Abstract
Evidence is accumulating that insect-specific plant responses are mediated by constituents in the oral secretions and regurgitants (R) of herbivores, however the relative importance of the different potentially active constituents remains unclear. Fatty acid-amino acid conjugates (FACs) are found in the R of many insect herbivores and have been shown to be necessary and sufficient to elicit a set of herbivore-specific responses when the native tobacco plant Nicotiana attenuata is attacked by the tobacco hornworm, Manduca sexta. Attack by this specialist herbivore results in a large transcriptional reorganization in N. attenuata, and 161 genes have been cloned from previous cDNA differential display-polymerase chain reaction and subtractive hybridization with magnetic beads analysis. cDNAs of these genes, in addition to those of 73 new R-responsive genes identified by cDNA-amplified fragment-length polymorphism display of R-elicited plants, were spotted on polyepoxide coated glass slides to create microarrays highly enriched in Manduca spp.- and R-induced genes. With these microarrays, we compare transcriptional responses in N. attenuata treated with R from the two most damaging lepidopteran herbivores of this plant in nature, M. sexta and Manduca quinquemaculata, which have very similar FAC compositions in their R, and with the two most abundant FACs in Manduca spp. R. More than 68% of the genes up- and down-regulated by M. sexta R were similarly regulated by M. quinquemaculata R. A majority of genes up-regulated (64%) and down-regulated (49%) by M. sexta R were similarly regulated by treatment with the two FACs. In contrast, few genes showed similar transcriptional changes after H2O2- and R-treatment. These results demonstrate that the two most abundant FACs in Manduca spp. R can account for the majority of Manduca spp.-induced alterations of the wound response of N. attenuata.
When herbivores attack plants, they cause wounding, but the response of a plant to herbivore attack cannot, in many cases, be mimicked by mechanical wounding (Baldwin, 1988; Baldwin et al., 2001; Kessler and Baldwin, 2002). Several different types of elicitors in the oral secretions and regurgitant (R) of herbivorous insects have been reported to alter the wound response of a plant. For example, a β-glucosidase in the R of Pieris brassica larvae elicits the release of volatile organic compounds that function as indirect defenses (Mattiacci et al., 1995). A second enzymatic elicitor found in the salivary glands of Helicoverpa zea larvae was identified as Glc oxidase (GOX; Musser et al., 2002). GOX activity was shown to inhibit wound-induced nicotine accumulation and induced resistance in tobacco (Nicotiana tabacum) and is thought to function by the production of H2O2 at the wound site. N-(17-Hydroxylinolenoyl)-l-Gln (volicitin), identified in R of Spodoptera exigua larvae (Alborn et al., 1997, 2000; Turlings et al., 2000), was the first fatty acid-amino acid conjugate (FAC) that showed biological activity in inducing volatile emissions in corn (Zea mays) plants. Subsequently, several FACs have been identified in R of other herbivorous insect larvae (Pohnert et al., 1999; Turlings et al., 2000; Halitschke et al., 2001).
As demonstrated in other papers in this series, attack by Manduca sexta larvae elicits a suite of direct and indirect defense responses in its native host plant Nicotiana attenuata. These induced defense responses are accompanied by a large-scale transcriptional reorganization (Hermsmeier et al., 2001; Schittko et al., 2001; Winz and Baldwin, 2001; Hui et al., 2003). Many of the defensive and transcriptional responses elicited by Manduca spp. attack can be mimicked by applying Manduca spp. R to mechanically produced puncture wounds (McCloud and Baldwin, 1997; Halitschke et al., 2000; Kahl et al., 2000; Schittko et al., 2001, 2000; Winz and Baldwin, 2001), and FACs in Manduca spp. R have been shown to be necessary and sufficient to elicit the release of terpenoid volatiles, an endogenous jasmonic acid burst, and changes in transcript accumulation of six herbivore-responsive genes in N. attenuata (Halitschke et al., 2001). Because herbivores transfer a bewildering array of potential elicitors to the plant during feeding and plants respond to herbivore attack with a bewildering array of responses, we provide a quantitative analysis of the proportion of transcriptional changes elicited by R from different Manduca spp. and two different constituents of R, namely: FACs and H2O2.
To conduct this analysis, we created a microarray enriched in M. sexta- and R-induced N. attenuata genes. Companion papers in this series using cDNA differential display (DDRT)-PCR and subtractive hybridization with magnetic beads (SHMB) techniques to identify differentially expressed genes in N. attenuata by comparing mRNA from M. sexta-attacked plants with that from developmentally synchronized unattacked control plants (Hermsmeier et al., 2001; Hui et al., 2003) did not discriminate between herbivore-specific and wound-induced transcript accumulations. The expression of a subset of the identified genes was shown to be specifically regulated by constituents of M. sexta R (Schittko et al., 2001). In this study, we compared the mRNA of plants mechanically wounded and had their wounds treated with either water or with M. sexta R with a cDNA-amplified fragment-length polymorphism (cDNA-AFLP; Bachem et al., 1998) analysis to identify additional genes exhibiting R-specific patterns of expression. We spotted PCR fragments of the newly identified differentials together with previously characterized N. attenuata expressed sequence tags and genes derived from DDRT-PCR, SHMB, and cDNA-AFLP display of Manduca spp.-attacked plants (Hermsmeier et al., 2001; Hui et al., 2003) on polyepoxide coated glass slides to create a cDNA microarray containing 241 genes.
The larvae of two Manduca spp. (M. sexta and Manduca quinquemaculata) have been responsible for the majority of leaf area lost from N. attenuata plants to insect herbivores in the past 15 years of field observations in Utah (I.T. Baldwin, unpublished data). To determine the relevance of FACs in organizing the transcriptional response of a plant, we first compared the transcriptional response of N. attenuata to R from these two species. Although FACs are a minor constituent of R, the two species have very similar FAC compositions (Halitschke et al., 2001), and if the transcriptional responses are not similar, we could infer that FACs play a minor role in determining herbivore-specific responses. After finding that more than 68% of the genes are similarly regulated by the R of the two Manduca spp., we analyzed the contribution of the two most abundant FACs in Manduca spp. R, N-linolenoyl-l-Gln and N-linolenoyl-l-Glu at concentrations equivalent to that found in R. We find that these two minor constituents of R account for 56% of the R-specific transcript accumulation. Furthermore, we examined the role of H2O2, a product of GOX activity, in the elicitation of R-elicited transcripts and find only 18% of the genes elicited by GOX treatment responded similarly to treatment with larval R, whereas a majority (42%) showed opposite changes in transcript accumulation. These results underscore the importance of FACs in determining N. attenuata's “recognition” of Manduca spp. attack.
