Abstract
In comparison with retrotransposons, which comprise the majority of the Triticeae genomes, very few class 2 transposons have been described in these genomes. Based on the recent discovery of a local accumulation of CACTA elements at the Glu-A3 loci in the two wheat species Triticum monococcum and Triticum durum, we performed a database search for additional such elements in Triticeae spp. A combination of BLAST search and dot-plot analysis of publicly available Triticeae sequences led to the identification of 41 CACTA elements. Only seven of them encode a protein similar to known transposases, whereas the other 34 are considered to be deletion derivatives. A detailed characterization of the identified elements allowed a further classification into seven subgroups. The major subgroup, designated the “Caspar ” family, was shown by hybridization to be present in at least 3,000 copies in the T. monococcum genome. The close association of numerous CACTA elements with genes and the identification of several similar elements in sorghum (Sorghum bicolor) and rice (Oryza sativa) led to the conclusion that CACTA elements contribute significantly to genome size and to organization and evolution of grass genomes.
All genomes contain repetitive elements and in some species, such elements comprise the majority of the nDNA. Repetitive elements can be divided into two main groups: class 1 and class 2 elements. Class 1 elements (also called retrotransposons) replicate via an mRNA intermediate that is reverse transcribed into DNA and integrated somewhere else in the genome. Retrotransposons contribute a large fraction to the total genomic DNA of plants with large genomes such as wheat, barley (Hordeum vulgare), or maize (Zea mays; SanMiguel and Bennetzen, 1998; Shirasu et al., 2000; Wicker et al., 2001; SanMiguel et al., 2002). Class 2 elements or transposons move via a DNA intermediate, which means that the elements are excised from the genome and integrated elsewhere. Excision and reintegration require an enzyme known as transposase. Transposons have been subdivided into several families. One of them, called the CACTA family, received its name because it is flanked by inverted repeats that terminate in a conserved CACTA motif. En-1 (also known as Suppressor-mutator or Spm) from maize was the first CACTA element that was analyzed at the molecular level (Pereira et al., 1986). En/Spm elements are present as autonomous elements that encode the proteins necessary for their transposition and deletion derivatives, which are nonautonomous. The nonautonomous elements depend for their transposition on enzymes encoded by the autonomous copies (Bennetzen, 2000). Active CACTA elements were isolated and characterized from a variety of species including CAC1 from Arabidopsis (Miura et al., 2001), PsI from petunia (Petunia hybrida; Snowden and Napoli, 1998), Tdc1 from carrot (Daucus carota; Ozeki et al., 1997), Tam-1 from snapdragon (Antirrhinum majus; Nacken et al., 1991), Tpn1 from Japanese morning glory (Ipomoea nil Inagaki et al., 1994), and Candystripe1 from sorghum (Sorghum bicolor; Chopra et al., 1999). Candystripe1 is believed to be a nonautonomous element because it does not encode a protein similar to known transposases.
The terminal regions of all identified CACTA elements show a similar sequence organization. They are flanked by short terminal inverted repeats (TIRs) of 10 to 28 bp in size that terminate in the CACTA motif. These serve as recognition sequences for the transposase protein (Lewin, 1997). In most cases, sequence conservation between the different families is limited to this short motif, which makes it virtually impossible to identify new elements based on the TIR sequences of known elements. In addition, CACTA elements contain sub-terminal repeats (TRs) that consist of 10- to 20-bp units that are repeated in direct and inverted orientation. As for the TIRs, these units also show no significant sequence conservation between different families. Therefore, CACTA transposons are difficult to identify and usually are only found because of the presence of a transposase-like protein.
Diploid Triticeae spp. such as barley or Triticum monococcum have genome sizes of more than 5,000 Mb and contain approximately 80% of repetitive DNA (Smith and Flavell, 1975; Bennet and Leitch, 1995). This high percentage of repetitive sequences has so far prevented them from becoming the focus of large-scale genomic sequencing projects. In recent years, however, a number of bacterial artificial chromosome (BAC) clones from Triticeae spp. were completely sequenced and to date, approximately 1.6 Mb of large contiguous stretches of genomic sequences are publicly available. Analysis of these sequences revealed that a large fraction of the repetitive DNA is comprised of retrotransposons (Shirasu et al., 2000; Wicker et al., 2001; Rostoks et al., 2002; SanMiguel et al., 2002; Wei et al., 2002), whereas class 2 elements were identified only in very few cases (Dubcovsky et al., 2001; Feuillet et al., 2001; Wei et al., 2002). So far, only one CACTA transposon from Triticeae has been described in detail (TAT-1; Feuillet et al., 2001). Therefore, it was assumed that this element class is present in a very limited copy number in the Triticeae genomes.
Recent analysis of the Glu-A3 loci in diploid and tetraploid wheat revealed the presence of 12 different CACTA transposons (Wicker et al., 2003). Interestingly, only four of these elements encode transposase proteins similar to those of previously described transposons. Eight of the 12 transposons were apparently deletion derivatives because they have no obvious coding capacity. Five of the deletion derivatives were designated as small nonautonomous CACTA (SNAC) transposons because their small size (700 bp–1.5 kb) clearly distinguished them from all other identified elements. The other three deletion derivatives range in size from 5 kb up to 11.3 kb.
The objective of our study was to characterize the previously described CACTA elements from wheat and to identify new Triticeae elements present in the public databases. Here, we report the identification and characterization of 41 novel CACTA transposons from Triticeae. Our results indicate that this transposon class is present at a high copy number in the wheat genome and that a large number are deletion derivatives. Elements similar to the ones in Triticeae were found in rice (Oryza sativa) and sorghum, indicating that also these genomes contain a wide variety of CACTA elements.
RESULTS
Identification of CACTA Transposons by BLAST Search and Dot-Plot Analysis
Because only a minority of the CACTA transposons were expected to actually encode a transposase-like protein, a first approach for the identification of new elements was based on their TR sequences. The TR regions that contain the TIRs and the sub-TRs usually have a size of 200 to 500 bp. In this study, the term “element with complete ends” was used for elements in which both TIRs contain an intact CACTA motif and are flanked by a 3-bp target site duplication. They were distinguished from elements truncated by deletions or elements with damaged ends (referred to as “truncated elements”).
Ten of the 12 CACTA elements with complete ends identified on the Glu-A3 contigs (Wicker et al., 2003) showed conserved sequence motifs within their TR regions. These 10 elements, the previously described TAT-1 (Feuillet et al., 2001) and another recently identified CACTA element from barley (Caspar_AF521177-1; Brunner et al., 2003) were used to derive a 127-bp TR consensus sequence. This was used as a query sequence for a BLAST search of public databases and the database for Triticeae repetitive sequences (Triticeae REPeat sequence database, http://wheat.pw.usda.gov/ITMI/Repeats; Wicker et al., 2002). Ten new CACTA elements were found in genomic DNA sequences from Triticeae, six of which are elements with complete ends, whereas the other four were either truncated or only partially covered by the sequence deposited in the databases. In addition, nine Triticeae expressed sequence tags (ESTs) that contain TR sequences were identified. The presence of TR sequences in ESTs was interpreted as the result of transposon insertions close to genes. These were distinguished from EST sequences of the actual transcripts of the coding sequences of transposase-like proteins (see below). The transcript of transposon genes starts some 100 bp downstream of the TR region; therefore, it does not include the TR sequences. For one EST (accession no. BF618436), BLASTX search revealed that the CACTA element has presumably inserted in the 3′-untranslated region. In two cases, the element was inserted into the coding region of a Gag-Pol polyprotein (accession nos. BJ247168 and BJ253225).
