Abstract
Apoptosis is essential for normal growth and development of multicellular organisms, including metazoans and higher plants. Although cell death processes have been reported in unicellular organisms, key elements of apoptotic pathways have not been identified. Here, we show that when placed in darkness, the unicellular chlorophyte alga Dunaliella tertiolecta undergoes a form of cell death reminiscent of apoptosis in metazoans. Many morphological criteria of apoptotic cell death were met, including an increase in chromatin margination, degradation of the nucleus, and DNA fragmentation. Biochemical assays of the activities of cell death-associated proteases, caspases, measured using highly specific fluorogenic substrates, increased with time in darkness and paralleled the morphological changes. The caspase-like activities were inhibited by caspase-specific inhibitors. Antibodies raised against mammalian caspases cross-reacted with specific proteins in the alga. The pattern of expression of these immunologically reactive proteins was correlated with the onset of cell death. The occurrence of key components of apoptosis, and particularly a caspase-mediated cell death cascade in a relatively ancient linage of eukaryotic photoautotrophs, argues against current theories that cell death evolved in multicellular organisms. We hypothesize that key elements of cell death pathways were transferred to the nuclear genome of early eukaryotes through ancient viral infections in the Precambrian Ocean before the evolution of multicellular organisms and were subsequently appropriated in both metazoan and higher plant lineages.
Programmed cell death (PCD) or “cell suicide” is a widespread process among metazoans and is essential for the proper development, function, and ultimately survival of the organism (Leist and Nicotera, 1997). An emerging topic in plant biology is whether plants display analogous elements of metazoan PCD (Greenberg, 1996; Pennell and Lamb, 1997). For example, in many plant-pathogen interactions, cell death occurs in both susceptible and resistant hosts. Specific recognition responses in plants trigger the hypersensitive response (HR) and activation of host defense mechanisms, resulting in restriction of pathogen growth and disease development. Several studies have provided evidence that cell death during HR involves activation of a plant-encoded pathway for cell death (e.g. Lam et al., 2001).
The identities of most of the key elements involved in plant PCD remain unknown. Iterative homology searches suggest that caspase-related proteases (metacaspases, first identified in yeasts) are encoded in the Arabidopsis genome (Uren et al., 2000). Recent evidence from inhibitor studies and biochemical approaches suggests that caspase-like proteases may also be involved in cell death control in higher plants (Korthout et al., 2000) and may constitute a common pathway for cell death in both metazoans and plant cells. However, the absence of homologs of metazoan cell death genes in Arabidopsis (see Arabidopsis Genome Initiative, 2000) suggests that cell death in higher plants has diverged from that found in the animal kingdom. This controversy can only be addressed by examining cell death in extant organisms that are representative of divisions that emerged before the divergence of metazoans and higher plants.
The origin of cell death programs in eukaryotes remains obscure (Aravind et al., 2001) but is probably ancient; apoptosis-like phenomena have been reported in unicellular organisms, including chlorophytes (Berges and Falkowski, 1998), dinoflagellates (Vardi et al., 1999), yeast (Frohlich and Madeo, 2000), and bacteria (Chen at al., 1998; Lewis, 2000). However, the pathways through which cell death proceeds have not yet been identified in these organisms.
Cells undergoing apoptosis suffer a series of typical changes, including chromatin condensation and margination as well as DNA cleavage into large and eventually small fragments (50 kb–50 bp). In the cytoplasm, organelles appear to remain intact (Cohen, 1997). Berges and Falkowski (1998) demonstrated that autocatalyzed cell death processes could also be elicited in the single-celled chlorophyte alga Dunaliella tertiolecta. This unicellular chlorophyte is an obligate photoautotroph that cannot use dissolved organic compounds and does not reproduce sexually in culture. Under normal culture conditions given simple inorganic nutrients and light, D. tertiolecta is immortal; that is, the cell divides by simple binary fission without any evidence of cell lysis, encystment, or spore formation. This organism belongs to a division of eukaryotes that evolved approximately 1.6 billion years ago, or about 900 million years before the first clearly identifiable metazoans appear in the fossil record (Lipps, 1993; Bhattacharya and Medlin, 1998).