RESULTS
cDNA-AFLP Analysis
We amplified the EcoRI/MseI cDNA fragments by PCR after subtractive hybridization, and we identified, isolated, and sequenced fragments of 73 genes, of which 22 had similarity to known genes in the databases (Table I). The microarray analysis of plants elicited by a single treatment of R to wounds (Fig. 1A) confirmed the differential expression of 53% of the 73 genes derived from the cDNA-AFLP analysis, but this may be an underestimation because the cDNA-AFLP analysis was conducted on mRNA from plants elicited multiple times, whereas in the microarray analysis, plants were only elicited by a single treatment. All sequences, with one exception, had not been identified in previous display analyses of Manduca spp.-attacked N. attenuata plants and confirm the pattern that a substantial proportion of the transcriptome of a plant is altered during attack. Comments on a selection of these genes follow.
Table I.
Sequence similarities (BLAST query) of N. attenuata cDNAs derived by cDNA-AFLP analysis of mRNA from plants that were wounded and immediately treated with water or treated with M. sexta oral secretions and regurgitants
| Clone | Accession No. | Sequence Similarity | E Value |
|---|---|---|---|
| DH02 | CA591820 | N. tabacum chloroplast genome DNA (Z00044) | 4e-68 |
| DH05 | CA591823 | Tomato (Lycopersicon esculentum) Hsc70 gene (L41253) | 6e-24 |
| DH17 | CA591835 | Tomato ripening-regulated protein DDTFR10 (AF204787) | 5e-33 |
| DH19 | CA591837 | Nicotiana plumbaginifolia metallothionein-like protein (U35225) | 5e-59 |
| DH23 | CA591841 | N. tabacum Rubisco SSU pseudogene (M32420) | 2e-10 |
| DH24 | CA591842 | N. tabacum chloroplast genome DNA (Z00044) | 4e-77 |
| DH25 | CA591843 | N. tabacum mRNA C-7 (X64399) | e-133 |
| DH31 | CA591849 | Tomato unknown mRNA (AF261140) | 3e-29 |
| DH32 | CA591850 | Nicotiana sylvestris ATPase β-subunit nsatp2.20.1 (U96498) | 3e-91 |
| DH40 | CA591858 | N. tabacum mRNA C-7 (X64399) | 2e-93 |
| DH43 | CA591861 | Sunflower (Helianthus annuus) NADPH thioredoxin reductase (L36129) | 2e-31 |
| DH44 | CA591862 | Popolus kitakamiensis gene for Phe ammonia-lyase (PAL; D43802) | 2e-17 |
| DH45 | CA591863 | N. tabacum (TSC40–4) 60S ribosomal protein L34 (L27107) | e-134 |
| DH47 | CA591865 | Pea (Pisum sativum) pore protein (Z73553) | 5e-19 |
| DH48 | CA591866 | N. tabacum chloroplast genome DNA (Z00044) | e-117 |
| DH49 | CA591867 | Potato (Solanum tuberosum) Ser/Gly hydroxymethyltransferase (Z25863) | e-107 |
| DH51 | CA591869 | Potato StLTSR low-temperature and salt-responsive protein (AB061265) | 3e-15 |
| DH54 | CA591872 | N. attenuata pathogen-inducible α-dioxygenase (AF229926) | 0.0 |
| DH57 | CA591875 | N. tabacum chloroplast genome DNA (Z00044) | e-107 |
| DH58 | CA591876 | Timothy grass (Phleum pratense) putative protein translation factor SUI (AJ249397) | 1e-35 |
| DH60 | CA591878 | Tomato Pto-responsive gene (Prg1) (AF146690) | 5e-59 |
| DH63 | CA591881 | Petunia (Petunia hybrida) triosephosphate isomerase (X83227) | 3e-29 |
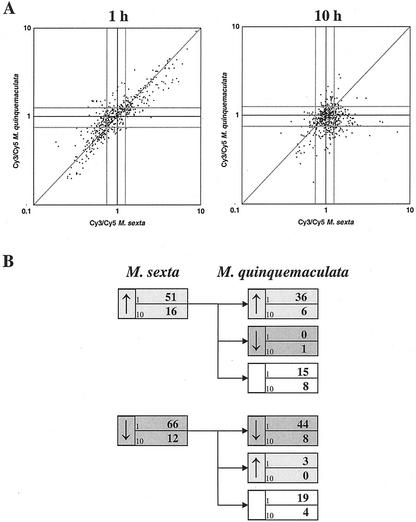
Figure 1.
A, Correlation between ERs of 250 N. attenuata genes 1 and 10 h after application of M. sexta or M. quinquemaculata oral secretions and regurgitant (R) to mechanically produced puncture wounds in fully expanded N. attenuata leaves. Each gene is represented by two data points corresponding to the mean ER of four replicate spots of the two probes for each gene (see “Materials and Methods”). Each array was hybridized with Cy3- or Cy5-labeled cDNA generated from plants that were wounded and immediately treated with either water (Cy5) or Manduca spp. R (Cy3). Expression limits used to define either up- and down-regulated expression are depicted as gray lines. B, Number of genes exhibiting significantly altered, up-regulated ( ) or down-regulated (↓), expression at either 1 or 10 h after application of R of M. sexta larvae (left panel) and expression patterns ( , ↓, or not regulated) of these genes in response to application of R of M. quinquemaculata (right panel) at the same analysis time. For example, 67 genes were up-regulated by M. sexta R treatment, of which 42 are also up-regulated, one was down-regulated, and 23 genes were not regulated by M. quinquemaculata R treatment at the same analysis time.