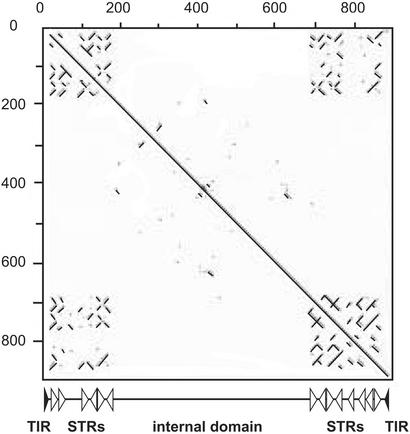
It was clear that the consensus TR would not identify CACTA elements that contain divergent TRs. Therefore, a second approach for the identification of new elements was based on their structural similarity rather than on sequence conservation: The subterminal direct and inverted repeats displayed a specific pattern when the transposon sequence is plotted against itself with dot plot (program DOTTER; Sonnhammer and Durbin, 1995). The example in Figure 1 shows a dot plot of an SNAC element from Triticum aestivum (Caspar_AF234649-1). Typically, the short TIRs are immediately followed by a variable number of sub-TRs. In the case of the Caspar_AF234649-1 element, they consist of direct and inverted repeats of a conserved 15-bp motif (CCTTTAGTCCCGGTT) that produce the characteristic “transposon signature.” All transposons analyzed in this study contain sub-TRs within their TR sequences. Most elements contain two to six repeat units. Usually, the number of repeat units at one end differs from the number at the other end. A set of 15 publicly available large genomic Triticeae sequences and the two sequences from T. monococcum and Triticum durum (Wicker et al., 2003) were collected in a local database with a total size of 1.9 Mb. This database was hereafter screened by dot plot for the occurrence of transposon signatures. This second approach led to the identification of six further CACTA elements with complete ends from genomic sequences.
Figure 1.
Dot plot of an SNAC transposon. The sequence of Caspar_AF234649-1 is graphically compared with itself. The main diagonal line corresponds to the 100% match when the sequence is plotted against itself. Direct repeats are lines parallel to the diagonal line, and inverted repeats are displayed as lines perpendicular to the diagonal line. The sub-TRs produce a very specific pattern (“transposon signature”) that can be easily recognized. The structure of the transposon is depicted below the dot plot. Black triangles, TIRs; white triangles, sub-TRs (STR).
In total, the database mining resulted in the identification of 16 new Triticeae CACTA transposons from genomic sequences and nine from EST sequences. None of the 16 new elements found in genomic sequences had been annotated as such. It is likely that they were not recognized because none of them encodes a transposase protein. As it was previously described for retrotransposons in Triticeae, the CACTA elements were often found as nested insertions in other class 1 or class 2 elements.
Two additional elements (Jorge_TREP766 and Caspar_TREP788) were kindly provided by Dr. Jorge Dubcovsky (University of California, Davis) and Dr. Nils Stein (Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany), respectively. Together with the initial 12 elements, TAT-1 (Feuillet et al., 2001) and Caspar_AF521177-1 (Brunner et al., 2003), a total of 32 CACTA elements from genomic sequences are now available. Twenty-six of them are elements with complete ends in which both TIRs are present and a 3-bp target site duplication could be identified. All elements were collected in a local database and subsequently submitted to the TREP database (accession nos. TREP746–TREP788; http://wheat.pw.usda.gov/ITMI/Repeats). The names and origins of the identified elements are summarized in Table I. The exact start and end positions of all identified elements in their source sequences are provided as supplemental material (see supplemental Table III at www.plantphysiol.org). As reference sequences, the previously described elements En-1 from maize (Pereira et al., 1986), Tam-1 from snapdragon (Nacken et al., 1991), Candystripe-1 from sorghum (Chopra et al., 1999), and an additional CACTA element from Lolium perenne (accession no. AY089999), which was found by keyword search in the EMBL database, were also included.
Table I.
List of all identified Triticeae CACTA transposons
| Name | Size | Organism | Comment | Reference |
|---|---|---|---|---|
| bp | ||||
| Caspar_107G22-1 | 1,305 | T. durum | Complete ends, SNAC | Wicker et al. (2003) |
| Caspar_107G22-2 | 5,096 | T. durum | Complete ends | Wicker et al. (2003) |
| Caspar_107G22-3 | 807 | T. durum | Fragment | Wicker et al. (2003) |
| Caspar_18B1-1ab | 9,913 | T. monococcum | Complete ends | Wicker et al. (2003) |
| Caspar_426K20-2 | 1,371 | T. monococcum | Complete ends, SNAC | Wicker et al. (2003) |
| Caspar_453N11-1ab | 12,664 | T. monococcum | Complete ends | Wicker et al. (2003) |
| Caspar_AF234649-2 | 750 | T. aestivum | Complete ends, SNAC | O.D. Anderson, C. Hsia, and V. Torres (unpublished data) |
| Caspar_AF325197-1 | 1,483 | T. aestivum | Fragment | Feuillet et al. (2001) |
| Caspar_AF325198-1 | 1,530 | T. aestivum | Complete ends, SNAC | Feuillet et al. (2001) |
| Caspar_AF427791-1b | 12,590 | Barley | Complete ends | Wei et al. (2002) |
| Caspar_AF446141-1 | 6,255 | Aegilops tauschii | Complete ends | Brooks et al. (2002) |
| Caspar_AF474072-1b | 10,540 | Barley | Complete ends | Rostoks et al. (2002) |
| Caspar_AF474373-1b | 12,446 | Barley | Complete ends | Rostoks et al. (2002) |
| Caspar_AF474373-2b | 7,376 | Barley | Complete ends | Rostoks et al. (2002) |
| Caspar_AF521177-1ab | 9,105 | Barley | Complete ends | Brunner el al. (2003) |
| Caspar_AL816166 | 284 | T. aestivum | Fragment (EST) | I. Wilson, R. Beswick, S. Shepherd, G. Barker, J. Parker, P. Owen, D. Edwards, J. Coghill, M. Holdsworth, J. Lenton et al. (unpublished data) |
| Caspar_AL816390 | 341 | T. aestivum | Fragment (EST) | I. Wilson, R. Beswick, S. Shepherd, G. Barker, J. Parker, P. Owen, D. Edwards, J. Coghill, M. Holdsworth, J. Lenton et al. (unpublished data) |
| Caspar_BF618436 | 143 | Barley | Fragment (EST) | R. Wing, T.J. Close, A. Kleinhofs, R. Wise, D. Begum, D. Frisch, Y. Yu, D. Henry, M. Palmer, T. Rambo et al. (unpublished data) |
| Caspar_TREP770b | 8,334 | A. tauschii | Complete ends | P. Langridge (unpublished data) |
| Caspar_TREP771 | 3,296 | A. tauschii | Fragment | P. Langridge (unpublished data) |
| Caspar_TREP788ab | 11,627 | Barley | Complete ends | N. Stein (unpublished data) |
| Caspar_X63357 | 569 | Barley | Fragment | Fernandez et al. (1993) |
| Caspar_Z66528-1 | 331 | Barley | Fragment | A. Molina, I. Diaz, F. Garcia-Olmedo (unpublished data) |
| Mandrake_107G22-1 | 930 | T. durum | Complete ends, SNAC | Wicker et al. (2003) |