D. tertiolecta is well known for its extraordinarily high tolerance to salt stress, high light, and relatively high temperatures. When placed in darkness, however, cell cultures undergo catastrophic cell death between the 4th and 6th d (Berges and Falkowski, 1998). Cell death is preceded by a reduction in photosynthetic capability and cell numbers. Upon triggering the cell death process, the cells literally dissolve, and the culture, which on the previous day had been green, becomes transparent. Zymograms and protein profiles, before and during the culture decline, reveal the induction of novel proteases in the cells (Berges and Falkowski, 1998).
Here, we report that the cell death program in D. tertiolecta is associated with caspase-like activity and that the morphology and biochemical features of the dying algal cells resemble apoptosis. Thus, we hypothesize that important parts of the apoptotic machinery appeared early in the evolution of eukaryotes; caspase-like proteases were strongly selected in metazoans and higher plant cells as a mechanism for development, differentiation, and defense, whereas in unicells they remained as relics.
RESULTS
When exponentially growing D. tertiolecta cultures were deprived of light, the cultures underwent a catastrophic decline in population density between the 4th and 6th d (essentially identical to that described by Berges and Falkowski, 1998). The cells literally dissolved, and the culture turned from green to clear (Fig. 1). This event is not virally mediated. J.A. Berges and C.D.P. Brussaard (unpublished) looked for evidence of viruses or changes in viral abundance in cultures of D. tertioloecta before, during, and after the cell death cascade using both SYBR-Green stain (Noble and Fuhrman, 1998) and by ultracentrifuging whole samples, staining, and examining using electron microscopy (Heldal and Bratbak, 1991). No viruses were detected.
Figure 1.
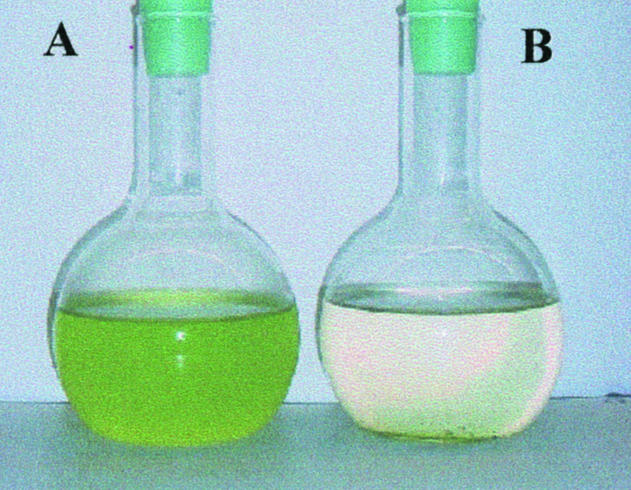
Effects of light deprivation in D. tertiolecta. A, Actively growing culture. B, Culture after 8 d in darkness.
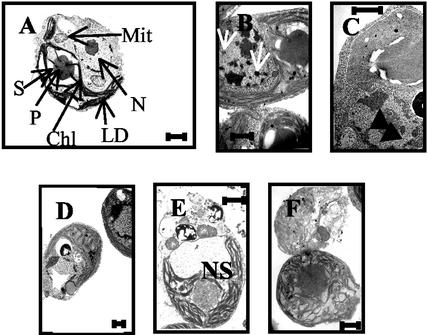
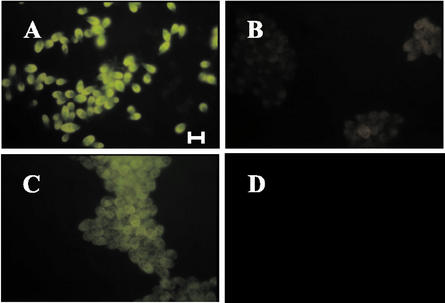
A time course of transmission electron micrographs over this period revealed ultrastructural changes bearing all the hallmarks of apoptosis. In actively growing cells in the light (Fig. 2A), the nuclear membrane was well defined, and chromatin was localized in the nucleolus. When placed in darkness, chromatin condensation and margination at the nuclear envelope could be observed (Fig. 2, B–D). After 4 d in darkness, nuclei were completely lost (Fig. 2E), and cell lysis quickly followed (Fig. 2F). Importantly, cell integrity was maintained, whereas the nucleus was lost; the mitochondria remained intact and although the starch layer surrounding the pyrenoid disappeared, the pyrenoid itself hardly changed. Although these morphological changes occurred, nuclear DNA was concurrently degraded. Free 3′OH ends of DNA, generated by activation of endonuclease activity in dying cells, were fluorescently labeled with a conventional TUNEL assay (Gavrieli et al., 1992). No labeling was observed in cells growing in light (Fig. 3A), but labeling increased from d 1 to 5 in darkness, indicating that some degree of DNA fragmentation occurred after only 1 d in darkness (Fig. 3B). Positive controls, consisting of cells treated with DNAse, showed strong staining, indicative of DNA degradation (Fig. 3C). Negative controls were analyzed by using cells that had been in darkness for 4 d and by substituting MilliQ water for the TdT enzyme. These cells did not stain (Fig. 3D). These results are consistent with caspase-like activity (Cohen, 1997).