DH54 provided a 438-bp fragment of the N. attenuata α-dioxygenase (α-DOX), which catalyzes the α-oxidation of fatty acids to hydroperoxy fatty acids and may be involved in signal generation (Sanz et al., 1998; Hamberg et al., 1999). α-DOX had been previously cloned twice by DDRT-PCR (Hermsmeier et al., 2001; Voelckel and Baldwin, 2003) with arbitrary primer R1, and a full-length sequence was isolated by a cDNA library screen (Hermsmeier et al., 2001). DH45 had sequence similarity to TSC40-4 from N. tabacum, a 60S ribosomal protein L34, which is known to be wound-induced (Gao et al., 1994). DH02, DH24, DH48, and DH57 had similarity to N. tabacum chloroplast genome DNA (Shinozaki et al., 1986). DH49 had similarity to a potato mRNA for a Ser/Gly hydroxymethyltransferase (Kopriva and Bauwe, 1995), which catalyzes the interconversion of Ser and Gly and is a component of the photorespiratory pathway, which recycles carbon and nitrogen lost from the Calvin cycle by the oxygenation of ribulose 1,5-bisphosphate. DH19 had similarity to a N. plumbaginifolia (P.C. LaRosa and A.C. Smigocki, unpublished data) and a tomato (Giritch et al., 1998) metallothionein-like protein. Some type 2 metallothioneins are thought to function as potent metal chelators, but others play roles in different cell death pathways, including senescence and the hypersensitive response after pathogen attack (Butt et al., 1998). DH60 had similarity to tomato PTO-responsive protein (rg1) mRNA (R.L. Thilmony and G.B. Martin, unpublished data) and again implicates pathogen recognition after herbivore attack. Prior work with DDRT-PCR of N. attenuata plants attacked by M. sexta larvae provided clone, RC144, which has similarity to a putative tomato pto gene (Hui et al., 2003). DH44 had similarity to a P. kitakamiensis PAL gene. PAL represents the keystone enzyme in synthesis of phenolics. Several phenolic compounds are induced in N. attenuata by feeding of M. sexta larvae (Keinanen et al., 2001; Roda et al., 2003). DH58 had similarity with a timothy grass mRNA for putative protein translation factor (R. Suck, S. Hagen, O. Cromwell, and H. Fiebig, unpublished data). DH63 had similarity with a petunia mRNA for triose phosphate isomerase (Bennissan and Weiss, 1995), which catalyzes the interconversion of dihydroxyacetone phosphate and d-glyceraldehyde 3-phosphate and plays an important role in gluconeogenesis, fatty acid biosynthesis, pentosephosphate pathway, and photosynthetic carbon fixation. In petunia corollas, this gene is induced by gibberellins, and its expression is highly correlated with respiration (Bennissan and Weiss, 1995). DH47 had similarity to a pea mRNA for chloroplast outer membrane pore protein (Pohlmeyer et al., 1997), which may mediate the transport of Gln and Glu to and from the choloroplast and thereby regulate the export of reduced nitrogen. DH32 had similarity to a N. sylvestris ATPase mitochondrial β-subunit, a family that is known to be stress responsive (Lalanne et al., 1998). DH51 had similarity with a potato mRNA for a low-temperature and salt-responsive protein (E. Nakane, H. Yoshioka, K. Kawakita, and N. Doke, unpublished data).
Quantitative Analyses
Prior analysis of R found the FAC profiles from the two Manduca spp. to be very similar (Halitschke et al., 2001). To determine the relevance of FACs in R-specific responses, we compared the responses to M. sexta R with those to M. quinquemaculata R. We spotted the fragments isolated by cDNA-AFLP together with PCR fragments of previously identified genes (Hermsmeier et al., 2001; Hui et al., 2003) to create a microarray highly enriched in N. attenuata genes with Manduca spp.-responsive expression.
To examine the suitability of the microarray for this analysis, we compared the expression ratios (ERs) of a set of genes (Thr deaminase, light-harvesting complex protein, α-DOX, and unknowns pDH68.1 and pDH39.1) that had been extensively characterized with northern-blot analyses in previous studies using similar experimental treatments (Halitschke et al., 2001; Schittko et al., 2001). All of the previously identified patterns of R- and FAC-elicited transcript regulation of these “control genes,” namely the up-regulation (α-DOX) or down-regulation (Thr deaminase, light-harvesting complex protein, pDH68.1, and pDH39.1) that M. sexta R-treatment alters in the wound response of N. attenuata were confirmed with the microarray, despite experimental differences in timing of harvests and treatment applications between the studies.
The microarray analysis revealed M. sexta-specific changes in expression levels of 134 (56%) of the 241 genes spotted on the array (ERs can be found in Supplementary Table II, which can be viewed at www.plantphysiol.org). The transcripts of 67 genes showed significant up-regulation, whereas 78 genes showed significant down-regulation at either of the two times (1 or 10 h) after elicitation (Fig. 1B, left panel). The majority (81%) of the transcripts were regulated at 1 h after elicitation and a minority (19%) at 10 h. Therefore elicitation by M. sexta R is a rapidly induced and rapidly waning response. This response was largely mimicked by elicitation by R of M. quinquemaculata (Fig. 1A). Forty-two (63%) up-regulated genes and 52 (67%) down-regulated genes showed the same transcriptional regulation with a similar kinetic after treatment with M. quinquemaculata R (Fig. 1B, right panel). Additionally, two of the up-regulated genes and three of the down-regulated genes showed the same directional regulation but with a different kinetic (data not shown). In summary, even though FACs compose only a minor fraction of the R of the two Manduca spp., which could differ in other unmeasured potential elicitors, the similarity of the response of the plant suggested that FACs were of fundamental importance.
