Abstract
Cold acclimation is a multigenic trait that allows hardy plants to develop efficient tolerance mechanisms needed for winter survival. To determine the genetic nature of these mechanisms, several cold-responsive genes of unknown function were identified from cold-acclimated wheat (Triticum aestivum). To identify the putative functions and structural features of these new genes, integrated genomic approaches of data mining, expression profiling, and bioinformatic predictions were used. The analyses revealed that one of these genes is a member of a small family that encodes two distinct groups of multispanning transmembrane proteins. The cold-regulated (COR)413-plasma membrane and COR413-thylakoid membrane groups are potentially targeted to the plasma membrane and thylakoid membrane, respectively. Further sequence analysis of the two groups from different plant species revealed the presence of a highly conserved phosphorylation site and a glycosylphosphatidylinositol-anchoring site at the C-terminal end. No homologous sequences were found in other organisms suggesting that this family is specific to the plant kingdom. Intraspecies and interspecies comparative gene expression profiling shows that the expression of this gene family is correlated with the development of freezing tolerance in cereals and Arabidopsis. In addition, several members of the family are regulated by water stress, light, and abscisic acid. Structure predictions and comparative genome analyses allow us to propose that the cor413 genes encode putative G-protein-coupled receptors.
To achieve their complete life cycle and reproduction in temperate regions, hardy plants like winter wheat (Triticum aestivum) have developed two major evolutionary adaptative mechanisms: vernalization and cold acclimation (CA). Overwintering plants sense the upcoming winter and delay flowering by postponing the transition from the vegetative to the cold-sensitive reproductive phase (Simpson et al., 1999). In addition, they develop the high degree of freezing tolerance (FT) needed for winter survival (Fowler et al., 1999). Following low temperature (LT) acclimation, some winter cereals can tolerate temperatures as low as −33°C. The regulatory mechanisms underlying these two processes and how they are interconnected are far from being fully understood. To gain further knowledge on the strategies that plants use for winter survival, the identification of cold-regulated (COR) genes is needed. A survey of the literature reveals that the expressions of a large number of genes are altered during the process of CA (Thomashow, 1999; Breton et al., 2000; Seki et al., 2002). These genes could be classified into four groups based on the presumed function of the encoded proteins. The first group comprises genes encoding structural proteins that may be involved in protecting the cell during LT stress. The second group represents those genes that regulate gene expression and signal transduction pathways, such as transcription factors, protein kinases, phosphatases, and the enzymes involved in phosphoinoside metabolism. The third group represents genes encoding enzymes involved in the biosynthesis of different osmoprotectants and membrane lipids and those of the antioxidative response. The fourth group contains cold-induced genes encoding proteins of unknown function.
To gain insight into the function of these novel proteins, a combination of expression profiling and bioinformatic analyses can be used to predict properties and features that may be important for their function. When a novel gene is found to be up-regulated by LT and its expression shows an association with the plants' capacity to develop FT, it is reasonable to assume that the encoded novel protein may play a role in FT. By taking advantage of intraspecies variability in FT, a second level of association can be determined. Previous studies have shown that, compared with winter varieties, the less hardy spring wheat varieties cannot maintain the expression of COR genes (e.g. the WCS120 family) at a high level and that this differential expression is closely associated with their low degree of FT (Sarhan et al., 1997). A third level of association can be further established by taking advantage of the natural diversity of plant species. For example, species such as rice (Oryza sativa) and maize (Zea mays) are highly sensitive to LT above the freezing point, whereas species such as winter wheat and rye can tolerate temperatures as low as −33°C. This association can help differentiate between cold-responsive genes related to cold performance from those related to the acquisition of FT.
As a subsequent step, each novel LT-regulated protein sequence can be analyzed using available bioinformatic tools. These tools help in the identification of sorting signals, conserved posttranslational modifications, transmembrane helices, and secondary and tertiary structures. The most recent prediction software incorporate machine-learning algorithms in the form of a neural network and a hidden Markov model (Blom et al., 1999; Krogh et al., 2001). Comparative studies have shown that their prediction accuracy is often superior to older programs (Möller et al., 2001; Tusnàdy and Simon, 2001) and is bound to improve further when more newly characterized proteins are included in their training data sets. Knowledge gained from analyzing novel proteins with such tools can lead to the identification of important functional domains, an element needed to design future experiments to confirm the predicted function.
In the present study, the integrated approaches of expression profiling, structural analysis, and bioinformatic predictions were used to study a novel unknown gene family named cor413. This family encodes two distinct groups of proteins containing five putative transmembrane domains (TMD). COR413-plasma membrane (COR413-PM) proteins are potentially targeted to the plasma membrane and COR413-thylakoid membrane (COR413-TM) proteins to the thylakoid. The use of intraspecies and interspecies comparative gene expression analysis shows that the regulation of this gene family is associated with the development of FT in cereals and Arabidopsis. A proposed structural and functional model for the COR413 protein family is discussed.
RESULTS
Identification of TaCOR413-PM1 Homologs
Differential screening of a wheat cold-acclimated cDNA library was used to isolate LT-responsive clones. One of these clones, Tacor413-pm1 (previously Wcor413), was selected for detailed molecular characterization (Danyluk, 1996). Sequence analysis revealed that the longest open reading frame (ORF) encodes a 210-amino acid (23 kD), highly hydrophobic protein with a predicted pI of 9.0 (Table I, TaCOR413-PM1).
Table I.
Characteristics of cor413-pm, -tm, and moss cor413 genes
| Type | Name | GenBank Accession No. | Amino Acid Residues | pI | Identity with TaCOR413-PM1 | Identity with TaCOR413-TM1 |
|---|---|---|---|---|---|---|
| % | ||||||
| COR413-PM | Tacor413-pm1 | AAB18207 | 210 | 9.0 | 100 | 29 |
| Tacor413-pm2 | AAL23724 | 208 | 9.7 | 78 | 27 | |
| Hvcor413-pm1 | BE421687 | 210 | 8.6 | 95 | 28 | |
| Hvcor413-pm2 | BF628071 | 208 | 9.3 | 79 | 27 | |
| Oscor413-pm1 | AF283006 | 210 | 9.4 | 71 | 26 | |
| Zmcor413-pm1 | AY181208 | 212 | 9.4 | 72 | 27 | |
| Sbcor413-pm1 | BI075784 | 213 | 9.1 | 72 | 28 | |
| Atcor413-pm1 | AF283004 | 197 | 9.1 | 55 | 26 | |
| Atcor413-pm2 | AF283005 | 203 | 9.4 | 55 | 25 | |
| Mtcor413-pm1 | BF003463 | 198 | 8.6 | 55 | 26 | |
| Mtcor413-pm2 | BG647116 | 199 | 8.6 | 55 | 25 | |
| Mtcor413-pm3 | BG456396 | 195 | 5.2 | 54 | 26 | |
| Gmcor413-pm1 | BE211677 | 198 | 9.1 | 59 | 27 | |
| Lecor413-pm1 | AW039062 | 198 | 9.2 | 56 | 23 | |
| Lecor413-pm2 | BG642925 | 202 | 8.6 | 61 | 24 | |
| COR413-TM | Tacor413-tm1 | AY181206 | 221 | 10.2 | 29 | 100 |
| Hvcor413-tm1 | AF465840 | 215 | 10.4 | 30 | 88 | |
| Oscor413-tm1 | AY181210 | 222 | 10.7 | 27 | 69 | |
| Zmcor413-tm1 | AY181209 | 226 | 10.5 | 27 | 64 | |
| Atcor413-tm1 | AAK76616 | 226 | 10.5 | 25 | 44 | |
| Atcor413-tm2 | AAL87293 | 225 | 10.6 | 24 | 45 | |
| Mtcor413-tm1 | BF639516 | 234 | 10.0 | 23 | 41 | |
| Lecor413-tm1 | AW034114 | 221 | 9.8 | 26 | 53 | |
| Stcor413-tm1 | BG047649 | 220 | 9.8 | 26 | 53 | |
| Cjcor413-tm1 | AY181207 | 241 | 10.3 | 27 | 39 | |
| Moss COR413 | Ppcor413-1 | AAL16410 | 207 | 5.7 | 44 | 26 |
| Ppcor413-2 | BJ200341 | 204 | 7.0 | 44 | 28 | |
| Ppcor413-3 | BJ169989 | 205 | 7.1 | 45 | 30 | |
Genes from wheat (Ta), barley (Hv), rice (Os), maize (Zm), sorghum (Sb), Arabidopsis (At), alfalfa (Mt), soybean (Gm), tomato (Le), potato (St), C. japonica (Cj), and P. patens (Pp) were translated, and the length in amino acids and pI of the longest ORF is presented. For genes sequenced in this study, accession numbers are provided. For sequences constructed from data available in the EST database, one accession number is provided and identified in italic. More information can be found in Supplemental Table VII on the source EST for in silico sequencing and on the Arabidopsis Genome Initiative number for Arabidopsis proteins.