| Mandrake_426K20-1 | 898 | T. monococcum | Complete ends, SNAC | Wicker et al. (2003) |
| Mandrake_AF234649-1 | 1,119 | T. aestivum | Complete ends, SNAC | O.D. Anderson, C. Hsia, and V. Torres (unpublished data) |
| Mandrake_AF446141-1 | 3,411 | A. tauschii | Complete ends | Brooks et al. (2002) |
| Mandrake_BG309793 | 736 | Barley | Fragment (EST) | R. Wing, T.J. Close, A. Kleinhofs, R. Wise, D. Begum, D. Frisch, Y. Yu, D. Henry, M. Palmer, T. Rambo et al. (unpublished data) |
| TAT-1_AF325196-1a | 7,190 | T. aestivum | Complete ends | Feuillet et al. (2001) |
| TAT-1_107G22-1 | 3,158 | T. durum | Fragment | Wicker et al. (2003) |
| TAT-1_AF427791−1 | 9,765 | Barley | Complete ends | Wei et al. (2002) |
| TAT-1_BJ247168 | 414 | T. aestivum | Fragment (EST) | Y. Ogihara, K. Murai (unpublished data) |
| TAT-1_BJ253225 | 514 | T. aestivum | Fragment (EST) | Y. Ogihara, K. Muirai (unpublished data) |
| Balduin_453N11-1a | 9,841 | T. monococcum | Complete ends | T. Wicker et al. (unpublished data) |
| Balduin_BJ218039 | 554 | T. aestivum | Fragment (EST) | Y. Ogihara, K. Murai (unpublished data) |
| Balduin_BQ469151 | 376 | Barley | Fragment (EST) | H. Zhang, W. Weschke, W. Michalek, N. Stein, A. Graner (unpublished data) |
| Balduin_BQ661452 | 376 | Barley | Fragment (EST) | W. Michalek, W. Weschke, K.-P. Pleissner, A. Graner (unpublished data) |
| Jorge_AF326781-1 | 16,497 | T. monococcum | Complete ends | Wicker et al. (2001) |
| Jorge_TREP766 | 14,733 | T. monococcum | Complete ends | J. Dubcovsky (unpublished data) |
| Enac_453N11-1 | 11,361 | T. monococcum | Complete ends | T. Wicker et al. (unpublished data) |
| Isaac_107G22-1a | 13,135 | T. durum | Complete ends | T. Wicker et al. (unpublished data) |
The elements are sorted according to their classification into families. The family name is given first. The source sequence (e.g. BAC clone address, GenBank no., or TREP accession no.) follows the name after an underscore. For elements from genomic sequences, a number for the individual copy of an element is indicated after a hyphen. SNAC, Small nonautonomous CACTA. The reference refers to the researcher who published the sequence in which the element was found.
Encodes a transposase.
Encodes a CTG-2 protein.
CACTA Transposons Can Be Classified Based on Their TR Sequences
Because the majority of the identified transposons have no apparent coding capacity and vary greatly in size, we decided to base their classification on the TR sequences, the only feature that all of them have in common. The 14 truncated elements contain only one intact TR each, whereas from the 26 element with complete ends, both TRs could be used. The total 66 TR sequences from Triticeae transposons were used for a multiple sequence alignment. The alignment was done with the terminal 200 bp of the elements. A phylogenetic analysis of the multiple sequence alignments allowed the classification of the TR sequences into seven distinct clades (Fig. 2A). Sequence conservation between members of different families is restricted basically to the terminal 20 to 30 bp containing the CACTA motif. The major group containing 28 TR sequences was designated the “Caspar ” family. One exclusive feature of the Caspar family is that the TR starts with a CACTAGT motif, whereas all others start with CACTAC(A/T). Three additional main families were designated Balduin, Mandrake, and TAT-1. Further similarities were discovered between Jorge_TREP766 and the previously described unclassified XB element (Wicker et al., 2001), which was called thereafter Jorge_AF326781-1. The TR sequences of Enac_453N11-1 and Isaac_107G22-1 are unique because they show no similarity to any of the other elements and groups in separate clades (Fig. 2A).
Figure 2.
Classification of Triticeae CACTA transposons based on their TR sequences. A, Classification based on multiple sequence alignment. The bootstrap values for the seven main families and the major subfamilies are indicated at the nodes of the tree. The TR sequences were aligned with PileUp and analyzed with the neighbor-joining method. B, Classification based on the dot-plot pattern. An array of six TR sequences is plotted against itself. The terminal 300 bp from one TR of an element are used for the array. Only TRs from elements belonging to the same family display the characteristic transposon signature when compared with each other, whereas TRs from different families display no signature when they are plotted against one another. C1 and C2, TRs of Caspar elements. M1 and M2, TRs of Mandrake elements. T1 and T2, TRs of TAT-1 elements.
To test this classification, a second approach for classification was based on the similarity of TR sequences displayed by dot-plot analysis: TRs from members of the same family display the characteristic transposon signature, whereas TRs of elements from different families show no signature. The terminal 300 bp of one TR from each element was used to generate a large array, which was then compared against itself by dot plot. Examples for dot-plot alignments of three different families are displayed in Figure 2B. In this approach, the classification into seven groups as it was obtained by the multiple sequence alignment could be confirmed for all elements. The results of the two classification approaches are summarized in Table I.
The CACTA Family Comprises Full-Length Elements and a Wide Variety of Deletion Derivatives
To investigate the range of diversity in size and sequence organization among members of the CACTA family, only the 26 elements with complete ends were used. Truncated elements were excluded because it is not possible to determine their actual size and coding capacity. Seven of the 26 elements with complete ends encode a transposase protein (Table I). However, all seven do not encode functional proteins because they all contain frameshifts or in-frame stop codons within their coding region (see below). In this study, we refer to elements that encode a transposase protein as “full-length elements,” even if the coding region of the transposase protein is apparently defective. Four of the seven elements encode a second protein (which we refer to as CTG-2) in addition to the transposase. The CTG-2 coding gene was only found in the members of the Caspar family (see below). All identified full-length elements are large in size, ranging from 9.9 up to 13.1 kb.
