf of plants at the 5-leaf stage. The midrib was shaved from the abaxial surface of each section and a 1.0-μCi (130 pmol) dose of [35S]Met dissolved in 3.3 μL of water was applied to the cut surface. After the labeled droplet was absorbed, 8 μL of water was added to wash the label into the tissue, and sections were incubated on moist filter paper for 2 h as above. Samples were processed as described (Ranocha et al., 2001) to measure [35S]SMM formation, as well as [35S]Met uptake and incorporation into protein. [35S]SMM was analyzed using TLC system 1 and thin layer electrophoresis systems 1 and 2 (James et al., 1995) and by its decomposition upon treatment with 2 n NH4OH at 95°C to 100°C for 3 h (Macnicol, 1986).
SMM Determination
Arabidopsis rosettes pooled from five plants and maize leaf samples from individual plants with five to six leaves were frozen in liquid N2 and stored at −80°C until analysis. SMM was assayed by MALDI-MS of cation fractions using [2H6]SMM as internal standard (Kocsis et al., 1998).
Ado-Met and AdoHcy Determination
Ado-Met and AdoHcy were quantified by HPLC as described previously (Capdevila and Wagner, 1998; Moffatt et al., 2002). Arabidopsis leaf samples (150–200 mg fresh weight) were extracted in 1 volume of 20% (w/v) trichloroacetic acid and clarified by centrifugation. After removal of trichloroacetic acid, the supernatant was separated on a C-8 column, followed by conversion of Ado-Met and AdoHcy to fluorescent isoindoles by derivatization with naphthalenedialdehyde and cyanide. The fluorescent derivatives were measured during isocratic elution from a C-18 column (Capdevila and Wagner, 1998). Ado-Met and AdoHcy peaks were identified and quantified by reference to standards. Data were corrected for recovery of Ado-Met and AdoHcy spikes (16 and 1.2 nmol, respectively) added to representative samples. Ado-Met recovery was 63% and AdoHcy recovery was 24%. These analyses were made by Dr. C. Wagner (Vanderbilt University, Nashville, TN).
Amino Acid Analysis
Samples of Arabidopsis rosette leaves pooled from five plants (0.4–0.5 g fresh weight) were frozen in liquid N2 at once after harvest and stored at −20°C until analysis. The samples were weighed exactly and extracted with 10 mL of methanol for 48 h at 4°C in darkness; each was spiked with 250 nmol of α-amino-n-buyrate as internal standard, plus 100 nmol [13C5]Met for Met quantification by isotope dilution MS. The methanol extract was phase-separated by adding 5 mL of chloroform plus 6 mL of water, and the aqueous phase was dried in an air stream. Dried aqueous phases were redissolved in 1 mL of water and applied to 4.5- × 1-cm columns of Dowex-1 [OH−]; after washing columns with water, amino acids were eluted with 6 mL of 2.5 n HCl and dried as above. The dried eluates were redissolved in 1 mL of water and applied to 4.5- × 1-cm columns of Dowex-50 [H+], which were washed with water and eluted with 6 mL of 6 m NH4OH. After drying, eluates were derivatized to N-(O,S)-heptafluorobutyryl isobutyl esters by reaction with isobutanol:acetyl chloride (5:1, v/v, 120°C, 20 min), followed by heptafluorobutyric anhydride (120°C, 10 min; Rhodes et al., 1986). After evaporating excess heptafluorobutyric anhydride, samples were redissolved in 200 μL of ethyl acetate:acetic anhydride (1:1, v/v) for analysis by GC and by electron ionization GC-MS as described (Rhodes et al., 1986), except that a GCQ mass spectrometer (Thermo Finnigan, San Jose, CA) was used.