A search in the GenBank nonredundant sequence database using the BLAST program revealed that TaCOR413-PM1 is a novel protein with no characterized homologs. Data mining of the GenBank EST database with TaCOR413-PM1 revealed that plants possess several homologs of this protein. A combination of EST sequencing and in silico reconstitution allowed the generation of 27 new COR413-related protein sequences from plants. Using pair wise sequence alignments with the initial TaCOR413-PM1, these proteins were clustered into two distinct groups (Table I). The first group is named COR413-PM and contains members sharing more than 54% overall identity with TaCOR413-PM1 (Table I). The second group is named COR413-TM and contains members sharing less than 30% overall identity with TaCOR413-PM1 (Table I). However, a region of 40 amino acids shows a higher degree of homology among all members of both groups (Supplemental Figs. 1–3, square brackets; they can be viewed at www.plantphysiol.org).
Data mining of cereal EST databases and rice genomic sequence helped in the identification of two different COR413-PM members in wheat, maize, and barley (Hordeum vulgare), whereas only one was identified in rice (O. sativa subsp. indica cv 93-11; Yu et al., 2002). In addition, four COR413-PM proteins were identified in the Arabidopsis genome. On the other hand, only one member belonging to the COR413-TM group was identified in the four cereal species analyzed. In Arabidopsis, two COR413-TM were found in tandem repeat on chromosome 2 (The Arabidopsis Genome Initiative, 2000). Furthermore, a search in the GenBank EST database revealed that other dicotyledonous plants such as tomato (Lycopersicon esculentum), soybean (Glycine max), ice plant (Mesembryanthemum crystallinum), poplar (Populus spp.), and cotton (Gossypium hirsutum) as well as the coniferales Cryptomeria japonica and Pinus taeda possess sequences encoding homologs of the COR413 groups (see Supplemental Table II; supplemental tables can be viewed at www.plantphysiol.org).
Other embryophytes such as the marchantiales Marchantia polymorpha and the moss Physcomitrella patens also have COR413 homologs. The deduced moss COR413 proteins share slightly higher identity with the COR413-PM group, suggesting that they are related to this group (Table I). Because of their lower degree of homology, they were classified separately in this study as moss COR413 (Table I). This lower homology may result from the evolutionary distance between moss and other plants listed in Table I. Because no entries encoding COR413 homologs were found in the green algae Chlamydomonas reinhardtii sequence database, it is possible that COR413 would be present only in multicellular Viridiplantae. COR413 homologous sequences were neither found in other eukaryotes nor in prokaryote databases, suggesting that this family is specific to the plant kingdom.
The cor413 Genes Encode Membrane Proteins Potentially Targeted to the Plasma and Thylakoid Membranes
The analyses of both COR413-PM and -TM sequences revealed that they are rich in hydrophobic amino acids, suggesting that they may be membrane proteins (Supplemental Figs. 1 and 2). The sequence alignments of both groups show many regions of high identity (shaded in black in Supplemental Figs. 1 and 2). In addition, many residues normally considered important for protein structure or activity such as Cys residues and Pro residues are conserved within COR413-PM or COR413-TM proteins (Supplemental Figs. 1–3, asterisks). Five of the Pro residues are even conserved between members of both groups (yellow-shaded asterisks). On the other hand, the sequence alignments also revealed that approximately the first 50 amino acids of COR413-PM and the first 80 amino acids of COR413-TM are poorly conserved. This observation prompted us to analyze these regions for subcellular targeting signals.
Analysis using the PSORT program revealed that there is no consensus targeting or retention signal present in COR413-PM sequences (Nakai and Kanehisa, 1992; Supplemental Table III). Although the program suggested different cellular localizations for each member, the average score was slightly higher for the plasma membrane localization. Because many proteins targeted to the plasma membrane possess a cleavable signal peptide, COR413-PM sequences were analyzed with SignalP (Nielsen and Krogh, 1998). The analysis of SignalP-HMM results revealed that six proteins have a high probability to possess a non-cleavable signal anchor for endoplasmic reticulum (ER) translocation (Supplemental Table III). These results are consistent with those obtained with PSORT and suggest that COR413-PM proteins are targeted to the plasma membrane. Moss COR413 shows the same features as the COR413-PM group, suggesting that they are also targeted to the plasma membrane (Supplemental Table IV). Using several secondary structure prediction programs available on the Network Protein Sequence Analysis server, we found that the N-terminal region of COR413-PM proteins contains a possible hinge-like structure consisting of two segments of 20 to 25 residues predicted to form α-helices that are separated by a Gly-rich region (Supplemental Fig. 1).
The use of the targeting signal programs PSORT, iPSORT, and TargetP for COR413-TM sequence analyses revealed that they are all likely to be targeted to the thylakoid membrane (Supplemental Table V; Nakai and Kanehisa, 1992; Emanuelsson et al., 2000; Bannai et al., 2002). Chloroplast targeting signals are generally highly basic and rich in Ser and Thr (Agarraberes and Dice, 2001). The N-terminal sequence of all COR413-TM members shows these two properties (Supplemental Fig. 2).