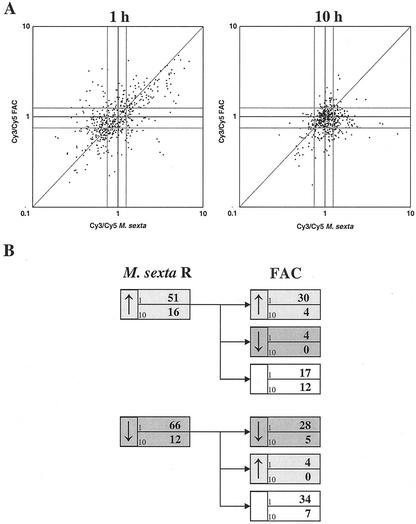
To directly determine the elicitor activity of FACs, we applied the two most abundant FACs in the R of both Manduca spp. and compared the response of the plant to that elicited by M. sexta R. Because purified FACs are not water-soluble, the two compounds were dissolved in water containing trace quantities of Triton detergent. To control for potential Triton-mediated effects, the array was hybridized against a cDNA derived from mRNA extracted from plants wounded and treated with only the Triton-containing solution. Because the M. sexta R was diluted in water, this array was hybridized against cDNA derived from mRNA extracted from plants wounded and treated with water. Elicitation by the two FACs was remarkably similar to the elicitation by M. sexta R (Fig. 2A). Thirty-four (51%) of the up-regulated genes and 33 (42%) of the down-regulated genes after application of M. sexta R showed the same transcriptional regulation with a similar kinetic after treatment with the FACs at spit-comparable concentration (Fig. 2B, right panel). Additionally, nine of the up-regulated genes and five of the down-regulated genes showed the same regulation but with a different kinetic (data not shown). In summary, despite the obvious differences between R and Triton matrices, N-linolenoyl-l-Gln and N-linolenoyl-l-Glu at concentrations found in R were sufficient to elicit a majority of the plant's complicated Manduca spp.-specific transcriptional response.
Figure 2.
A, Correlation between ERs of 250 N. attenuata genes 1 and 10 h after application of M. sexta oral secretions and regurgitant (R) or a mixture of the two most abundant FACs of Manduca spp. R dissolved in a Triton-containing solution at concentrations equivalent to that of R to mechanically produced puncture wounds in fully expanded N. attenuata leaves. Each gene is represented by two data points corresponding to the mean ER of four replicate spots of the two probes for each gene (see “Materials and Methods”). The arrays were hybridized with Cy3- or Cy5-labeled cDNA generated from plants treated with water (Cy5) or M. sexta spp. R (Cy3) and a Triton control (Cy5) or FAC solution (Cy3), respectively. Expression limits defining up- and down-regulated expression are depicted as gray lines. B, Number of genes exhibiting significantly altered (up-regulated [ ] or down-regulated [↓]) expression either 1 or 10 h after application of R of M. sexta larvae (left panel) and expression patterns ( , ↓, or not regulated) of these genes in response to application of FACs (right panel) at the same analysis time.
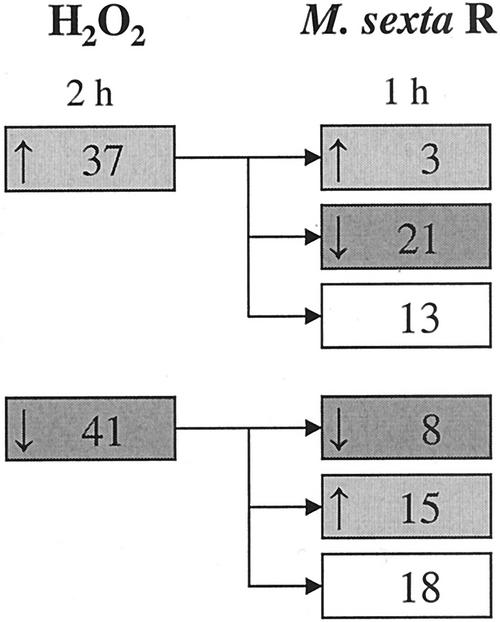
GOX-derived H2O2 has been implicated in the differential regulation of tobacco defense responses (Musser et al., 2002), and we elicited plants with GOX-derived H2O2 and determined the similarity to M. sexta R-elicited responses. Note that we identified genes elicited by the GOX treatment and compared these with genes elicited by R treatment. Both microarrays include the appropriate controls (wound + buffer or water controls) and are therefore normalized for the respective wound response for their treatment. H2O2 treatment up-regulated 37 (15%) and down-regulated 41 (17%) transcripts of the 241 genes spotted on the microarray within 2 h (Fig. 3, left panel). Only three (8%) of the up-regulated and eight (20%) of the down-regulated genes showed comparable changes in transcript accumulation 1 h after treatment with M. sexta R (Fig. 3, right panel). We conclude that GOX-derived H2O2 plays only a minor role in elicitation of Manduca spp.-specific plant responses.
Figure 3.
Number of N. attenuata genes exhibiting significantly altered expression 2 h after fully expanded N. attenuata leaves were infused with a solution of Glc and GOX to produce H2O2 in situ (left panel), and expression pattern of these genes 1 h (right panel) after application of R of M. sexta larvae.
Qualitative Analyses
A complete listing of the mean (± se) ERs of all spotted genes can be found in Supplementary Table II. The observed expression patterns elicited by R- and FAC-treatments reflect a basic shift in plant metabolism in response to herbivore attack. Overall, genes coding for photosynthetic enzymes (small subunit of Rubisco, light-harvesting complex protein) were down-regulated by all treatments including the application of GOX-derived H2O2. The exception to this trend was seen with photosystem II (PSII) O2-evolving complex, which was up-regulated at 1 h by Manduca spp. R, but down-regulated by FACs and H2O2. Two additional subunits of PSII, similar to NtPII10 and a spinach (Spinacia oleracea) PSII polypeptide, were down-regulated by Manduca spp. R and FACs but not by H2O2. Furthermore, a majority of these photosynthesis-related genes retained their patterns of regulation 10 h after elicitation.
Genes coding for enzymes of the oxylipin-signaling cascade (13-lipoxygenase [LOX]; allene oxide synthase [AOS]; hydroperoxide lyase [HPL]) were strongly up-regulated by Manduca spp. R and FAC treatments. Surprisingly, these genes were down-regulated in response to H2O2 treatment. Furthermore, a gene coding for a germin homolog, an enzyme involved in production of endogenous H2O2, was up-regulated in response to the H2O2 treatment but down-regulated by Manduca spp. R.
The same expression pattern observed for genes of the oxylipin-signaling cascade, namely up-regulation by Manduca spp. R and FACs but down-regulation by H2O2, was found in transcripts coding for WRKY transcription factors, a luminal binding protein, a Glu synthase, and an ATPase β-subunit. A set of genes involved in herbivore-induced activation of secondary metabolism, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, α-DOX, and NADPH thioredoxin reductase, was up-regulated by R and FAC but did not respond to H2O2.