The other 19 CACTA transposons are considered to be deletion derivatives that have lost some or all of their coding capacity and depend for their transposition on enzymes encoded elsewhere in the genome. These deletion derivatives vary drastically in size: At one end of the spectrum, there are seven SNAC transposons that encode no proteins and range in size from 750 bp to 1.5 kb. The TR regions of these seven SNAC elements have sizes of 200 to 300 bp and are separated by an internal domain.
Three SNAC elements belonging to the Caspar family (Caspar_107G22-1, Caspar_426K20-2, and Caspar_AF325198-1) plus a fragment of a putative SNAC element (Caspar_107G22-3) contain a 64-bp region that is 75% to 81% identical to a part of the 5S rDNA gene (120 bp) from T. monococcum (accession no. Z11461). This region is embedded in an approximately 400-bp region that is more strongly conserved than the rest of the elements. In the 400-bp region, all four are 91% to 95% identical, whereas their overall sequence identity is 79% to 91%. The 5S derivative conserved in the four elements corresponds to the internal RNA polymerase III promoter that is involved in the recruitment of transcription factors. It includes the highly conserved motifs BoxA, IE, and BoxC (Cloix et al., 2000). In addition, three of the four elements contain a 191-bp region that is 63% to 90% identical to the spacer region of the 5S rDNA gene in Hordeum cordobense (accession no. AY034735). In total, 61 5S rDNA from barley gave strong BLASTN hits with this 191-bp region. The other three SNAC elements belong to the Mandrake family and show no obvious structure within their internal domain.
The 12 large deletion derivatives range in size from 3,411 bp up to 16.5 kb. Seven are members of the Caspar family, five of which encode a CTG-2 protein. All seven large Caspar deletion derivatives contain regions of tandem repeated DNA (see below). The other five deletion derivatives do not contain any sequences similar to known repetitive elements or genes. They also do not contain obvious structures like direct repeats, which would explain their large size. The largest deletion derivative identified is Jorge_AF326781-1, which has a size of 16,497 bp.
Elements of the Caspar Family Encode a Transposase and a Protein of Unknown Function
Four Caspar elements (Caspar_453N11-1, Caspar_18B1-1, Caspar_AF521177-1, and Caspar_TREP788) gave strong BLASTX hits with numerous transposase-like proteins from rice and sorghum. The coding region for the transposase is located in the 5′ region of the elements. All four are likely to be nonfunctional because they all contain frameshifts or in-frame stop codons within their coding regions. However, because they show a high degree of sequence conservation within the coding region of the transposase, a multiple sequence alignment allowed to determine at which positions frameshifts have to be introduced in an individual element to obtain a contiguous open reading frame. All four elements contain between one and three frameshifts and Caspar_453N11-1 and Caspar_TREP788 contain one and two in-frame stop codons, respectively. Comparison with transposase proteins from public databases helped to determine the positions of the putative start and stop codons. The four deduced transposase proteins have sizes ranging from 1,044 to 1,122 amino acids and are 73% to 79% similar to one another. The coding region does not contain any introns. The four putative proteins are 68% to 74% similar to TNP2-like proteins from rice (accession no. Q9AUX7) and from sorghum (accession no. Q9XEQ1) but only 40% to 45% similar to the transposase of En/Spm (accession no. AAA66266). The transposase genes of Caspar elements are expressed as more than 30 ESTs from Triticeae corresponding to the transposase region were found in public databases.
Nine Caspar elements contain a coding region for a second protein we refer to as CTG-2 (Caspar transposon gene 2). BLAST search of the CTG-2 region revealed similarity to 12 hypothetical proteins from rice and one from sorghum. In contrast to the transposase, which is well conserved among the different Caspar elements, the CTG-2 protein is highly variable. Therefore, it was difficult to predict a protein sequence. Based on sequence conservation between different Caspar elements and on the similarity to the proteins identified by BLASTX, putative protein sequences of eight Caspar CTG-2 proteins were deduced. The proteins have sizes of 968 to 1,292 amino acids. In all cases, they consist of one large putative first exon, which varies strongly in size between the different copies. The differences are caused by a region that contains multiple repeats of short 3- to 30-bp units, and the number of repeat units differs in the different elements. This putative first exon is followed by five short exons (25–50 amino acids) that show a higher degree of sequence conservation. The exon/intron structure of the last five exons was determined by comparison with the amino acid sequences of the 12 hypothetical proteins from rice that were identified by BLASTX. The predicted exon/intron structure of CTG-2 is strongly conserved in all analyzed elements. Eight ESTs similar to the CTG-2 region were found in public databases, indicating that the CTG-2 proteins are also expressed.
The predicted CTG-2 protein sequences show no clear homology to previously described transposon proteins. A weak similarity to previously described proteins could be shown if sequences were aligned with the GCG program BESTFIT (Genetics Computer Group, Madison, WI), and gap creation and gap extension penalties were decreased to 4 and 1, respectively. Using these parameters, all CTG-2 proteins are between 42% and 50% similar over most of their length to the TNP1 protein of Tam-1 (accession no. CAA40554) and TNPA of En/Spm (accession nos. AAG17044). However, the sequence alignments contain a large number of gaps; therefore, one can only speculate that the CTG-2 protein may represent a highly diverged homolog to TNP1 and TNPA.
CACTA Elements Contain Large Amounts of Low-Complexity DNA
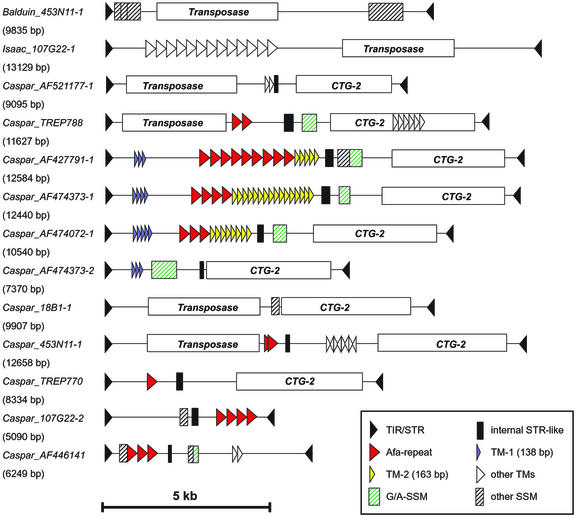
Dot-plot analysis of the identified transposons revealed that several elements contain patterns of tandem repeats of variable length and sequence. The repeated sequence units range in size from 2 to 30 up to 380 bp. A selection of 13 CACTA elements with complete ends that contain multiple different repeat structures were chosen for further analysis (Fig. 3). Eleven of them are members of the Caspar family, and the two others are Balduin_453N11-1 and Isaac_107G22-1. SNAC transposons, the large deletion derivatives Jorge_TREP766, Jorge_AF326781-1 and Enac_453N11-1, and truncated elements were excluded because they do not contain comparable repeat patterns.