COR413 Proteins Contain Five TMD
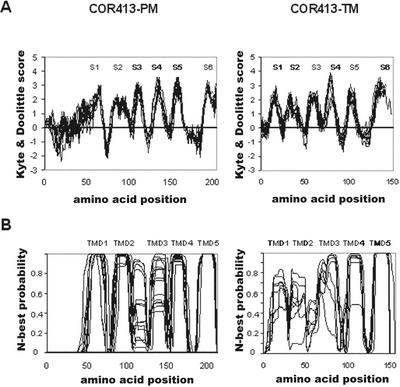
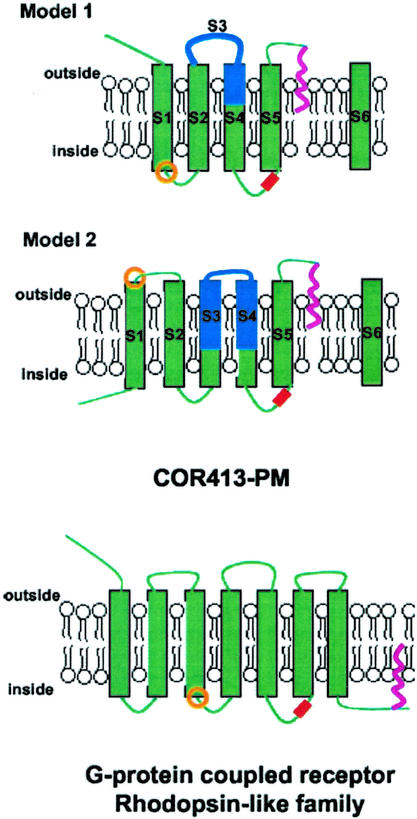
As expected from the overall amino acid composition, the Kyte and Doolittle hydrophobicity plot of TaCOR413-PM1 shows a highly hydrophobic pattern with six clear spikes (Supplemental Fig. 1, S1–S6; Kyte and Doolittle, 1982). Superposition of the Kyte and Doolittle plot of the 15 available COR413-PM sequences showed that the overall hydrophobicity is well conserved among the different members (Fig. 1A). To analyze the number of TMD and the possible topology of COR413-PM proteins, the newly developed and accurate membrane topology prediction program TMHMM was used (Krogh et al., 2001; Möller et al., 2001). The final prediction generated by the program for each COR413-PM members is listed in Supplemental Table III, and the compilation of all TMD predictions is presented in Figure 1B. These analyses allowed us to propose two structural models (Fig. 2). In the first, COR413-PM proteins would have five TMD with the N-terminal end outside and the C-terminal end inside (Fig. 2, model 1). This model is supported by the following observations: (a) The final prediction of 11 of 15 proteins have this topology; and (b) the compilation of the N-probability graphs for TMD shows that the five-TMD topology is favored. In this compilation, spike 4 was chosen as the third TMD in nine of the 11 proteins. The data analysis of the inside/outside probability graphs generated by TMHMM and the calculation of the median probability revealed that the N-terminal topology is favored (64%). Although model 1 is the software's preferred topology, it does not take into account the following points: (a) All 15 proteins contain six hydrophobic spikes suggesting six TMD; and (b) four proteins of 15 are predicted to have six TMD with TMHMM. Therefore, an alternative model could be proposed where group COR413-PM members would have six TMD with both the N-terminal and C-terminal ends inside (Fig. 2, model 2). The main difference with the first model is the inverted topology in the first one-half of the protein.
Figure 1.
Hydropathy and transmembrane predictions. A, Compilation of Kyte and Doolittle profiles of all group COR413-PM and -TM members. S1 to S6, Spikes 1 to 6. B, Compilation of profiles generated by TMHMM 2.0 for all group COR413-PM and -TM members. TMD1 to TMD5, Transmembrane helices 1 to 5. For group COR413-TM, the two profiles were generated without the N-terminal chloroplastic targeting signal (cut after the conserved Cys residue identified in Supplemental Fig. 2).
Figure 2.
Proposed models for COR413-PM proteins and comparison with the GPCR Rhodopsin-like family. Green boxes and green lines, TMD and interconnecting loops, respectively. Boxes and lines shaded in blue, Region containing the highest similarity with COR413-TM proteins. S1 to S6 correspond to Kyte and Doolittle spikes from Figure 1. In model I and II, S6 is separated from the rest of the proteins due to the addition of a GPI anchor. Red box, Phosphorylation sites. Pink line, Lipidic anchor. Orange circle, Position of the highly conserved DRT (COR413-PM) or DRY (GPCR) triplet motif.
COR413-TM sequences were analyzed before and after removal of the putative N-terminal chloroplastic targeting signal. The comparison of the 10 Kyte and Doolittle profiles clearly shows that COR413-TM proteins possess six hydrophobic spikes (S1–S6 in Supplemental Fig. 2 and Fig. 1A). Despite the clear hydrophobic pattern, TMHMM had difficulties generating clear topology predictions (Supplemental Table V). However, the compilation of the 10 TMHMM graphs suggests that group II may also have a five-TMD structure (Fig. 1B). Because no clear consensus can be deduced for the TMHMM inside-outside topology, it is impossible to predict which loops are exposed on the lumenal and stromal side of the thylakoids. The extracellular loop 1 of COR413-PM model 1 (Fig. 1, S3) falls in the region that is conserved between COR413-PM and -TM, and corresponds to the second predicted loop and the third TMD of COR413-TM (Supplemental Figs. 1–3; identified in blue in Fig. 2).
COR413-PM Proteins Contain Conserved Putative Phosphorylation and Glycosylated Phosphatidylinositol (GPI)-Anchoring Sites
Motif searches against the PROSITE, Pfam, and Smart databases, after exclusion of patterns with a high probability of occurrence, did not detect known motifs. However, the neural network-based NetPhos phosphorylation site prediction software generated several interesting findings (Blom et al., 1999). Even though the TaCOR413-PM1 sequence contains eight Ser residues, 10 Thr residues, and four Tyr residues, only one Thr residue is predicted to be a phosphorylation site (Supplemental Fig. 1 in yellow). Analysis of the other COR413-PM members with the NetPhos software always identified a putative phosphorylation site at this position (Supplemental Fig. 1). Interestingly, in both of our models, this phosphorylation site is located on the internal side of the membrane where it may be the target for intracellular kinases (see Fig. 2). For the chloroplastic COR413-TM proteins, NetPhos predicted a phosphorylation site in the same region (between TMD3 and TMD4) for eight of the 10 proteins. The other two proteins are those from Arabidopsis, which raises the possibility that the prediction of the phosphorylation site for the chloroplastic proteins may be incorrect (Supplemental Fig. 2).
The DGPI program predicted a GPI-anchoring site at the C-terminal end of all COR413-PM family members (D. Buloz and J. Kronegg, unpublished data). The conserved features are a highly hydrophobic C-terminal end and a consensus cleavage site needed for the addition of the GPI anchor (Supplemental Fig. 1). This second posttranslational modification fits well with our structural models because GPI anchors are modifications located on the external side of the membrane (Fig. 2). This modification will result in the cleavage of the second extracellular loop as schematized in Figure 2. The chloroplastic COR413-TM proteins' C-terminal tail is also very hydrophobic but the potential cleavage sites are less conserved.