Similarly, the majority of genes down-regulated by Manduca spp. R and FACs was either down-regulated by (histone H3, major intrinsic protein MIP2, and metallothionein-like proteins) or not responsive to (GAL83 and Ser carboxypeptidase) H2O2 treatment. In addition to these genes, for which the R-induced regulation could be attributed to FACs in the Manduca spp. R, some genes showed R-induced accumulation that could be elicited by neither FACs nor H2O2. Thr deaminase, a C-7 mRNA, membrane channel protein, protein translation factor sui1, PTO-responsive gene 1, RNA polymerase II, and triosephosphate isomerase were down-regulated by Manduca spp. R but not by FACs and H2O2. Genes up-regulated by Manduca spp. R, but not by FACs or H2O2 code for Mg protopophyrin IX chelatase, thiazole, and Gly/Ser hydroxymethyltransferase.
DISCUSSION
By comparing the transcriptome of N. attenuata plants elicited by either wounding alone or wounding plus the addition of M. sexta R to the wounds, we identified fragments of 73 genes (Table I). The list of genes identified by this cDNA-AFLP display analysis contained only one previously identified gene from prior analyses: α-DOX (Hermsmeier et al., 2001). The lack of overlap is consistent with earlier predictions of a large herbivore-induced transcriptome (Hermsmeier et al., 2001; Hui et al., 2003; Voelckel and Baldwin, 2003). Moreover, the microarray proved to be an efficient tool for the verification of differential gene expression, and when combined with various display procedures, it provides a powerful means of analyzing ecological questions in non-model systems without the attendant bioinformatics overload.
We investigated the eliciting mechanism of the transcriptional reorganization in N. attenuata by analyzing two characterized types of elicitors identified in larval R, GOX enzyme activity (Musser et al., 2002), and FACs (Alborn et al., 1997; Turlings et al., 2000; Halitschke et al., 2001). A large proportion of the specific alteration in transcript accumulation could be attributed to the activity of FACs in Manduca spp. R. Treatment of N. attenuata with R of two closely related herbivore species, M. sexta and M. quinquemaculata, which had similar FACs profile (Halitschke et al., 2001), induced similar expression patterns (Fig. 1). Treatment of wounds with only two of the eight FACs found in Manduca spp. R, elicited more than 55% of the response (Fig. 2). The total influence of FACs in the herbivore-induced transcriptional reorganization is likely underestimated in our study, because the applied FACs represent only a limited portion of the total FAC bouquet of R (Halitschke et al., 2001). Furthermore, the synthetic FACs were applied in a chemical environment differing (in pH, matrix constituents, etc.) from that found in natural R collected from larvae. The large number of coregulated genes in response to Manduca spp. R and FACs points to the existence of an unknown trans-activating factor in N. attenuata that responds to FACs and organizes the transcriptional response of the plant. The identification of the putative Manduca spp.-recognition element would represent a major milestone in understanding plant-insect interactions.
Changes in transcript accumulation elicited by GOX-produced H2O2 do not correlate with R-induced responses (Fig. 3). The majority of genes elicited by H2O2 treatment had the opposite patterns of regulation as was elicited by Manduca spp. R and FAC treatments, suggesting the activation of different signaling cascades. Therefore, a signaling cascade suggested for activation of defense genes in tomato, involving H2O2 downstream of octadecanoid and systemin signaling (Orozco-Cardenas et al., 2001) is unlikely to mediate the activation of Manduca spp.-specific defense responses in N. attenuata.
MATERIALS AND METHODS
Plant Growth
Nicotiana attenuata Torr. Ex W. (seven times inbred line of seeds collected from the DI ranch, Santa Clara, UT) seeds were germinated in smoke-treated soil and plants were grown in individual 1-L hydroponic chambers as previously described (Hermsmeier et al., 2001). After 10 to 14 d of growth in 1-L hydroponic chambers, plants received additional 7 mg of N as KNO3 and were randomly assigned to treatment groups 24 h before starting treatments. All plants were grown in a growth room under the following conditions: 28°C/16 h light, 25°C/8 h dark, and 800 to 1,000 μmol m−2 s−1 PAR at plant height from high-pressure sodium lamps.
Treatments
To simulate herbivore damage, the second fully expanded leaf of rosette stage N. attenuata plants was treated in all experiments. For plants used in the cDNA-AFLP analysis, one row of puncture wounds was created on each leaf half with a pattern wheel (Dritz, Spartanburg, SC), and 5 μL of either deionized water or a 1:10 (v:v) dilution of M. sexta oral secretion and regurgitant (R) was applied to the fresh wounds. The treatment was repeated two times at 20-min intervals to create a total of three rows of puncture wounds on each leaf half. The treated leaf of 15 individual plants was harvested 20 min after the final treatment and pooled together for RNA extraction.
For the microarray analysis, the second fully expanded leaf of 10 individual rosette stage N. attenuata plants was wounded by creating three rows of puncture wounds on each leaf half as described for the cDNA-AFLP treatment, and 20 μL of the different treatment solutions was applied to the fresh wounds. Treatment solutions were the following: deionized water, 1:1 (v:v) dilutions of R collected from 3rd to 4th instar M. sexta and M. quinquemaculata larvae, or a synthetic mixture of the two most abundant FACs in Manduca spp. R, N-linolenoyl-l-Gln and N-linolenoyl-l-Glu (Halitschke et al., 2001). The FACs were dissolved at R-equivalent concentrations in an aqueous solution of 0.005% (w/v) Triton X-100 (Fluka, Buchs, Switzerland) and diluted 1:1 (v:v) with deionized water before the treatment. To control for possible Triton effects, we applied a 0.0025% (w/v) Triton solution to wounds as a control. The treated leaves from 10 replicate plants were harvested at 1 and 10 h after the treatment and flash frozen in liquid nitrogen.