Figure 3.
Repeat structures within different CACTA elements. Direct repeats larger than 100 bp are displayed as triangles. Repeat regions with shorter units are indicated as shaded boxes. TM, Tandem repeat; SSM, tandem repeats of short sequence motifs; STR, sub-TR.
The repeat regions in Balduin_453N11 and Isaac_107G22 showed no similarity to each other or to the ones from the Caspar family, whereas nine of the 11 Caspar elements share common repeat units. A surprising finding was that eight Caspar elements contain the previously described Afa repeats (Rayburn and Gill, 1986; Nagaki et al., 1998a). Afa repeats are a class of tandem repeats of approximately 340 bp in size that are believed to be present in all Triticeae spp. Their copy number, however, was shown to vary up to 100-fold in different Triticeae spp., and they were found in various, genome-specific locations in Triticeae genomes (Nagaki et al., 1998a). Copy numbers of the Afa repeats in the identified Caspar elements range from one (Caspar_TREP770) to nine (Caspar_AF427791; Fig. 3). Two further repeat types (TM-1 and TM-2) occur in three and four elements, respectively. In addition, most of the Caspar elements contain large regions (200–500 bp) of tandem repeated short sequence motifs (most often G/A-rich regions) and a region of 100 to 250 bp that is 70% to 85% identical to their sub-TRs (Fig. 3).
The tandem repeats within CACTA elements obviously can undergo rapid changes in copy number: Four Caspar elements from barley (Caspar_AF427791-1, Caspar_AF474373-1, Caspar_AF474373-2, and Caspar_AF474072-1) appear to be very closely related because they are approximately 92% to 95% identical on the DNA level. However, the most striking difference between them is the number of direct repeats (Fig. 3). Caspar_AF427791-1, for example, contains three copies of TM-1, nine Afa repeats, and five copies of TM-2, whereas Caspar_AF474373-1 contains four TM-1 units, four Afa units and 16 TM-2 units. In contrast, Caspar_AF747373-2 contains only four TM-1 repeats but neither Afa nor TM-2 repeats (Fig. 3).
The Caspar Family Is Present at a High-Copy Number in the Wheat Genome
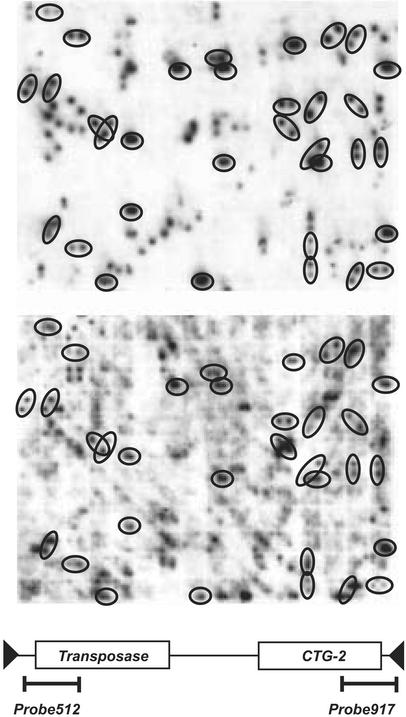
The fact that the transposons of the Caspar family were found in several copies in the publicly available sequences suggested that this elements may occur very frequently in Triticeae genomes. To estimate the copy number of the Caspar transposons, one high-density filter (Filter C) from the T. monococcum BAC library (Lijavetzky et al., 1999) was hybridized with two different probes. One high density filter contains 18,432 BAC clones that cover approximately 0.4 genome equivalents. The first probe (Probe512) was chosen in the 5′ region of the transposase-coding region of the Caspar_453N11-1 element, and the second one (Probe917) covers the 3′ region of CTG-2 of Caspar_453N11. These two probes allowed the determination of how many elements contain both proteins and how many contain only one of the two. The hybridization pattern of both probes from a small region of filter C is shown in Figure 4. Probe512 and Probe917 identified 672 and 795 BAC clones, respectively, and 292 BACs gave signals with both probes. These numbers were extrapolated to one genome equivalent (multiplied by 2.5). From these data, we estimate that the wheat genome contains a minimum of 2,900 copies of the Caspar elements. About 25% of them contain both the transposase and the CTG-2 region. Approximately 950 copies contain only a transposase, and 1,250 copies contain only CTG-2. If one takes the average size of the nine Caspar transposons that encode one of the two proteins (10.5 kb), the roughly 3,000 Caspar elements might contribute approximately 0.6% to the T. monococcum genome. As shown above, many Caspar elements contain neither of the two proteins and are excluded from this estimate. It also has to be considered that the estimated copy number from the hybridization data was based on the assumption that each BAC clone that gave a signal contains only one Caspar element. Therefore, the actual number of Caspar-like transposons in the wheat genome might be considerably higher.
Figure 4.
Estimation of the copy number of Caspar elements in the T. monococcum genome. One BAC filter was hybridized with two different probes corresponding to the transposase (top) and CTG-2 regions (bottom) from Caspar_453N11-1, respectively. The fraction of the filter shown corresponds to approximately 3.3% of a genome equivalent. BAC clones that hybridized with both probes are indicated with circles.
Caspar-Like Elements Are Also Frequently Found in Other Grass Genomes
The apparently high copy number of Caspar elements in Triticeae genomes inspired the search for similar elements in other grass genomes. Three BACs from rice and one from sorghum encoding the proteins that gave the strongest BLASTX hits with CTG-2 from Caspar were screened for the presence of transposon-like sequences. In all four cases, an annotated transposase protein was found upstream of the protein that gave the BLASTX hit with CTG-2, but transposase and CTG-2 were not annotated as belonging to the same element. In all four cases, CTG-2 was annotated as a putative gene. The predicted exon/intron structure as it was annotated in the publicly available sequences differed slightly from our prediction of the structure of CTG-2. However, comparison with our predicted proteins from the Triticeae elements showed that that the same exon/intron structure also can be found in the elements from rice and sorghum, although the proteins from the different species were only about 46% to 50% similar to one another.
Two proteins from rice BACs AP002484 and AP003020 and one from sorghum BAC AF114171 were deduced by applying our predicted exon/intron structure and used as query sequences for a TBLASTN search. The number of hits was striking: CTG-2_AP002484 and CTG-2_AP003020 gave 218 and 214 hits in rice, respectively, with E values below 3E-4. CTG-2_AF114171 identified five putative CTG-2 proteins in sorghum (E value = 0.0).
Using dot plot, the actual borders of the elements on the rice and sorghum BACs were identified, and four Caspar-like elements with complete ends could be characterized. In addition, the four BAC clones were searched for further transposon signatures by dot plot, which led to the identification of two additional SNAC transposons (one from rice BAC AP002484 and one from sorghum BAC AF114171), both of which were not annotated. The positions of the elements on their respective BAC clones are shown in Table II.