cor413 Genes Are Regulated by Environmental Stresses
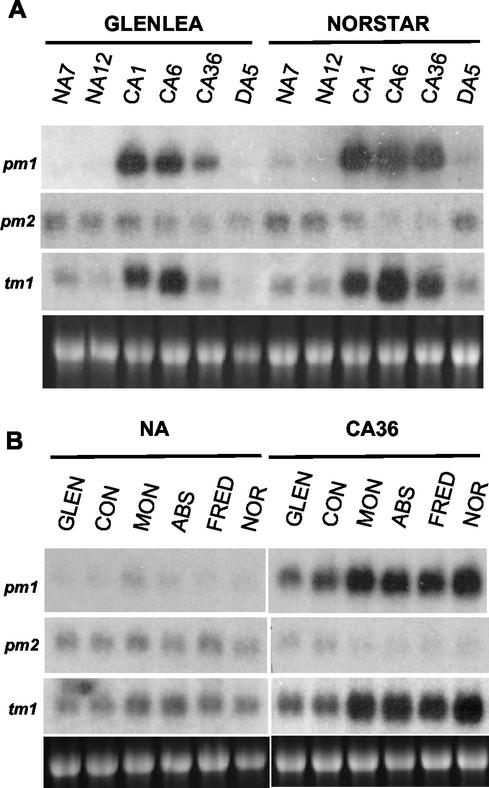
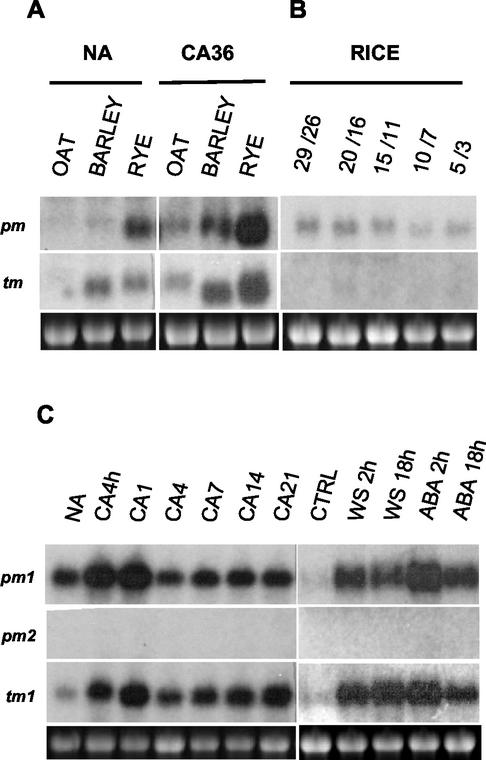
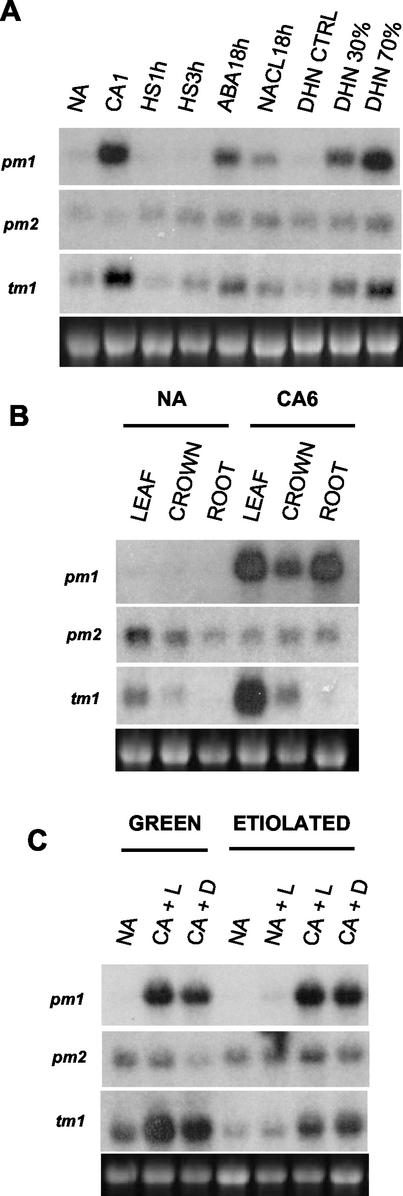
Northern-blot analyses indicated that Tacor413-pm1 and Tacor413-tm1 transcripts are strongly up-regulated by LT in leaf tissues (Fig. 3A). In contrast, the Tacor413-pm2 transcript was down-regulated. The LT kinetics study in winter wheat cv Norstar leaves shows that the Tacor413-pm1 and tm1 transcripts accumulate rapidly within 24 h and remain at high levels throughout the acclimation period (Fig. 3A). In comparison, the transcripts accumulation in the less freezing-tolerant spring wheat cv Glenlea peaks at 24 h and then declines (Fig. 3A). When the plants are deacclimated at 24°C for 5 d, Tacor413-pm1 and tm1 transcripts decline to the nonacclimated control levels in both cultivars. The intra- and interspecies comparative expression analyses are shown in Figures 3B and 4. Tacor413-pm1 and tm1 mRNA levels are higher in winter wheat cultivars compared with the less FT spring wheat cultivars (Glenlea and Concorde). These results suggest that the accumulation of Tacor413-pm1 and tm1 transcripts is associated with the capacity of the plants to develop FT. This figure also shows that Tacor413-pm2 level is slightly down-regulated by long term LT treatments because transcript levels are higher in nonacclimated wheat leaves than in the 36-d-acclimated ones. The use of the wheat Tacor413-pm1 and tm1 full-length probes revealed that LT-sensitive oat and LT-tolerant barley and rye also possess cold-inducible homologs of the cor413 family (Fig. 4A). The wheat probes did not detect any signal in rice, but the use of rice-specific probes showed that the transcript level of Oscor413-pm1 is detectable but not LT-regulated under the four temperature regimes used. In contrast, Oscor413-tm1 transcripts are practically undetectable (Fig. 4B). Results obtained with the maize Zmcor413-pm1 and -tm1 probes using similar treatments have shown that both transcripts are undetectable (data not shown). In Arabidopsis, the Atcor413-pm1 and Atcor413-tm1 transcripts accumulate in response to the LT treatments, but Atcor413-pm2 transcripts are undetectable (Fig. 4C).
Figure 3.
Accumulation of Tacor413-pm and -tm mRNAs during CA in spring and winter wheat. A, Accumulation of Tacor413-pm and -tm mRNAs during CA in spring wheat cv Glenlea and winter wheat cv Norstar. NA7, NA12, nonacclimated control plants grown for 7 and 12 d; CA1, CA6, and CA36, 7-d-old plants were cold-acclimated plants for 1, 6, and 36 d; DA5, cold-acclimated plants (36 d) were deacclimated for 5 d. B, Accumulation of Tacor413-pm and -tm mRNAs during CA in spring and winter wheat cultivars. Total RNA (7.5 μg) from shoots of two spring wheat genotypes (cv Glenlea [Glen], LT50 [lethal temperature that kills 50% of the seedlings] of −8°C; and cv Concorde [Con], LT50 of −8°C), four winter wheat genotypes (cv Monopole [Mon], LT50 of −15°C; cv Absolvent [Abs], LT50 of −16°C; cv Fredrick [Fred], LT50 of −16°C; and cv Norstar [Nor], LT50 of −19°C. NA, Nonacclimated plants grown for 13 d; CA36, 7-d-old plants were cold acclimated for 36 d. The 28S ribosomal band stained with ethidium bromide is included to show RNA loads.
Figure 4.
Accumulation of Cor413-pm and -tm mRNAs during CA in cereals and Arabidopsis. A, Differential accumulation in various species. In this study, total RNA (7.5 μg) from oat (Avena sativa L. cv Laurent, LT50 of −6°C), barley (cv Winchester, LT50 of −7°C), and winter rye (Secale cereale L. cv Musketeer, LT50 of−21°C) were used. NA, Nonacclimated plants grown for 13 d; CA36, 7-d-old plants were cold acclimated for 36 d. B, Accumulation in rice (O. sativa subsp. indica cv IR36). Plants grown for 24 h under the corresponding day/night temperatures in degrees Celsius. C, Accumulation in Arabidopsis. NA, Nonacclimated plants grown for 40 d under short photoperiod; CA4h, CA1, CA4, CA7, CA14, and CA21, cold-acclimated plants for 4 h and 1, 4, 7, 14, and 21 d; CTRL, dehydration control plants were removed from pots and placed in water; WS, water-stressed plants were water-stressed by removing them from pots and allowing them to dry for the indicated periods of time; ABA, plants treated with 0.1 mm ABA (Sigma-Aldrich, St. Louis) for 2 and 18 h. The 28S ribosomal band stained with ethidium bromide is included to show RNA loads.