Figure 2.
Transmission electron micrographs showing the morphological changes in D. tertiolecta placed in the darkness. Chl, Chloroplast; LD, lipid drops; Mit, mitochondria; N, nucleus; NS, nuclear space; P, pyrenoid; S, starch. A, Normal vegetative cells grown in the light. B and C, Cells after 1 and 2 d in darkness, respectively. Note the movement of chromatin from the nucleolus (arrows). D, After 3 d in darkness, chromatin has moved to the nuclear membrane, and the nucleolus has disappeared. Chloroplastic thylakoids are disorganized, but none of the organelles show evidence of disintegration or swelling. E, After 4 d in darkness, the nucleus is no longer observed. F, After 5 d in darkness, cells have lysed. Horizontal bar = 1 μm.
Figure 3.
DNA fragmentation in D. tertiolecta in darkness revealed by TUNEL staining. A, Cells after 1 to 5 d in darkness. B, Actively growing cells in the light. C, Positive control: cells pretreated with DNAse I. D, Negative control. TdT enzyme substituted with distilled water. Horizontal bar = 10 μm.
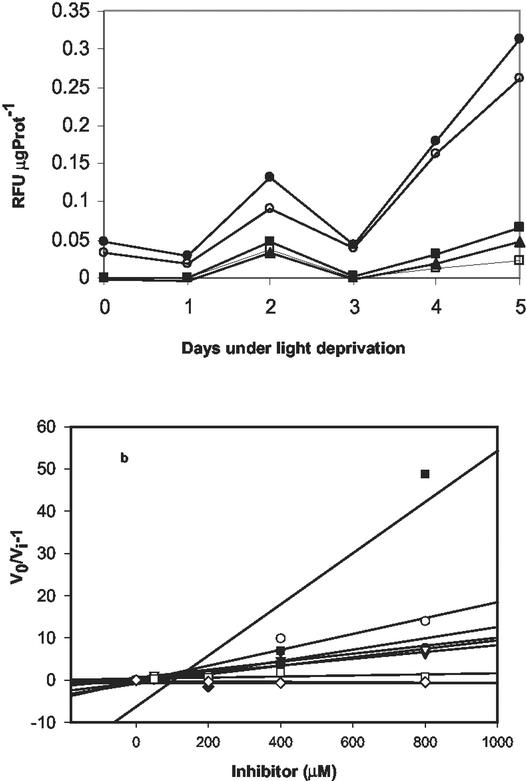
During the cell death process, cleavage of fluorescent substrates specific for caspases 1, 3, 6, 8, and 9 was observed (Fig. 4). These activities followed Michaelis-Menten kinetics (Km values of about 50 μm and saturation at 200 μm; data not shown) and were inhibited by the irreversible caspase inhibitors Boc-D-FMK, Ac-VAD-FMK, and Ac-YEVD-CMK (dissociation constant of an enzyme-inhibitor complex values ranged from 17–124 μm; Fig. 4B). We observed an increase in caspase activities beginning on the 2nd d of darkness and culminating on the 5th d, as the cells lysed (Fig. 4A). The highest activities, observed on d 2, corresponded to substrates for caspases 8 and 9. However, the activities corresponding to substrates for caspases 1, 3, and 6 were somewhat lower.