To generate H2O2 in situ, GOX and Glc solutions were injected into unwounded leaves (Orozco-Cardenas et al., 2001). Glc (25 mm) and GOX (from Aspergillus niger; 50 units mL−1) were introduced to the leaf in phosphate buffer P (20 mm sodium phosphate, pH 6.5) by pressing a 1-mL syringe onto the leaf surface and twice injecting 200 μL. Control plants received 2× 200-μL injections of buffer P without enzyme and Glc. The treated leaf of 10 individually treated plants in the rosette stage of growth was harvested and flash frozen in liquid nitrogen. Pooled samples were stored at −80°C until RNA extraction.
cDNA-AFLP
RNA was extracted as described by Hermsmeier et al. (2001) and mRNA isolated from 100 μg of total RNA using magnetic beads (Hui et al., 2003). RNA from R-treated plants was used as the tester sample, whereas RNA from wounded and water-treated plants was used as the driver. First strand was synthesized with SuperScript II RNase H− reverse transcriptase (Invitrogen, Groningen, The Netherlands), and the second strand synthesized with DNA Pol I (NEB, Beverly, MA) and dsDNA blunt-ended with T4 DNA polymerase (NEB). The blunt-ended DNA was extracted with phenol/chloroform, precipitated, and processed following the procedures of Bachem et al. (1998), with the modification that EcoRI/MseI restriction enzymes were used. The adaptors were created by annealing the following primer pairs: 5′-CTAACAAGATCTACTCTAGGGCCTCGTAGACTGCGTACC-3′ and 3′-CATCTGACGCATGGTTAA-5′ for EcoRI; 5′-CTAACAAGATCTACTCTAGGGCGACGATGAGTCCTGAG-3′ and 3′-TACTCAGGACTCAT-5′ for MseI. Samples were double digested with EcoRI/MseI (NEB) and used according to ligation, hybridization, and amplification procedures described by Bachem et al. (1998). The PCR fragments amplified with EcoRI/MseI adaptor-specific primers (5′-CTCGTAGACTGCGTACCAATT-3′ and 5′-GACGATGAGTCCTGAGTAA-3′) were cloned into pCR2.1-TOPO vector (Invitrogen) and sequenced on a ABI Prism 377 XL DNA sequencer with the Big Dye terminator kit (PE-Applied Biosystems, Weiterstadt, Germany), and analyzed with the Lasergene software package (DNASTAR, Madison WI).
Fabrication of cDNA Microarray
The cDNAs cloned in the pCR2.1-TOPO and pUC18 vectors (Hermsmeier et al., 2001; Hui et al., 2003) were PCR amplified using the following primers derived from vector sequences close to the insert: TOP9-22 (5′-CTAGTAACGGCCGCCAGTGTGC-3); TOP10-24 (5′-CGCCAGTGTGATGGATATCTGCAG-3′); SMA1-19 (5′-GAATTCGAGCTCGGTACCC-3′); SMA4-23 (5′-CAGGTCGACTCTAGAGGATCCCC-3′); SMA3-22 (5′-TACGAATTCGAGCTCGGTACCC-3′); and SMA2-20 (5′-GTCGACTCTAGAGGATCCCC-3′). For pCR2.1-TOPO, TOP10-24 and TOP9-22 were used. For pUC18, primer pairs SMA3-22 and SMA2-20 and SMA1-19, and SMA4-23 were used. The N. attenuata control gene PCR products to be spotted onto the chip were synthesized as follows (primer sequences and templates were described previously [Hui et al., 2003]): pi, hpl, pmt1, aos, xet, and wrky with primers ASV5-21, ASV6-22, templates pNATPI1, pNATHPL1, pNATPMT1, pNATAOS1, pNATXET1, and pNATTFN1, respectively; 3′ region of lox with primers LOX4-22, ASV6-22, and template pNATLOX1; and 5′ region of lox with primers ASV5-21, LOX3-21, and template pNATLOX1. For each cDNA, two PCR fragments, with 5′-Aminolink C6 modification (Sigma-ARK, Darmstadt, Germany) on either strand, were synthesized. Even numbered fragments (Table I) carry the Aminolink modification at primers TOP9-22, SMA4-23, or ASV6-22, whereas odd numbered fragments carry the modification at primers TOP10-24, SMA3-22, or ASV5-21. PCR products were purified by a PCR purification kit (QIAquick, Qiagen, Hilden, Germany) following the manufacturer's instructions. Agarose gel electrophoresis was performed to confirm the purity and to determine the concentration of the amplified products. Commercially available epoxy-coated slides (Quantifoil Micro Tools GmbH, Jena, Germany) were used. Before spotting, all of the cDNA samples were purified through a micron-MultiScreen-PCR (Millipore, Bedford, MA) and concentrated to approximately 0.3 to 0.6 μg μL−1 in 1× QMT Spotting Solution I (Quantifoil Micro Tools GmbH). All cDNA samples, including the seven well-characterized Manduca spp.-induced genes as controls, were commercially spotted four times by Quantifoil Micro Tools GmbH according to their procedure on the slides using a robot equipped with six printing tips (Biorobotics MicroGrid II Microarrayer, Genemachine, Apogent Discoveries, Hudson, NH). Hence each gene was represented on the microarray by two independent PCR fragments that, in turn, were spotted in quadruplicate. A complete list of identities and positions of spotted PCR products on the microarray can be found at www.plantphysiol.org. After processing, sample slides were hybridized with 9-mer random primers 5′-labeled with Cy3 and Cy5, respectively, to examine qualitative and quantitative characteristics of the microarrays.
Microarray Hybridization and Quantification
Pooled leaf samples were ground under liquid nitrogen, and total RNA was extracted as described by Winz and Baldwin (2001). To exclude the unspecific wound response, we hybridized cDNA probes derived from plants that received the same mechanical damage but different treatment solutions. The R-, FAC-, and H2O2- treated samples served as treatment (Cy3) and the water- and Triton-treated samples, respectively, were labeled and hybridized as controls (Cy5).
Poly(A+) RNAs were isolated from 400 μg of total RNA with Dynabeads Oligo(dT)25 (Dynal Biotech, Oslo) and used for reverse transcription. To synthesize the first strand, 2 μg of poly(A+) RNAs was mixed with 4 μg of random hexamer oligonucleotide and 2 μg of oligonucleotide (dT)21 in 15.5 μL and incubated at 65°C for 10 min. Subsequently, 0.6 μL of 50× 5-(3-aminoallyl)-2′-dUTP sodium salt/dNTPs (42.5 μL of each 100 mm dATP, dGTP, and dCTP; 25.5 μL of 100 mm dTTP; 17 μL of 100 mm 5-(3-aminoallyl)-2′-dUTP sodium salt (Sigma); 6 μL of 5× first-strand buffer (Invitrogen); 3 μL of dithiothreitol (0.1 m); 1.9 μL of SuperScript II RNase H− reverse transcriptase (Invitrogen); and 3 μL of water were added to a volume of 30 μL and incubated at 42°C for 2 h. cDNA/mRNA hybrids were hydrolyzed with 10 μL of NaOH (1 n) and 10 μL of EDTA (0.5 m) and incubated at 65°C for 15 min, followed by neutralization with 25 μL of 1 m Tris, pH 7.4.