To determine whether the wheat and Arabidopsis cor413 gene families are specifically regulated by LT, plants were subjected to different stress treatments (Figs. 4C and 5A). RNA gel-blot analysis indicated that water stress induces the accumulation of Atcor413-pm1 and -tm1 as well as Tacor413-pm1 and -tm1 transcripts to a level comparable to 1 d of LT exposure. Exogenous application of the stress-associated growth regulator abscisic acid (100 μm) also induced the accumulation of the four transcripts. Taken together, these results suggest that the AtCOR413-PM1 and TM1 proteins could be dicotyledonous orthologs of the wheat COR413-PM1 and TM1 proteins.
Figure 5.
Accumulation of Tacor413-pm and -tm mRNAs under stress conditions and tissue specificity in winter wheat cv Norstar. A, Accumulation of Tacor413-pm and -tm mRNAs under different stress conditions. NA, Nonacclimated plants grown for 7 d; CA1, plants cold acclimated for 1 d; HS1 and 3 h, plants exposed to 40°C for 1 and 3 h (heat shock); ABA18h, plants treated with 0.1 mm ABA (Sigma-Aldrich) for 18 h; NaCl, plants treated with 300 mm NaCl for 18 h; DHN CTRL, dehydration control plants grown for 7 d; DHN30% and 70%, water stressed plants with a relative water content of 30% and 70%. B, Tissue specificity in winter wheat cv Norstar. Leaf, crown, and roots of nonacclimated plants (NA) and 6-d cold-acclimated (CA6) plants. C, Tacor413-tm1 expression is dependent on the chloroplast differentiation stage. NA, Nonacclimated plants grown for 7 d in the presence of a light cycle (8 h of light:16 h of dark; green) or in the dark (etiolated); NA+L, 7-d-old etiolated plants after one light cycle; CA, 7-d-old green or etiolated plants cold acclimated for 24 h in the presence of light (L) or in the dark (D). The 28S ribosomal band stained with ethidium bromide is included to show RNA loads (7.5 μg).
Tissue Specificity and Light Regulation of cor413 Genes
The expression data in Figure 5B shows that, under LT conditions, Tacor413-pm1 is expressed more abundantly in leaves and roots, whereas the chloroplastic protein-encoding Tacor413-tm1 accumulated only in the photosynthetic tissues. To further investigate the association of the Tacor413-tm1 expression profile with photosynthetic tissue, we analyzed the regulation of the cor413 family members under different light conditions. The results in Figure 5C show that the LT-induced expression of Tacor413-pm1 is not light dependent and is not associated with the chloroplast differentiation stage. In contrast, Tacor413-tm1 LT accumulation is dependent on the chloroplast differentiation stage because Tacor413-tm1 accumulation is higher in light-grown plants than in etiolated plants.
To take advantage of the large body of information generated from the different plant EST projects, we analyzed systematically each GenBank cor413-related entry for information regarding tissue specificity. The result of our survey is presented in Supplemental Table II. In addition to the leaf and roots tissues, cor413-pm and cor413-tm transcripts were found in wheat pre-anthesis spike, maize glume, and rice panicle. In dicotyledonous plants, they are found in Arabidopsis flower buds, cotton post-anthesis fiber bolls, potato (Solanum tuberosum) sprouting eyes, alfalfa (Medicago sativa) root tips, soybean immature flowers, and tomato flower buds and maturing fruits. Cor413 members were also found in P. taeda bark tissue, P. patens protonemata, and M. polymorpha immature sex organs. This survey reveals that cor413-pm and -tm expression is not restricted to the plant vegetative stage but also occurs in the final phase of the reproductive stage.
DISCUSSION
A combination of comparative expression profiling and bioinformatic tools was used to identify and to characterize a novel family of plant multispanning transmembrane proteins. Data mining of various nucleotide databases and protein sequence alignments revealed that the higher plant COR413 family can be clustered into two distinct groups. Sequence analyses revealed four important conserved features on COR413-PM and two on COR413-TM. These features are related to the cellular localization, the structure, and the presence of a phosphorylation site and of a GPI-anchoring site.
The bioinformatic approach used allowed us to propose that COR413-PM proteins are targeted to the plasma membrane and that COR413-TM proteins are targeted to the thylakoid membrane. The predicted localization of COR413-TM proteins is further corroborated by the fact that their corresponding transcripts are more abundant in photosynthetic tissues and are regulated by the chloroplast differentiation stage. The existence of some EST entries from non-photosynthetic tissues such as alfalfa developing flowers, Arabidopsis flower buds, potato sprouting eyes, and barley etiolated tissues suggests that the proteins may also be associated with other plasts.
The hidden Markov-based TMHMM software predicted that both COR413-PM and -TM proteins are likely to possess five transmembrane helices and that the N-terminal end of group COR413-PM may be located on the extracellular side of the plasma membrane. Although this is the most probable topology for the moment, TMHMM had a difficulty reaching a consensus topology, especially with COR413-TM proteins. This may be due to the fact that TMHMM, which uses a machine-learning algorithm, was tested using a data set containing very few plant membrane proteins (Krogh et al., 2001). This hypothesis suggests that the prediction of plant membrane protein structures will certainly become more accurate with time when more plant data becomes available for data set generation. This particular program was used for two main reasons: first, it is considered the most accurate prediction software (Möller et al., 2001), and second, because it is based on a hidden Markov algorithm. A recent review on membrane protein topogenesis concluded that the current knowledge makes it impossible to establish consensus rules because too many different processes seem to influence simultaneously the insertion of the protein into the membrane (Goder and Spiess, 2001). On the basis of this conclusion, we believe that machine learning algorithms such as the hidden Markov algorithm can better take into account the subtle differences in amino acids that cannot be deduced by any other method.
The use of the neural network-based predictor NetPhos 2.0 (Blom et al., 1999) suggested that COR413-PM members are likely to possess a different phosphorylation site on the second intracellular loop. It is worth mentioning that the software predicted a phosphorylation site in the loop between TMD3 and TMD4 of the wheat TaCOR413-PM1 and -PM2 proteins, and this loop is a region that is highly divergent between the two proteins. This suggests that the two proteins may be regulated by different kinases (Supplemental Fig. 1).
The last consensus prediction was obtained from the anchoring site predictor DGPI. This program found the presence of a cleavage site and a favorable environment for the addition of a GPI anchor (proper hydrophobic tail length and hydrophilic region length) at the second extracellular loop of COR413-PM proteins. The only feature that DGPI did not detect on COR413-PM sequences is the presence of an N-terminal cleavable signal peptide for translocation to the ER, and neither was this feature detected by the accurate SignalP signal sorting predictor (Nielsen and Krogh, 1998). However, SignalP did predict the presence of a non-cleavable signal anchor for ER translocation in several COR413-PM. It is thus possible that in our case, a signal anchor may replace the signal peptide. Although no multispanning transmembrane proteins are currently known to be GPI-anchored (Borner et al., 2002), the results obtained from our bioinformatic analyses do not at this point rule out the possibility that COR413-PM proteins could be GPI-anchored. The GPI-modified proteins are usually identified by their presence in the soluble fraction after GPI cleavage by specific lipases. Thus, multispanning transmembrane proteins will always remain attached to the membrane fraction and will not be identified as GPI-containing proteins. Thus a special experimental procedure needs to be developed to confirm our hypothesis.