Thiol Analysis
Samples comprised the rosette leaves or roots from two or three pooled Arabidopsis plants. Thiols were determined as monobromobimane derivatives by HPLC with fluorescence detection (Fahey and Newton, 1987). Tissues were extracted in 5 or 10 volumes of 0.01 m HCl. Three microliters of 5 mm dithiothreitol and 50 μL of CHES buffer (100 mm, pH 9.3) were added to 20-μL portions of extracts, followed by incubation at 37°C for 20 min to allow reduction to occur. Five microliters of 30 mm monobromobimane was then added, and incubation at 37°C was continued for 45 min. The derivatization reaction was stopped by adding 30 μL of glacial acetic acid, and the volume was brought to 160 μL with water. Aliquots (20 μL) were analyzed by HPLC, using a Mightysil RP-18GP (5 μm) 250- × 4.6-mm column (Kanto, Portland, OR) eluted from 0 to 15 min and from 22 to 30 min with 99% buffer A + 1% buffer B, and from 15 to 22 min with buffer B. Buffer A was 8% (v/v) methanol:92% (v/v) water containing 0.25% (v/v) acetic acid (pH 3.9); buffer B was 90% (v/v) methanol:10% (v/v) water containing 0.25% (v/v) acetic acid (pH 3.9). The flow rate was 1.2 mL min−1. Monobromobimane derivatives were detected by fluorescence (excitation at 390 nm, emission at 482 nm).
Determination of Sulfur Content of Seeds
Seed samples (100 mg) were extracted by treatment with 4 mL of 65% (w/w) HNO3 and 3 mL of 30% (w/w) H2O2 in high-pressure Teflon vessels in a microwave oven for 20 min at 195°C and 1.2 MPa. After cooling, bidistilled water (50 mL) was added and extracts were transferred to polyethylene bottles. The extracts were diluted (1:4) with 7% (w/w) HNO3, and their sulfur contents were determined by inductively coupled plasma optical emission spectroscopy using a Liberty 150 instrument at a wavelength of 182.034 nm (Varian Medical Systems, Palo Alto, CA).
ACKNOWLEDGMENTS
We thank L.C. Hannah and J. Baier for advice and help with maize crosses, S. Roje for help with Southern analysis, T. Jack for the pD991 plasmid, and T. Leustek for insightful discussion.
Footnotes
This work was supported in part by the National Science Foundation (grant nos. IBN–981399 and MCB-0114117 to A.D.H. and IBN-9904263 to D.A.G.), by the Department of Energy (grant no. DE–FG02–99ER20344 to D.R.), by the Japan Society for the Promotion of Science (grants to K.S. and M.A.), by Core Research for Evolutional Science and Technology of Japan Science and Technology Corporation (to K.S.), by an endowment from the C.V. Griffin, Sr. Foundation, and by the Florida Agricultural Experiment Station (journal series no. R–09217).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.018846.
LITERATURE CITED
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP. Cloning and characterization of the maize An1 gene. Plant Cell. 1995;7:75–84. doi: 10.1105/tpc.7.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, Fisher DB, Tarczynski MC, Li C, Herschbach C, Rennenberg H, Pimenta MJ, Shen TL et al. S-Methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell. 1999;11:1485–1498. doi: 10.1105/tpc.11.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi L, Yang Y, Yi Y, Heilig E, Herman B, Cassista AJ, Allen DW, Xiang H, Jack T. Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 1999;17:699–707. doi: 10.1046/j.1365-313x.1999.00409.x. [DOI] [PubMed] [Google Scholar]
- Cantoni GL. S-Adenosylmethionine: present status and future perspectives. In: Salvatore F, Borek E, Williams-Ashman HG, Schlenk F, editors. The Biochemistry of Adenosylmethionine. New York: Columbia University Press; 1977. pp. 557–577. [Google Scholar]
- Cantoni GL, Richards HH, Chiang PK. Inhibitors of S-adenosylhomocysteine hydrolase and their role in the regulation of biological methylation. In: Usdin E, Borchardt RT, Creveling ER, editors. Transmethylation. New York: Elsevier; 1979. pp. 155–164. [Google Scholar]