The cDNA mixtures were cleaned with a Microcon 30 concentrator (YM-30, Millipore) and dried in a vacuum concentrator (Eppendorf, Hamburg, Germany). The pellets of both induced and control sample were resuspended in 9 μL of NaHCO3 buffer (0.5 m, pH 9.0), added to the dried aliquot of monofunctional N-hydroxysuccinimide-ester Cy3 dye and to Cy5 dye (Amersham Biosciences, Little Chalfont, UK), respectively, for labeling at room temperature in darkness. After 1.5 h, the Cy3 and Cy5 reactions were quenched with 4.5 μL of hydroxylamine (4 m). After purification with QIAquick PCR purification kit (Qiagen), concentration and labeling efficiency of the purified cDNA was checked spectrophotometrically, and samples were dried in a vacuum concentrator (Eppendorf).
The probe solution was prepared by resuspending the dried pellets in 3 μL of water, mixing them, and adding 20 μL of polyadenylic acid (Sigma) and 2.5 μg of yeast tRNA (Sigma). After heating at 95°C for 2 min, 90 μL of Quantifoil Hybridization buffer was added. The probe solution was placed onto the chip prepared according to the Quantifoil protocol. Hybridization was carried out for 16 h in a wet hybridization chamber at 55°C. After hybridization, the slides were immediately washed at room temperature, initially with a solution of 2× SSC and 0.2% (w/v) SDS for 10 min, and then with 2× SSC and 0.2× SSC solutions for 10 min each, before being dried in a 3.5-bar nitrogen stream.
An array scanner (428, Affymetrix, Inc., Santa Clara, CA) was used to scan the hybridized microarrays with sequential scanning for Cy5- and then for Cy3-labeled cDNA at a maximum resolution of 10 μm pixel−1 with a 16-bit depth. The images were evaluated using the program AIDA Image Analyzer (Raytest Isotopenmessgräte GmbH, Straubenhardt, Germany). Each image was overlaid with a grid to assess the signal strength for both dyes from each spot. The background correction was calculated with the “non spot” mode of the AIDA software package.
To calculate a microarray-specific normalization factor, the measured Cy5 and Cy3 fluorescence intensities were ranked independently, and after discarding of the 12.5% maximum and minimum values, the remaining 75% of the values were summed. The array-specific normalization factor was obtained by dividing the calculated sum of Cy3 values by those of the Cy5 values. This procedure excludes the confounding effects of massively down- and up-regulated transcripts (the 12.5% at either end of the distribution) from the normalization procedure. The ratios of normalized fluorescence values for Cy3 and Cy5 of each individual spot (ER) and the mean of the four replicate spots for each cDNA (two for each gene = ER1, ER2) were calculated. Subsequently, log-transformed ERs with a hypothetical mean of 0, corresponding to an ER of 1, were subjected to an untailed t test (P < 0.05). A transcript was defined as being differentially regulated if both of the following criteria were fulfilled: (a) Both individual ERs (ER1 and ER2) were equal to or exceeded the arbitrary thresholds for differential expression (0.75 and 1.25) representing 25% down- and up-regulation, respectively; and (b) both individual ERs were significantly different from 1 as evaluated by t-tests to control for ER-variance and ER-sample size.
The use of statistically rigorous criteria to evaluate the within-array variance allowed us to use lower thresholds with this polyepoxide microarray in comparison with the poly-Lys microarray used in the companion paper (Hui et al., 2003). In addition, the analysis of within-array variance provides valuable information about the quality of the mRNA used in the hybridization and the effects of microarray age (M. Held, K. Gase, and I.T. Baldwin, unpublished data). Moreover, an ER calculated as a mean of replicate ERs (rather than a single value) allows one to use lower arbitrary thresholds with greater confidence.
To evaluate these criteria, we hybridized two arrays with the same two cDNA pools (R. Halitschke and I.T. Baldwin, unpublished data) and found that 210 of 241 genes (84%) had the same regulation identified by the criteria described above. Of the 41 genes that did not show consistent regulation in the two repeat hybridizations, 24 had the same direction in mean ER, but did not meet the statistical requirements for a significant change. A complete list of all signal ratios (± se) can be found in Supplementary Table II.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matthias Held, Thomas Hahn, and Susan Kutschbach for invaluable assistance in microarray hybridization, reading and data analysis.
Footnotes
This work was supported by the Max Planck Gesellschaft.
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.018184.