Table II.
Examples of CACTA elements from rice and sorghum
| BAC Clone | Size | Positions | Organism |
|---|---|---|---|
| bp | |||
| AC079029ab | 10,544 | 51,199–64,657 | Rice |
| AP002484a | 10,545 | 15,795–26,339 | Rice |
| AP002484ac | 1,210 | 9,911–11,120 | Rice |
| AP003020a | 11,280 | 111,118–122,397 | Rice |
| AP003446c | 274 | 57,711–57,984 | Rice |
| AF114171a | 12,722 | 109,076–121,797 | Sorghum |
| AF114171ac | 1,716 | 82,698–84,413 | Sorghum |
Positions of the elements on the BAC clone are indicated.
Used for the initial BLASTN search.
Contains insertions of repetitive elements.
SNAC transposon.
All sequences identified in this way were used for a next round of BLASTN search against the National Center for Biotechnology Information nonredundant database to obtain a rough estimate of the abundance of these elements in the rice and sorghum genomes. This search revealed the presence of a very high number of similar elements in the genomes of rice and sorghum, ranging from 493 hits for SNAC_ AP002484-1 up to 824 hits for the CACTA element from rice BAC AP003020 that contains both a transposase and CTG-2. E values for all these BLASTN hits were below 3E-4. The CACTA element from sorghum BAC AF114171 identified four elements in sorghum (E value = 0.0). Because the focus of this study was not a complete survey of rice CACTA elements but to study their structure and sequence organization, we focused our attention on the isolation of a small number of elements with complete ends. The result of the database mining was a set of 18 CACTA elements from rice and six elements from sorghum. The precise location of all identified rice and sorghum elements on their source sequences is provided as supplemental material (Table III). Interestingly, only one additional element that encodes proteins was identified, and all others were SNAC transposons. None of the SNAC transposons had been annotated as such. These data suggest that the rice genome might contain a very large number of yet undiscovered CACTA elements and that the majority of them might be small nonautonomous elements. A very interesting finding in this context is SNAC_AP003446-1 from rice, which at 274 bp is the smallest element identified in this study (Table II). It is the only element that does not contain an internal domain but consists exclusively of terminal and sub-TR sequences.
DISCUSSION
Why Were the CACTA Elements in Triticeae Not Discovered Earlier?
The high density of CACTA elements observed at the Glu-A3 loci from T. monococcum and T. durum was a fortunate constellation (Wicker et al., 2003). It allowed the characterization of a large number of elements belonging to different families and conclusions to be drawn about their general features and structures. The main reason why CACTA elements have remained undiscovered for so long is that not enough sequence data was available for the identification of these elements. From the handful of CACTA elements that were described so far in other species, only limited conclusions could be drawn as to what types of elements could be expected to be present in Triticeae. As we show in this study, sequence conservation at the DNA level is very low even between Triticeae elements and limited to the very TR regions among different grass species. A second reason for them being hidden so well is the unexpected finding that most CACTA elements are deletion derivatives and do not encode transposase proteins. Several elements containing the CTG-2 proteins actually had been described before but due to the misleading BLASTX results had been interpreted as putative genes. In one case, the sub-TR structures flanking the CTG-2 were interpreted as arrays of very small miniature inverted-repeat transposable elements (MITEs; Wei et al., 2002).
CACTA Sequences in Grass Genomes Are Mainly Deletion Derivatives
All identified CACTA elements appear to be defective or nonautonomous because they either lack sufficient coding capacity, or their coding sequences are interrupted by frameshifts or in-frame stop codons. For En/Spm and Ac/Ds elements from maize, it was shown that numerous deletion derivatives exist that are only able to transpose in the presence of a functional element (for review, see Gierl and Saedler, 1989). One can speculate that the initial autonomous Caspar transposon had a size of approximately 10 kb and encoded both a transposase and an CTG-2 protein. During evolution of these elements, a large number of deletion derivatives were established, which themselves evolved and diverged further. Obviously, a large number of elements have lost their transposase region but have maintained the CTG-2, whereas other elements have lost both proteins and were reduced basically to their TR regions, which are in most cases separated by a small internal domain (SNAC transposons). A possible final product of this tendency of size reduction is the SNAC_AP003446-1 transposon from rice that does not even contain an internal domain but consists exclusively of TR sequences. Therefore, SNAC_AP003446-1 might represent the “minimal transposon” that is reduced to its very basic functional components. All SNAC elements identified in this study contain both TIR and sub-TR sequences. This differentiates them from the previously described mobile element-like sequences, which also contain a conserved CACTA motif but only have TIR sequences (for review, see Hoshino et al., 2001). Both SNAC elements and mobile element-like sequences resemble MITEs (Bureau and Wessler, 1994), which are also considered to be nonautonomous elements.
However, during the evolution of nonautonomous elements, there was obviously no selection pressure that would favor smaller sized elements, as is illustrated by the numerous large elements such as Jorge_AF326781-1. An even more impressive example is the 23-kb Candystripe1 transposon from sorghum. This CACTA element was shown to be active in sorghum, although it is also considered to be nonautonomous (Chopra et al., 1999). This concept can be expanded to other classes of repetitive elements. For example, the Sabrina retrotransposon (Shirasu et al., 2000) is one of the most abundant retroelements in Triticeae, but only few copies that actually encode a protein similar to reverse transcriptase were identified so far (SanMiguel et al., 2002; Wei et al., 2002). Thus, we conclude that nonautonomous repetitive elements are widely present in grass genomes and possibly include the majority of all mobile DNA sequences. Therefore, the Triticeae genomes may contain an enormous number of such nonautonomous elements, and many of them have not yet been discovered because they lack obvious coding sequences.
The Presence of Afa Repeats in Caspar Elements Explains Some of the Features of These Repeats But Also Raises New Questions
Because Afa repeats were found in several members of the Caspar family but never isolated outside of Caspar elements, we conclude that all Afa repeats are actually compounds of such transposons. This “transposon hypothesis” explains three properties of this repeat family as they were described by Nagaki et al. (1998a). First, it was reported that the copy number of Afa repeats is highly variable in different Triticeae spp. On one hand, a transposon can be more active in one species than in another and, therefore, produce more copies. On the other hand, we showed that the number of Afa repeats can vary drastically even within very closely related elements, indicating a very rapid evolution of these sequences. Second, the mobility of a transposon explains why no chromosome specificity within one species was observed. Third, Nagaki et al. (1998a) suggested the presence of a specific mechanism to remove Afa repeats from the genome. The transposon hypothesis can provide this specific mechanism.