Figure 4.
Caspase activity in D. tertiolecta. Changes in darkness. a, Caspase activity was measured as hydrolysis of 7-amino-4-fluoromethyl coumarin-labeled substrates specific for caspases 1 (WEHD, ▴), 3 (DEVD, □), 6 (VEID, ▪), 8 (IETD, ●), and 9 (LEHD, ○) during the dark period. Symbols are means of triplicate measurements. The difference between the triplicates was less than 10%. RFU μg Prot−1, relative fluorescence units per microgram of protein. b, Inhibition plots of caspase activity in presence of different concentrations of the irreversible broad spectrum inhibitor Boc-D-FMK. v0, Uninhibited rate of reaction; vi, Rate in the presence of an inhibitor. Plotting v0/vi against the concentration of the inhibitor (I) gives a line with a slope equal to 1/dissociation constant of an enzyme-inhibitor complex (Salvesen and Nagase, 1989); thus, greater slopes indicate smaller inhibition constants and greater effects. Activity decreased as the concentration of the inhibitor increased. Complete inhibition was reached at 400 μm inhibitor. Symbols as in a.
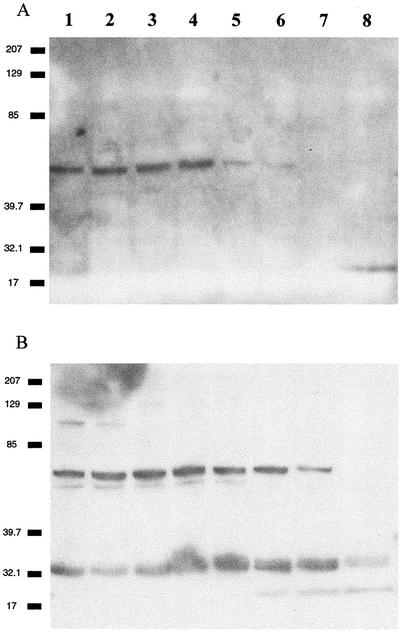
To determine whether the proteases detected with activity assays were related to metazoan caspases, we challenged D. tertiolecta extracts with six polyclonal antibodies derived from mammalian caspase antisera. Antibodies for caspases 1, 3, and 9 cross-reacted with multiple Mr bands (Fig. 5; note that representative blot images have been presented; to save space, we have condensed the results of many blots into Table I), whereas antibodies raised against caspases 6 to 8 recognized only one band. Antibodies raised against caspases 2 and 10 did not cross-react. The apparent Mrs of the antigens were generally larger than what would be expected for the respective metazoan caspases, assuming a higher degree of evolutionary conservation (Aravind et al., 1999). In addition, there was evidence of posttranslational modification; antibodies against caspase 1, 3, and 9 detected high-Mr proteins that declined during dark incubation and lower Mr bands (presumably corresponding to the activated proteases) that increased during the incubation.
Figure 5.
Western blot showing cross-reactions of whole-cell protein extracts from D. tertiolecta with antibodies raised against human caspases. Lanes 1 to 8, Zero to7 d in darkness, respectively. A, Caspase 9; B, caspase 3.
Table I.
Cross-reactions of D. tertiolecta proteins challenged with caspase antibodies
| Days in Darkness | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|---|
| Caspase 1 | ||||||||
| 85 kD | ++ | ++ | ++ | ++ | ||||
| 50 kD | ++ | +++ | ++ | ++ | ++ | ++ | +++ | +++ |
| 30 kD | ++ | +++ | +++ | ++ | ||||
| Caspase 3 | ||||||||
| 100 kD | ++ | + | ||||||
| 70 kD | +++ | +++ | ++++ | ++++ | +++ | +++ | ++ | |
| 33 kD | ++ | ++ | ++ | ++ | +++ | +++ | +++ | + |
| Caspase 6 | ||||||||
| 75 kD | ++ | +++ | +++ | +++ | ||||
| Caspase 7 | ||||||||
| 75 kD | ++ | + | + | |||||
| Caspase 8 | ||||||||
| 25 kD | ++ | ++ | ++++ | ++++ | ++++ | ++ | +++ | +++ |
| Caspase 9 | ||||||||
| 60 kD | +++ | +++ | +++ | +++ | + | + | ||
| 20 kD | ++ |
Intensities of cross reactions correspond to absorbance units: +, 0.1 to 0.5; ++, 0.5 to 1.0; +++, 1.0 to 1.5; and ++++, >1.5.