Gene Expression Studies
To understand the function of the COR413 family, expression patterns were determined during several environmental stresses. The expression of one member of group COR413-PM and one from group COR413-TM was closely associated with the acquisition of FT in several plant species such as wheat, rye, and Arabidopsis. This observation is in agreement with the recent microarray analysis that identified Atcor413-pm1 as an LT-inducible gene in Arabidopsis (Seki et al., 2001; clone FL3–5A3). On the other hand, group Cor413-pm and -tm transcripts were not induced in the LT-sensitive species rice and maize. Together, these results suggest that the cor413 expression is not associated with a general metabolic response to LT. Furthermore, the wheat and Arabidopsis cor413-pm1 and -tm1 genes were also induced by water stress and abscisic acid. Interestingly, a cor413 homolog was found in an EST survey of ABA-treated protonemata cells of the moss P. patens (Machuka et al., 1999). Furthermore, recent results have shown that a P. patens homolog of cor413 is induced by ABA and slightly by LT, and these increases were associated with the development of FT of the protonemata cells (Nagao et al., 2001).
Putative COR413 Function
It is known that the plasma membrane is the primary site of freezing injury (Steponkus, 1984). To date, only highly soluble amphipatic proteins have been proposed to act as membrane-stabilizing proteins (Artus et al., 1996; Danyluk et al., 1996). As an integral membrane protein, COR413-PM could play a structural role by stabilizing the plasma membrane lipid bilayer. If the proposed function is exact, the existence of a thylakoid COR413 may suggest that this membrane also needs structural reinforcement.
The second proposed hypothesis is that the COR413 protein family is associated with environmental stress signaling. This hypothesis is based on the comparison between our structural model (Fig. 2, model 1) and that of the mammalian Rhodopsin-like G-protein-coupled receptor (GPCR) family (Bockaert and Pin, 1999). GPCR is the largest family of receptors in animals, and sequence alignment studies have helped classify them into five large clusters. The largest cluster is named the Rhodopsin-like class A GPCR and contains at least 1,000 different members (Horn et al., 1998). All of these proteins share little sequence identity, but one triplet motif (E/D-R-Y) is highly conserved and has been the subject of numerous mutational studies (Scheer et al., 1996; Alewijnse et al., 2000; Chung et al., 2002, and refs. therein). It is located on the internal side of the membrane at the border of the third TMD and second intracellular loop of GPCR (Fig. 2). The aspartic/Glu residues contribute to maintain the receptor in its quiescent state (Chung et al., 2002). A similar motif (D-R/K-T) was found in the most conserved region of COR413-PM and moss COR413 (Supplemental Figs. 1 and 3), and it is also located on the internal side of the membrane at the border of TMD1 and the first intracellular loop (Fig. 2). Although there is compelling biochemical and molecular evidence for the existence of GPCR-based signaling in plants (the three components of the heterotrimeric G-protein are identified), no receptor has been clearly shown to act as a GPCR (Millner, 2001). Two plant proteins are actually considered GPCR. The first is the MLO protein family that is related to the animal GPCR family because it also possesses seven TMD (Devoto et al., 1999). The second GCR1, was isolated by its sequence homology with the Dictyostelium spp. cAMP GPCR (Josefsson and Rask, 1997; Plakidou-Dymock et al., 1998). These cAMP receptors are not clustered with the Rhodopsin-like family, but GCR1 possesses a motif similar to the D-R-Y triplet (H-R-T). On the basis of transgenic studies Colucci et al. (2002) recently suggested that GCR1 may be the gibberellic acid receptor.
Three other features support the assumption that group COR413-PM proteins are related to the Rhodopsin-like GPCR family. GPCR are regulated by kinases, and the phosphorylation sites are often located on the last intracellular loop (Pitcher et al., 1998). It is on this loop that the conserved putative phosphorylation site was predicted for the COR413-PM proteins. The second feature is the presence of a lipid-anchoring site. It is known that saturated acyl chains are sometimes added on GPCR to link the C-terminal tail to the inner leaflet of the plasma membrane (Fig. 2; Bouvier et al., 1995). Therefore, a hydrophobic molecule is added to an already highly hydrophobic protein, as is predicted for COR413-PM members with the addition of a GPI anchor. In animal cells, GPI anchors are used to target the modified proteins into special cholesterol and sphingolipid-rich membrane domains named lipid rafts (Brown and London, 2000). These membrane domains were shown to be the site of intense signaling events. Similar domains were recently identified in the plant plasma membrane (Peskan et al., 2000). The third feature linking COR413-PM to GPCR is the presence of seven highly conserved Pro residues (Supplemental Fig. 1). In transmembrane proteins such as GPCR, some highly conserved Pro residues located inside the TMD are known to be important for correct folding and function (Sansom and Weinstein, 2000). The rigid body motion of the two portions of a Pro-kinked TMD is proposed as a key dynamic component in the rearrangement of GPCR structure upon activation by ligand binding. Interestingly, five of the seven Pro residues in COR413-PM sequences are conserved in COR413-TM sequences, suggesting that they may play the same role in both subgroups (Supplemental Figs. 1–3).
On the basis of these analyses, one may ask what is the specific ligand for COR413-PM? Several molecules can act as GPCR ligands in animal cells, and molecules sharing chemical characteristics with some of these ligands do exist in plants (Supplemental Table VI; Wink, 1997). Knowing that the larger extracellular loop of COR413-PM members is the region with the highest homology with the chloroplastic COR413-TM, it is possible that both proteins bind the same ligand. The exact biochemical properties and function of this new protein family during LT acclimation remains to be determined. Nevertheless, the combination of data mining, bioinformatic analyses, and expression profiling presented here will help us in the design of experimental procedures aimed at answering those questions.
MATERIALS AND METHODS
Plant Material and Growth Conditions
In this study, we used two spring wheat genotypes (Triticum aestivum L. cv Glenlea, LT50 of −8°C; and cv Concorde, LT50 of −8°C); four winter wheat genotypes (T. aestivum L. cv Monopole, LT50 of −15°C; cv Absolvent, LT50 of −16°C; cv Fredrick, LT50 of −16°C; and cv Norstar, LT50 of −19); winter rye (Secale cereale L. cv Musketeer, LT50 of −21°C); oat (Avena sativa L. cv Laurent, LT50 of −6°C); barley (Hordeum vulgare L. cv Winchester, LT50 of −7°C); rice (Oryza sativa subsp. indica cv IR36, LT50 of 4°C); maize (Zea mays, LT50 of 4°C); and Arabidopsis ecotype Columbia (LT50 of −9°C). Growth of plants and stress treatments were as previously described (Frenette Charron et al., 2002).
Cloning and Data Mining
The Tacor413-pm1 clone (previously pWcor413) was isolated by differential screening of a Lambda Zap II library constructed from poly(A+) RNA isolated from 1-d cold-acclimated winter wheat (cv Norstar; Houde et al., 1992). The Tacor413-pm1 clone was purified and excised as a pBluescript vector following the library supplier's protocol (Stratagene, La Jolla, CA).