- Capdevila A, Wagner C. Measurement of plasma S-adenosylmethionine and S-adenosylhomocysteine as their fluorescent isoindoles. Anal Biochem. 1998;264:180–184. doi: 10.1006/abio.1998.2839. [DOI] [PubMed] [Google Scholar]
- Caudill MA, Wang JC, Melnyk S, Pogribny IP, Jernigan S, Collins MD, Santos-Guzman J, Swendseid ME, Cogger EA, James SJ. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. J Nutr. 2001;131:2811–2818. doi: 10.1093/jn/131.11.2811. [DOI] [PubMed] [Google Scholar]
- Curien G, Job D, Douce R, Dumas R. Allosteric activation of Arabidopsis threonine synthase by S-adenosylmethionine. Biochemistry. 1998;37:13212–13221. doi: 10.1021/bi980068f. [DOI] [PubMed] [Google Scholar]
- Dellaporta S. Plant DNA miniprep and microprep: versions 2.1–2.3. In: Freeling M, Walbot V, editors. The Maize Handbook. New York: Springer-Verlag; 1994. pp. 522–525. [Google Scholar]
- Fahey RC, Newton GL. Determination of low-molecular weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol. 1987;143:85–96. doi: 10.1016/0076-6879(87)43016-4. [DOI] [PubMed] [Google Scholar]
- Fell DA. Understanding the Control of Metabolism. London: Portland Press; 1997. [Google Scholar]
- Fitzgerald MA, Ugalde TD, Anderson JW. Sulphur nutrition affects delivery and metabolism of S in developing endosperms of wheat. J Exp Bot. 2001;52:1519–1526. doi: 10.1093/jexbot/52.360.1519. [DOI] [PubMed] [Google Scholar]
- Galili G, Höfgen R. Metabolic engineering of amino acids and storage proteins in plants. Metab Eng. 2002;4:3–11. doi: 10.1006/mben.2001.0203. [DOI] [PubMed] [Google Scholar]
- Giovanelli J, Mudd SH, Datko AH. Sulfur amino acids in plants. In: Miflin BJ, editor. The Biochemistry of Plants. Vol. 5. New York: Academic Press; 1980. pp. 453–505. [Google Scholar]
- Giovanelli J, Mudd SH, Datko AH. Regulatory structure of the biosynthetic pathway for the aspartate family of amino acids in Lemna paucicostata Hegelm. 6746, with special reference to the role of aspartokinase. Plant Physiol. 1989;90:1584–1599. doi: 10.1104/pp.90.4.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsted CH, Villanueva JA, Devlin AM, Niemela O, Parkkila S, Garrow TA, Wallock LM, Shigenaga MK, Melnyk S, James SJ. Folate deficiency disturbs hepatic methionine metabolism and promotes liver injury in the ethanol-fed micropig. Proc Natl Acad Sci USA. 2002;99:10072–10077. doi: 10.1073/pnas.112336399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn GW, Davin L, Giblin M, Underhill EW. Biochemical genetics of plant secondary metabolites in Arabidopsis thaliana: the glucosinolates. Plant Physiol. 1991;97:217–226. doi: 10.1104/pp.97.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Fujiwara T, Hayashi H, Chino M, Komeda Y, Naito S. Isolation of an Arabidopsis thaliana mutant, mto1, that overaccumulates soluble methioine: temporal and spatial patterns of soluble methionine accumulation. Plant Physiol. 1994;104:881–887. doi: 10.1104/pp.104.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James F, Paquet L, Sparace SA, Gage DA, Hanson AD. Evidence implicating dimethylsulfoniopropionaldehyde as an intermediate in dimethylsulfoniopropionate biosynthesis. Plant Physiol. 1995;108:1439–1448. doi: 10.1104/pp.108.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz ML, Gerhardt KO. Storage protein in hereditary ceroid-lipofuscinosis contains S-methylated methionine. Mech Ageing Dev. 1990;53:277–290. doi: 10.1016/0047-6374(90)90045-h. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee M, Chalam R, Martin MN, Leustek T, Boerjan W. Constitutive overexpression of cystathionine γ-synthase in Arabidopsis leads to accumulation of soluble methionine and S-methylmethionine. Plant Physiol. 2002;128:95–107. [PMC free article] [PubMed] [Google Scholar]