The presence of Afa and other repeat structures such at TM-1, TM-2, and the extensive regions comprising short sequence repeats raises new questions. First, the amplification mechanism is still obscure. Template slippage during DNA replication or unequal crossing over can explain the rapid change in copy number, but it does not explain why only some conserved repeat sequences are amplified. A rolling circle amplification, as was suggested by Nagaki et al. (1998a), also seems unlikely because it would require a template to be excised from the genome and the amplified product to be reintegrated back into the same element. Second, what is the function of these tandem repeated regions? The presence of such structures in different families of CACTA transposons suggests that they are functional components of these elements rather than the result of random DNA rearrangements.
Despite these open questions, the mere knowledge that tandem repeats are often found within transposons might be important for future analysis of genomic regions. The presence of such arrays can be an indication for the presence of a novel diverged transposon family that could not be detected otherwise. In addition, it is possible that in future studies, tandem repeats from other species such as saccharum CENtromeric sequence repeats from sugarcane (Saccharum officinarum; Nagaki et al., 1998b) can be associated with transposons.
The Contribution of CACTA Elements to Genome Evolution
The function and possible benefit of repetitive elements for the “host” plant is a hotly debated question. MITEs, for example, are often found in close association with genes, and they are believed to contribute regulatory sequences that may alter gene expression (Zhang et al., 2000). A similar role can be suggested for CACTA elements. Nine of the total 41 elements were found in EST sequences, suggesting that they may also be found frequently in close proximity to genes. In addition, one Mandrake element was found a few kilobase pairs upstream of the Td-Glu-A3-1 gene in T. durum (Wicker et al., 2003). Interestingly, a different Mandrake element was identified at a similar distance to an alpha-gliadin gene in T. aestivum (accession no. AF234649). Glutenins and gliadins are genes that belong to the same family. The position of insertion and the degree of sequence conservation between the two genes indicates that both insertions have been independent events rather than an insertion that occurred already in the common ancestor of the two genes. Therefore, it is possible that certain types of CACTA elements can be involved in specific interactions with certain genes in the Triticeae genomes.
The finding that the four Caspar SNAC elements contain sequences similar to 5S rDNA genes is intriguing. The fact that the region that contains the 5S derivative is more conserved among the four elements than the rest of the elements suggests that a selection pressure has been acting on these sequences. It is possible that these sequences have been acquired by a CACTA element during evolution and that they have gained a function that was beneficial for the plant, eventually leading to their fixation within the genome. Acquisition of fragments of cellular genes by CACTA elements has been reported before (Takahashi et al., 1999).
Concluding Remarks
Repetitive DNA, which is still often referred to as “junk DNA,” is rarely the focus of a detailed analysis. Our results demonstrate the importance of detailed characterization of repetitive elements and database mining of public databases. Because of their high amount of repetitive DNA, genomic sequences from Triticeae are an essential resource for the identification of novel repetitive elements. The information gained about these elements then can be used for a targeted search for similar elements in other plant genomes. This was demonstrated by the discovery of the rice SNAC transposons, which were not annotated in the publicly available rice sequences. Another important result of our study is the finding that the CTG-2 protein is actually a part of the Caspar transposon. This information suggests that numerous sequences that were interpreted as genes could actually belong to repetitive elements. This has an important implication for future estimates of the total gene contents of entire genomes and also for the calculation of local gene densities in large genome plants such as wheat or maize. Finally, the identification of novel CACTA elements could eventually lead to the discovery of active wheat transposons that could be used for transposon-tagging systems similar to those based on En/Spm and Ac/Ds elements.
MATERIALS AND METHODS
Southern Hybridization of High-Density BAC Filters
Two copies of Filter C from the Triticum monococcum BAC library (Lijavetzky et al., 1999) were incubated over night at 65°C with radioactively labeled Probe512 and Probe179, respectively. The filters were washed three times for 20 min at 65°C in 0.5× SSC and 0.1% (w/v) SDS and exposed to BIOMAX MS films (Eastman-Kodak, Rochester, NY) overnight.
Database Mining and Sequence Analysis
Public databases and the database for Triticeae repetitive elements (TREP, http://wheat.pw.usda.gov/ITMI/Repeats) were screened with the BLASTN and BLASTX algorithms (Altschul et al., 1997). For the identification of TR sequences, a 127-bp consensus sequence was used as a query for BLASTN search (consensus TR sequence: CACTACTAGGGAAAAGGCCT-ACTAATAGCGCACCGGATTGCTACTAATGGCGCCCAGGGGTGCGCC-ACTAGCGCTACCACGCCAGTACTATATCTTACTAATGGCGCACCAGG-GTGGTATAAACCC). Detailed sequence analysis was performed with the GCG Sequence Analysis Software Package version 10.1 (Devereux et al., 1984) and by dot-plot analysis (program DOTTER; Sonnhammer and Durbin, 1995). Sequence alignments were done with the GCG programs BESTFIT and PILEUP. The multiple alignment of the TR sequences was done with PILEUP (gap creation penalty = 2, gap extension penalty = 0). Phylogenetic analysis was performed with ClustalW (Thompson et al., 1994). Distances between pairs of TRs were calculated using the neighbor-joining method. Confidence values for the nodes were calculated using 1,000 bootstraps. For efficient processing of large sets of sequences, programs were written using the language PERL. Identified transposons were named as follows: The name of the transposon is separated by an underscore from the address of the BAC clone or the GenBank accession number of the sequence in which the element was discovered. Copy numbers of individual elements from the same source sequence are separated from the name by a hyphen.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Jorge Dubcovsky (University of California, Davis) and Dr. Nils Stein (Genomanalyse im biologischen System Pflanze grant no. 0312280A, Bundesministerium für Bildung und Forschung, Berlin, Germany) for making their unpublished transposon sequences available for our study. We are also grateful to Dr. Catherine Feuillet (Institute of Plant Biology, University of Zurich, Switzerland) and Clair Wicker for critical reading of the manuscript.
Footnotes
This work was supported by the Swiss National Science Foundation (grant no. 31–65114.01).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.015743.