Database searches to identify Tacor413-pm1 homologs were performed using the Canadian Bioinformatics Resource (Halifax, Nova Scotia, Canada; http://www.cbr.nrc.ca) and National Center for Biotechnology Information (Bethesda, MD; http://www.ncbi.nlm.nih.gov/BLAST) Web implementation of BLAST (Altschul et al., 1990) against the GenBank nonredundant sequence database and GenBank EST database (Benson et al., 2002). In the first round of data mining, COR413 homologs were identified by using the TaCOR413-pm1 protein sequence as query with TBLASTN against the GenBank EST database. In the second round, the identified EST from each different plant species containing the longest 5′ or 3′ end were used as query to search the same database. Overlapping ESTs were assembled, and a consensus cDNA was deduced when two or more identical sequence could be aligned. To obtain the largest number of complete COR413 sequences, available clones were ordered, sequenced, and submitted to GenBank, and the others were deduced from the available genomic and EST sequences. The ORF of the assembled gene was identified using ORFinder on the NCBI Web site (T. Tatusov and R. Tatusov, unpublished data; http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The longest ORF was always chosen (except for CjCOR413γ where the third ATG was chosen). The complete in silico assembled nucleotide sequence and its encoded protein were then used to screen back the EST database. This other round of data mining was useful for the identification of very near homologs with subtle amino acid differences. Only the complete COR413 homologs were used for subsequent structural and functional domain prediction analyses. Survey of all the homologs identified can be found in Supplemental Table VII. AtCOR413-pm3 and AtCOR413-pm4 were not used in the bioinformatic analysis. Although AtCOR413-pm3 and pm4 seem to be related to group COR413-PM, their sequences are slightly different, and no other similar plant proteins were found.
The degree of sequence identity in Table I was determined using ALIGN (Pearson, 1990) on the Biology Workbench workstation (http://workbench.sdsc.edu/). Group I and II sequences were aligned and analyzed by using ClustalW (Thompson et al., 1994) on the Biology Workbench (http://workbench.sdsc.edu/) and Network Protein Sequence Analysis servers (Combet et al., 2000; http://pbil.ibcp.fr/). Shading of amino acids was performed with the BOXSHADE program at the BOXSHADE Web server at the University of Lausanne (Switzerland; http://ulrec3.unil.ch/software/boxshade/boxshade.html).
Structural Analyses
For detection of specific targeting sequences, we used PSORT, iPSORT (Nakai and Kanehisa, 1992; Bannai et al., 2002; http://psort.nibb.ac.jp/), and TargetP v1.01 (Emanuelsson et al., 2000; http://www.cbs.dtu.dk). For detection of signal peptides, SignalP v2.0 was used (Nielsen and Krogh, 1998; http://www.cbs.dtu.dk). Before performing structural prediction, a Kyte and Doolittle hydropathic plot was generated by using the Protscale program (http://ca.expasy.org/cgi-bin/protscale.pl) with the Kyte and Doolittle option and a window of nine amino acids (Kyte and Doolittle, 1982). The superposition of COR413 hydropathic plot was generated by transferring the raw data to Microsoft Excel (Microsoft, Redmond, WA). The graph was constructed by aligning the data table to the last C-terminal amino acid therefore compensating for the various protein N-terminal lengths. For TMD prediction, TMHMM (http://www.cbs.dtu.dk) was used (Krogh et al., 2001). The TMHMM data tables were processed with Microsoft Excel as for hydropathic plot. α-Helical regions were identified with secondary structure prediction programs integrated in the Web implementation of ClustalW at the Network Protein Sequence Analysis Web site (Combet et al., 2000; http://pbil.ibcp.fr).
Other Prediction Servers
For functional domain identification, we first used ScanPROSITE on the Expasy Web server for PROSITE motif database screening (http://ca.expasy.org/tools/scanprosite/) and NCBI RPS-BLAST for Pfam and Smart conserved domain databases screening (Altschul et al., 1997; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). In a subsequent search, we used most of the software available on the Expasy server (http://ca.expasy.org/). Two types of the software gave interesting results. NetPhos was used for consensus phosphorylation site detection (http://www.cbs. dtu.dk; Blom et al., 1999), and DGPI was used for GPI-anchoring site detection (http://129.194.186.123/GPI-anchor/index_en.html; D. Buloz and J. Kronegg, unpublished data).
Expression Studies
Cereal RNA extraction and RNA gel-blot analysis were performed as already described (Houde et al., 1992). Total RNA from Arabidopsis and etiolated wheat were extracted using Tri Reagent (Molecular Research Center, Cincinnati) according to the manufacturer protocol. To prevent cross-hybridization between Tacor413-pm1 and -pm2 probe in northern analysis, specific probes for the 3′-non-coding region of each cDNA were used. These probes were generated by PCR with the following primers: Tacor413- pm1, 5′-ttcatctacccggtctgggccgtc and 5′-ccaggaaacaaactaagacgtgacacc; and Tacor413-pm2, 5′-agtctgggtcctggtgctc and 5′-tcataccagaactacaacaaatcg (Tacor413-pm2 and Tacor413-tm1 clones were kindly provided by Dr Anderson [U.S. Department of Agriculture-Agricultural Research Service-Plant Gene Expression Conter, Albany, CA). Northern blots of rice and maize samples were performed using the complete Oscor413-pm1 or Oscor413-tm1 clones (kindly provided by Dr. Sasaki as part of the Japanese Rice Genome Research Program of the National Institute of Agrobiological Sciences and the Institute of the Society of Techno-Innovation in Agriculture, Forestry and Fisheries; Yamamoto and Sasaki, 1997) and Zmcor413-pm1 or Zmcor413-tm1 clones (kindly provided by Dr. Singh [Agriculture and Agri-Food Canada]). The Arabidopsis Atcor413-pm1 and pm2 probes also showed cross-hybridization. Therefore, probes specific to the 5′ non-coding regions were generated by PCR and used in the hydridizations. The first primer hybridized with the vector cloning site left border (5′-atagagctcactagtccggaattcccgggtcga) and the second hybridized specifically to the Atcor413-pm1 or -pm2 sequences (Atcor413-pm1, 5′-gtatatggcggcgattgaagcaacc; and Atcor413-pm2, 5′-tggcagcgaaagaagcgaggaatttga). For Atcor413-tm1 the complete cDNA was used as probe. Dr. Newman (Department of Energy-Plant Research Laboratory, East Lansing, MI) kindly provided the three Arabidopsis clones from Arabidopsis Biological Resource Center (Ohio State University, Columbus) distribution services. Northern analyses for each sample were performed at least three times from two biological replicates. For other COR413 sequence analysis, Dr. Ujino-Ihara from the Forestry and Forest Products Research Institute kindly provided the C. japonica clone, Dr. Bashiardes as part of the Physcomitrella EST Program at the University of Leeds (UK) and Washington University (St. Louis) kindly provided the P. patens clone, and Dr. Anderson from Clemson University Genomic Institute kindly provided the barley clone. All distributed clones are identified in Supplemental Table VII.
Supplementary Material
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada, by Genome Canada, by Genome Québec, and by Fonds pour la Formation des Chercheurs et l'Aide à la recherche (research grants to F.S.).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.015255.