LITERATURE CITED
- Alborn HT, Jones TH, Stenhagen GS, Tumlinson JH. Identification and synthesis of volicitin and related components from beet armyworm oral secretions. J Chem Ecol. 2000;26:203–220. [Google Scholar]
- Alborn T, Turlings TCJ, Jones TH, Stenhagen G, Loughrin JH, Tumlinson JH. An elicitor of plant volatiles from beet armyworm oral secretion. Science. 1997;276:945–949. [Google Scholar]
- Bachem CWB, Oomen RJFJ, Visser RGF. Transcript imaging with cDNA-AFLP: a step-by-step protocol. Plant Mol Biol Rep. 1998;16:157–173. [Google Scholar]
- Baldwin IT. The alkaloidal responses of wild tobacco to real and simulated herbivory. Oecologia. 1988;77:378–381. doi: 10.1007/BF00378046. [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Halitschke R, Kessler A, Schittko U. Merging molecular and ecological approaches in plant-insect interactions. Curr Opin Plant Biol. 2001;4:351–358. doi: 10.1016/s1369-5266(00)00184-9. [DOI] [PubMed] [Google Scholar]
- Bennissan G, Weiss D. Developmental and hormonal-regulation of a triosephosphate isomerase gene in Petuniacorollas. J Plant Physiol. 1995;147:58–62. [Google Scholar]
- Butt A, Mousley C, Morris K, Beynon J, Can C, Holub E, Greenberg JT, Buchanan-Wollaston V. Differential expression of a senescence-enhanced metallothionein gene in Arabidopsis in response to isolates of Peronospora parasitica and Pseudomonas syringae. Plant J. 1998;16:209–221. doi: 10.1046/j.1365-313x.1998.00286.x. [DOI] [PubMed] [Google Scholar]
- Gao JW, Kim SR, Chung YY, Lee JM, An GH. Developmental and environmental-regulation of 2 ribosomal- protein genes in tobacco. Plant Mol Biol. 1994;25:761–770. doi: 10.1007/BF00028872. [DOI] [PubMed] [Google Scholar]
- Giritch A, Ganal M, Stephan UW, Baumlein H. Structure, expression and chromosomal localisation of the metallothionein-like gene family of tomato. Plant Mol Biol. 1998;37:701–714. doi: 10.1023/a:1006001701919. [DOI] [PubMed] [Google Scholar]
- Halitschke R, Kessler A, Kahl J, Lorenz A, Baldwin IT. Ecophysiological comparison of direct and indirect defenses in Nicotiana attenuata. Oecologia. 2000;124:408–417. doi: 10.1007/s004420000389. [DOI] [PubMed] [Google Scholar]
- Halitschke R, Schittko U, Pohnert G, Boland W, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: III. Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol. 2001;125:711–717. doi: 10.1104/pp.125.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberg M, Sanz A, Castresana C. Alpha-oxidation of fatty acids in higher plants: identification of a pathogen-inducible oxygenase (PIOX) as an alpha-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. J Biol Chem. 1999;274:24503–24513. doi: 10.1074/jbc.274.35.24503. [DOI] [PubMed] [Google Scholar]
- Hermsmeier D, Schittko U, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: I. Large-scale changes in the accumulation of growth- and defense-related plant mRNAs. Plant Physiol. 2001;125:683–700. doi: 10.1104/pp.125.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D, Javeed I, Lehmann K, Gase K, Saluz HP, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: V. Microarray analysis and further characterization of large-scale changes in the accumulations of herbivore-induced mRNAs. Plant Physiol. 2003;131:1877–1893. doi: 10.1104/pp.102.018176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl J, Siemens DH, Aerts RJ, Gabler R, Kuhnemann F, Preston CA, Baldwin IT. Herbivore-induced ethylene suppresses a direct defense but not a putative indirect defense against an adapted herbivore. Planta. 2000;210:336–342. doi: 10.1007/PL00008142. [DOI] [PubMed] [Google Scholar]
- Keinanen M, Oldham NJ, Baldwin IT. Rapid HPLC screening of jasmonate-induced increases in tobacco alkaloids, phenolics, and diterpene glycosides in Nicotiana attenuata. J Agric Food Chem. 2001;49:3553–3558. doi: 10.1021/jf010200+. [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT. Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol. 2002;53:299–328. doi: 10.1146/annurev.arplant.53.100301.135207. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Bauwe H. Serine hydroxymethyltransferase from Solanum tuberosum. Plant Physiol. 1995;107:271–272. doi: 10.1104/pp.107.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne E, Mathieu C, Vedel F, De Paepe R. Tissue-specific expression of genes encoding isoforms of the mitochondrial ATPase β-subunit in Nicotiana sylvestris. Plant Mol Biol. 1998;38:885–888. doi: 10.1023/a:1006088308544. [DOI] [PubMed] [Google Scholar]
- Mattiacci L, Dicke M, Posthumus MA. Beta-glucosidase: an elicitor of herbivore-induced plant odor that attracts host-searching parasitic wasps. Proc Natl Acad Sci USA. 1995;92:2036–2040. doi: 10.1073/pnas.92.6.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloud ES, Baldwin IT. Herbivory and caterpillar regurgitants amplify the wound-induced increases in jasmonic acid but not nicotine in Nicotiana sylvestris. Planta. 1997;203:430–435. [Google Scholar]
- Musser RO, Hum-Musser SM, Eichenseer H, Peiffer M, Ervin G, Murphy JB, Felton GW. Herbivory: Caterpillar saliva beats plant defences. A new weapon emerges in the evolutionary arms race between plants and herbivores. Nature. 2002;416:599–600. doi: 10.1038/416599a. [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas ML, Narvaez-Vasquez J, Ryan CA. Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell. 2001;13:179–191. [PMC free article] [PubMed] [Google Scholar]
- Pohlmeyer K, Soll J, Steinkamp T, Hinnah S, Wagner R. Isolation and characterization of an amino acid-selective channel protein present in the chloroplastic outer envelope membrane. Proc Natl Acad Sci USA. 1997;94:9504–9509. doi: 10.1073/pnas.94.17.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohnert G, Jung V, Haukioja E, Lempa K, Boland W. New fatty acid amides from regurgitant of lepidopteran (Noctuidae, Geometridae) caterpillars. Tetrahedron Lett. 1999;55:11275–11280. [Google Scholar]
- Roda A, Oldham NJ, Svatos A, Baldwin IT (2003) Allometric analysis of the induced flavonols on the leaf surface of wild tobacco (Nicotiana attenuata). Phytochemistry (in press) [DOI] [PubMed]
- Sanz A, Moreno JI, Castresana C. PIOX, a new pathogen-induced oxygenase with homology to animal cyclooxygenase. Plant Cell. 1998;10:1523–1537. doi: 10.1105/tpc.10.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittko U, Hermsmeier D, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: II. Accumulation of plant mRNAs in response to insect-derived cues. Plant Physiol. 2001;125:701–710. doi: 10.1104/pp.125.2.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittko U, Preston CA, Baldwin IT. Eating the evidence? Manduca sexta larvae cannot disrupt specific jasmonate induction in Nicotiana attenuataby rapid consumption. Planta. 2000;210:343–346. doi: 10.1007/PL00008143. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchishinozaki K et al. The complete nucleotide-sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlings TCJ, Alborn HT, Loughrin JH, Tumlinson JH. Volicitin, an elicitor of maize volatiles in oral secretion of Spodoptera exigua: isolation and bioactivity. J Chem Ecol. 2000;26:189–202. [Google Scholar]
- Voelckel C, Baldwin IT (2003) Detecting herbivore-specific transcriptional responses in plants with multiple DDRT-PCR and subtractive library procedures. Physiol Plant (in press)
- Winz RA, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: IV. Insect-induced ethylene reduces jasmonate-induced nicotine accumulation by regulating putrescine N-methyltransferase transcripts. Plant Physiol. 2001;125:2189–2202. doi: 10.1104/pp.125.4.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.