- Kocsis MG, Nolte KD, Rhodes D, Shen TL, Gage DA, Hanson AD. Dimethylsulfoniopropionate biosynthesis in Spartina alterniflora: evidence that S-methylmethionine and dimethylsulfoniopropylamine are intermediates. Plant Physiol. 1998;117:273–281. doi: 10.1104/pp.117.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laber B, Maurer W, Hanke C, Grafe S, Ehlert S, Messerschmidt A, Clausen T. Characterization of recombinant Arabidopsis thaliana threonine synthase. Eur J Biochem. 1999;263:212–221. doi: 10.1046/j.1432-1327.1999.00487.x. [DOI] [PubMed] [Google Scholar]
- Macnicol PK. Analysis of S-methylmethionine and S-adenosylmethionine in plant tissue by a dansylation, dual-isotope method. Anal Biochem. 1986;158:93–97. doi: 10.1016/0003-2697(86)90594-4. [DOI] [PubMed] [Google Scholar]
- Meeley RB, Briggs SP. Reverse genetics for maize. Maize Genet Coop Newslett. 1995;69:67–82. [Google Scholar]
- Moffatt BA, Stevens YY, Allen MS, Snider JD, Pereira LA, Todorova MI, Summers PS, Weretilnyk EA, Martin-McCaffrey L, Wagner C. Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol. 2002;128:812–821. doi: 10.1104/pp.010880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd SH, Datko AH. The S-methylmethionine cycle in Lemna paucicostata. Plant Physiol. 1990;93:623–630. doi: 10.1104/pp.93.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale R, Simile M, De Miglio M, Feo F. Chemoprevention of hepatocarcinogenesis: S-adenosyl-l-methionine. Alcohol. 2002;27:193–198. doi: 10.1016/s0741-8329(02)00227-6. [DOI] [PubMed] [Google Scholar]
- Pokorny M, Marèenko E, Kegleviè D. Comparative studies of l- and d-methionine metabolism in lower and higher plants. Phytochemistry. 1970;9:2175–2188. [Google Scholar]
- Raamsdonk LM, Teusink B, Broadhurst D, Zhang N, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19:45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- Ranocha P, Bourgis F, Ziemak MJ, Rhodes D, Gage DA, Hanson AD. Characterization and functional expression of cDNAs encoding methionine-sensitive and -insensitive homocysteine S-methyltransferases from Arabidopsis. J Biol Chem. 2000;275:15962–15968. doi: 10.1074/jbc.M001116200. [DOI] [PubMed] [Google Scholar]
- Ranocha P, McNeil SD, Ziemak MJ, Li C, Tarczynski MC, Hanson AD. The S-methylmethionine cycle in angiosperms: ubiquity, antiquity and activity. Plant J. 2001;25:575–584. doi: 10.1046/j.1365-313x.2001.00988.x. [DOI] [PubMed] [Google Scholar]
- Rhodes D, Handa S, Bressan RA. Metabolic changes associated with adaptation of plant cells to water stress. Plant Physiol. 1986;82:890–903. doi: 10.1104/pp.82.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roje S, Chan SY, Kaplan F, Raymond RK, Horne DW, Appling DR, Hanson AD. Metabolic engineering in yeast demonstrates that S-adenosylmethionine controls flux through the methylenetetrahydrofolate reductase reaction in vivo. J Biol Chem. 2002;277:4056–4061. doi: 10.1074/jbc.M110651200. [DOI] [PubMed] [Google Scholar]
- Schlenk F. Biochemical and cytological studies with sulfonium compounds. In: Shapiro SK, Schlenk F, editors. Transmethylation and Methionine Biosynthesis. Chicago: University of Chicago Press; 1965. pp. 48–65. [Google Scholar]
- Shen B, Li CJ, Tarczynski MC. High free-methionine and decreased lignin content result from a mutation in the Arabidopsis S-adenosyl-l-methionine synthetase 3 gene. Plant J. 2002;29:371–380. doi: 10.1046/j.1365-313x.2002.01221.x. [DOI] [PubMed] [Google Scholar]
- Tagmount A, Berken A, Terry N. An essential role of S-adenosyl-l-methionine:l-methionine S-methyltransferase in selenium volatilization by plants: methylation of selenomethionine to Se-methyl-l-Se-methionine, the precursor of volatile selenium. Plant Physiol. 2002;130:847–856. doi: 10.1104/pp.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trethewey RN. Gene discovery via metabolic profiling. Curr Opin Biotechnol. 2001;12:135–138. doi: 10.1016/s0958-1669(00)00187-7. [DOI] [PubMed] [Google Scholar]