LITERATURE CITED
- Agarraberes FA, Dice JF. Protein translocation across membranes. Biochim Biophys Acta. 2001;1513:1–24. doi: 10.1016/s0304-4157(01)00005-3. [DOI] [PubMed] [Google Scholar]
- Alewijnse AE, Timmerman H, Jacobs EH, Smit MJ, Roovers E, Cotecchia S, Leurs R. The effect of mutations in the DRY motif on the constitutive activity and structural instability of the histamine H(2) receptor. Mol Pharmacol. 2000;57:890–898. [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäfer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3381–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin C, Thomashow MF. Constitutive expression of the cold-regulated Arabidopsis thalianaCOR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA. 1996;93:13404–13409. doi: 10.1073/pnas.93.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. Genbank. Nucleic Acids Res. 2002;30:17–20. doi: 10.1093/nar/30.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Bockaert J, Pin JP. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P. Prediction of glycosylphosphatidylinositol-anchored proteins in ArabidopsisA genomic analysis. Plant Physiol. 2002;129:486–499. doi: 10.1104/pp.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier M, Moffett S, Loisel TP, Mouillac B, Hebert T, Chidiac P. Palmitoylation of G-protein-coupled receptors: a dynamic modification with functional consequences. Biochem Soc Trans. 1995;23:116–120. doi: 10.1042/bst0230116. [DOI] [PubMed] [Google Scholar]
- Breton G, Danyluk J, Ouellet F, Sarhan F. Biotechnological applications of plant freezing associated proteins. Biotechnol Annu Rev. 2000;6:59–101. doi: 10.1016/s1387-2656(00)06019-1. [DOI] [PubMed] [Google Scholar]
- Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- Chung DA, Wade SM, Fowler CB, Woods DD, Abada PB, Mosberg HI, Neubig RR. Mutagenesis and peptide analysis of the DRY motif in the alpha2A adrenergic receptor: evidence for alternate mechanisms in G protein-coupled receptors. Biochem Biophys Res Commun. 2002;293:1233–1241. doi: 10.1016/S0006-291X(02)00357-1. [DOI] [PubMed] [Google Scholar]
- Colucci G, Apone F, Alyeshmerni N, Chalmers D, Chrispeels MJ. GCR1, the putative ArabidopsisG protein-coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc Natl Acad Sci USA. 2002;99:4736–4741. doi: 10.1073/pnas.072087699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: Network Protein Sequence Analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- Danyluk J. Identification et caractérisation moléculaire de gènes induits au cours de l'acclimatation au froid chez le blé (Triticum aestivum). PhD thesis. Montréal: Université de Montréal; 1996. [Google Scholar]
- Danyluk J, Perron A, Houde M, Limin A, Fowler B, Benhamou N, Sarhan F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell. 1996;10:623–638. doi: 10.1105/tpc.10.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, Schulze-Lefert P. Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem. 1999;274:34993–35004. doi: 10.1074/jbc.274.49.34993. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Fowler DB, Limin AE, Ritchie JT. Low-temperature tolerance in cereals: model and genetic interpretation. Crop Sci. 1999;39:626–633. [Google Scholar]
- Frenette Charron JB, Breton G, Badawi M, Sarhan F. Molecular and structural analyses of a novel temperature stress-induced lipocalin from wheat and Arabidopsis. FEBS Lett. 2002;517:129–132. doi: 10.1016/s0014-5793(02)02606-6. [DOI] [PubMed] [Google Scholar]
- Goder V, Spiess M. Topogenesis of membrane proteins: determinants and dynamics. FEBS Lett. 2001;504:87–93. doi: 10.1016/s0014-5793(01)02712-0. [DOI] [PubMed] [Google Scholar]
- Horn F, Weare J, Beukers MW, Horsch S, Bairoch A, Chen W, Edvardsen O, Campagne F, Vriend G. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res. 1998;26:275–279. doi: 10.1093/nar/26.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde M, Danyluk J, Laliberté J-F, Rassart E, Dhindsa RS, Sarhan F. Cloning, characterization and expression of a cDNA encoding a 50-kilodalton protein specifically induced by cold acclimation in wheat. Plant Physiol. 1992;99:1381–1387. doi: 10.1104/pp.99.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson LG, Rask L. Cloning of a putative G-protein-coupled receptor from Arabidopsis thaliana. Eur J Biochem. 1997;249:415–420. doi: 10.1111/j.1432-1033.1997.t01-1-00415.x. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Machuka J, Bashiardes S, Ruben E, Spooner K, Cuming A, Knight C, Cove D. Sequence analysis of expressed sequence tags from an ABA-treated cDNA library identifies stress response genes in the moss Physcomitrella patens. Plant Cell Physiol. 1999;40:378–387. doi: 10.1093/oxfordjournals.pcp.a029553. [DOI] [PubMed] [Google Scholar]
- Millner PA. Heterotrimeric G-protein in plant cell signaling. New Phytol. 2001;151:165–174. doi: 10.1046/j.1469-8137.2001.00172.x. [DOI] [PubMed] [Google Scholar]
- Möller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Nagao M, Minami A, Takezawa D, Arakawa K, Fujikawa S. ABA-induced freezing tolerance in Physcomitrella patensand gene expression (abstract no. 354[F455]) Plant Cell Physiol. 2001;42:s121. [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. In: Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1998. pp. 122–130. [PubMed] [Google Scholar]
- Pearson WR. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- Peskan T, Westermann M, Oelmuller R. Identification of low-density Triton X-100-insoluble plasma membrane microdomains in higher plants. Eur J Biochem. 2000;267:6989–6995. doi: 10.1046/j.1432-1327.2000.01776.x. [DOI] [PubMed] [Google Scholar]
- Pitcher JA, Freedman NJ, Lefkowitz RJ. G protein-coupled receptor kinases. Annu Rev Biochem. 1998;67:653–692. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- Plakidou-Dymock S, Dymock D, Hooley R. A higher plant seven-transmembrane receptor that influences sensitivity to cytokinins. Curr Biol. 1998;8:315–324. doi: 10.1016/s0960-9822(98)70131-9. [DOI] [PubMed] [Google Scholar]
- Sansom MS, Weinstein H. Hinges, swivels and switches: the role of prolines in signalling via transmembrane alpha-helices. Trends Pharmacol Sci. 2000;21:445–451. doi: 10.1016/s0165-6147(00)01553-4. [DOI] [PubMed] [Google Scholar]
- Sarhan F, Ouellet F, Vazquez-Tello A. The wheat wcs120 gene family: a useful model to understand the molecular genetics of freezing tolerance in cereals. Physiol Plant. 1997;101:439–445. [Google Scholar]
- Scheer A, Fanelli F, Costa T, De Benedetti PG, Cotecchia S. Constitutively active mutants of the alpha 1β-adrenergic receptor: role of highly conserved polar amino acids in receptor activation. EMBO J. 1996;15:3566–3578. [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsisgenes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Gendall AR, Dean C. When to switch to flowering. Annu Rev Cell Dev Biol. 1999;15:519–550. doi: 10.1146/annurev.cellbio.15.1.519. [DOI] [PubMed] [Google Scholar]
- Steponkus PL. Role of the plasma membrane in freezing injury and cold acclimation. Annu Rev Plant Physiol. 1984;35:543–584. [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnàdy GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Wink M. Special nitrogen metabolism. In: Dey PM, Harbourne JB, editors. Plant Biochemistry. San Diego: Academic Press; 1997. pp. 439–486. [Google Scholar]
- Yamamoto K, Sasaki T. Large-scale EST sequencing in rice. Plant Mol Biol. 1997;35:135–144. [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al. A draft sequence of the rice genome (Oryza sativaL. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.