LITERATURE CITED
- Altschul S, Madden TL, Schaeffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennet MD, Leitch IJ. Nuclear DNA amounts in angiosperms. Ann Bot. 1995;76:113–176. [Google Scholar]
- Bennetzen JL. Transposable element contributions to plant genome evolution. Plant Mol Biol. 2000;42:251–269. [PubMed] [Google Scholar]
- Brooks SA, Huang L, Gill BS, Fellers JP. Analysis of 106 kb of contiguous DNA sequence from the D genome of wheat reveals high gene density and a complex arrangement of genes related to disease resistance. Genome. 2002;45:963–972. doi: 10.1139/g02-049. [DOI] [PubMed] [Google Scholar]
- Brunner S, Keller B, Feuillet C (2003) A large rearrangement involving genes and low copy DNA interrupts the microlinearity between rice and barley at the Rph7 locus. Genetics (in press) [DOI] [PMC free article] [PubMed]
- Bureau T, Wessler SR. Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Proc Natl Acad Sci USA. 1994;9:1411–1415. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra S, Brendel V, Zhang J, Axtell JD, Peterson T. Molecular characterisation of a mutable pigmentation phenotype and isolation of the first active transposable element from Sorghum bicolor. Proc Natl Acad Sci USA. 1999;96:15330–15335. doi: 10.1073/pnas.96.26.15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloix C, Tutois S, Mathieu O, Cuvillier C, Espagnol MC, Picard G, Tourmente S. Analysis of 5S rDNA arrays in Arabidopsis thaliana: physical mapping and chromosome-specific polymorphisms. Genome Res. 2000;10:679–690. doi: 10.1101/gr.10.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Ramakrishna W, SanMiguel PJ, Busso CS, Yan LL, Shiloff BA, Bennetzen JL. Comparative sequence analysis of colinear barley and rice bacterial artificial chromosomes. Plant Physiol. 2001;125:1342–1353. doi: 10.1104/pp.125.3.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez JA, Moreno M, Carmona MJ, Castagnaro A, Olmedo F. The barley alpha-thionin promoter is rich in negative regulatory motifs and directs tissue-specific expression of a reporter gene in tobacco. Biochem Biophys Acta. 1993;1172:346–348. doi: 10.1016/0167-4781(93)90229-7. [DOI] [PubMed] [Google Scholar]
- Feuillet C, Penger A, Gellner K, Mast A, Keller B. Molecular evolution of receptor-like kinase genes in hexaploid wheat: independent evolution of orthologs after polyploidization and mechanisms of local rearrangements at paralogous loci. Plant Physiol. 2001;125:1304–1313. doi: 10.1104/pp.125.3.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl A, Saedler H. Maize transposable elements. Annu Rev Genet. 1989;23:71–85. doi: 10.1146/annurev.ge.23.120189.000443. [DOI] [PubMed] [Google Scholar]
- Hoshino A, Johzuka-Hisatomi Y, Iida S. Gene duplication and mobile genetic elements in the morning glories. Gene. 2001;265:1–10. doi: 10.1016/s0378-1119(01)00357-2. [DOI] [PubMed] [Google Scholar]
- Inagaki Y, Hitsatomi Y, Suzuki T, Kasahara K, Iida S. Isolation of a Suppressor-Mutator/Enhancer-like transposable element, Tpn1, from Japanese morning glory bearing variegated flowers. Plant Cell. 1994;6:375–383. doi: 10.1105/tpc.6.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin B. Transposons. In: Lewin B, editor. Genes VI. New York: Oxford University Press, Inc.; 1997. pp. 563–595. [Google Scholar]
- Lijavetzky D, Muzzi G, Wicker T, Keller B, Wing RA, Dubcovsky J. Construction and characterization of a bacterial artificial chromosome (BAC) library for the A genome of wheat. Genome. 1999;42:1176–1182. [PubMed] [Google Scholar]
- Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- Nacken WKF, Piotrowiak R, Saedler H, Sommer H. The transposable element TAM-1 of A. majus shows structural homology to the maize transposon En/Spm and has no sequence specificity of insertion. Mol Gen Genet. 1991;228:201–208. doi: 10.1007/BF00282466. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Tsujimoto H, Sasakuma T. Dynamics of tandem repetitive Afa-family sequences in Triticeae, wheat-related species. J Mol Evol. 1998a;47:183–189. doi: 10.1007/pl00006375. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Tsujimoto H, Sasakuma T. A novel repetitive sequence of sugar cane, SCEN family, locating on centromeric regions. Chromosome Res. 1998b;6:295–302. doi: 10.1023/a:1009270824142. [DOI] [PubMed] [Google Scholar]
- Ozeki Y, Davies E, Takeda J. Somatic variation during long term subculturing of plant cells caused by insertion of a transposable element in a phenylalanine ammonia-lyase (PAL) gene. Mol Gen Genet. 1997;254:407–416. doi: 10.1007/s004380050433. [DOI] [PubMed] [Google Scholar]
- Pereira A, Cuypers H, Gierl A, Sommer ZS, Saedler H. Molecular analysis of the En/Spm transposable element system of Zea mays. EMBO J. 1986;5:835–841. doi: 10.1002/j.1460-2075.1986.tb04292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayburn AL, Gill BS. Isolation of a G-genome specific sequence repeated DNA sequence from Aegilops squarrosa. Plant Mol Biol Rep. 1986;4:102–109. [Google Scholar]
- Rostoks N, Park Y, Ramakrishna W, Ma J, Druka A, Shiloff BA, Jiang Z, Brueggeman R, Sandhu D, Gill K et al. Genomic sequencing reveals gene content, genomic organization, and recombination relationships in barley. Funct Integr Genomics. 2002;2:51–59. doi: 10.1007/s10142-002-0055-5. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Bennetzen JL. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann Bot. 1998;82:37–44. [Google Scholar]
- SanMiguel PJ, RamaKrishna W, Bennetzen JL, Busso C, Dubovsky J. Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5A(m) Funct Integr Genomics. 2002;2:70–80. doi: 10.1007/s10142-002-0056-4. [DOI] [PubMed] [Google Scholar]
- Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P. A contiguous 66 kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 2000;10:908–915. doi: 10.1101/gr.10.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Flavell RB. Characterisation of the wheat genome by renaturation kinetics. Chromosoma. 1975;50:223–242. [Google Scholar]
- Snowden KC, Napoli CA. PsI: a novel Spm-like transposable element from Petunia hybrida. Plant J. 1998;14:43–54. doi: 10.1046/j.1365-313x.1998.00098.x. [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Reprinted from. Gene Combis. 1995;167:GC1–GC10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Inagaki Y, Hoshino A, Iida S. Capture of a genomic HMG domain sequence by the En/Spm-related transposable element Tpn1 I the Japanese moring glory. Mol Gen Genet. 1999;261:447–451. doi: 10.1007/s004380050987. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Wing RA, Wise RP. Genome dynamics and evolution of the Mla(powdery mildew) resistance locusRT in barley. Plant Cell. 2002;14:1903–1917. doi: 10.1105/tpc.002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Matthews DE, Keller B. TREP: a database for Triticeae repetitive elements. Trends Plant Sci. 2002;7:561–562. [Google Scholar]
- Wicker T, Stein N, Albar L, Feuillet C, Schlagenhauf E, Keller B. Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcumL.) reveals multiple mechanism of genome evolution. Plant J. 2001;26:307–316. doi: 10.1046/j.1365-313x.2001.01028.x. [DOI] [PubMed] [Google Scholar]
- Wicker T, Yahiaoui N, Guyot R, Schlagenhauf E, Liu Z-D, Dubcovsky J, Keller B (2003) Rapid genome divergence at orthologous LMW glutenin loci of the A and Am genomes of wheat. Plant Cell (in press) [DOI] [PMC free article] [PubMed]
- Zhang Q, Arbuckle J, Wessler SR. Recent, extensive, and preferential insertion of members of the miniature inverted-repeat transposable element family Heartbreaker into genic regions. Proc Natl Acad Sci USA. 2000;97:1160–1165. doi: 10.1073/pnas.97.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]