DISCUSSION
Previous workers have speculated about the origins of PCD and suggested that “true” metazoan caspases evolved from earlier forms that diverged into paracaspases (in metazoans and Dictyostelium discoideum) and metacaspases (in plants, fungi, and protozoa) (Uren et al., 2000), and which probably played very different roles in unicellular organisms (Aravind et al., 2001). Despite the fact that no genes homologous to animal caspases have been isolated from plants, caspase-like activity has been reported in higher plants (Solomon et al., 1999; Delorme et al., 2000; Korthout et al., 2000; Lam and del Pozo, 2000). Iterative homology searches suggest there are two types of metacaspases in Arabidopsis. Type I metacaspases contain a predicted caspase-like proteolytic domain lacking the death effector domain typical in metazoan caspases. Type II metacaspases contain a zinc pro-domain, which is also found in LSD-1, a protein involved in the control of the HR in plants. The functions of these genes have thus far only been inferred from sequence homologies; the enzymatic activity of these metacaspases has not been assayed. The activation of a plant-encoded pathway for cell death has been demonstrated in transgenic tobacco plants expressing human Bcl-2 and Bcl-xl, nematode CED-9, or baculovirus Op-IAP transgenes that negatively regulate apoptosis (Dickman et al., 2001). The ability of Bax, a protein belonging to the Bcl-2 family proteins and known to induce cell death and a defense reaction in plants, suggests that some features of animal and plant cell death processes may be widely shared among multicellular eukaryotes (Lacomme and Cruz, 1999).
There are three different phases in the cell death process (Jabs, 1999): a stimulus-dependent induction phase, an effector phase, and a degradation phase. The degradation phase we describe in D. tertiolecta consists of the activation of caspase-like proteases, leading to complete cell disintegration and lysis. The evidence for caspase-like enzymes is the cross-reactivity with specific antibodies against “true” mammalian caspases, biochemical substrate specificity and inhibition, and morphological processes are consistent with apoptosis. The biochemical substrates with the highest activities on d 2 correspond to caspases 8 and 9, consistent with their role as upstream-regulated proteases. The activity for substrates of caspases 6, 3, and 1, which are effector caspases acting downstream of the apoptotic event, is somewhat lower (Cohen, 1997), indicating the activation cascade of the apoptotic event. The increase in activity observed from d 1 to 2 parallels the results obtained by transmission electron microscopy (TEM) and TUNEL, suggesting that the cells were poised to die. Increases in caspase activities should proceed DNA degradation; however, this is not clear in the present study. Part of the problem may be that the temporal resolution of events is relatively coarse (days versus hours), but it may also be that detection limits of caspase activities are higher than limits on TUNEL assays.
Caspases are among the most specific proteases, having an unusual and stringent requirement for cleavage after Asp. Recognition of at least four amino acids, NH2-terminal to the cleavage site, is also a requirement for efficient catalysis; very few proteolytic enzymes can cleave the fluorogenic substrates used to detect caspases (Thornberry, 1999). Therefore, it is unlikely that our activity assays are biased by the presence of other proteases, especially considering the effectiveness of caspase inhibitors. Despite the fact that no genes homologous to animal caspases have yet been isolated from plants, caspase activity itself has been reported in a number of studies (del Pozo and Lam, 1998; Korthout et al., 2000). In yeast (Saccharomyces cerevisiae), a type-I-like metacaspase protein has been shown to have caspase-like activity and to mediate PCD in aging cells (Madeo et al., 2002).
Our results strongly suggest that the family of caspase-like proteins and the origin of cell death programs predates multicellular life. Moreover, our results suggest that cellular morphology matches that of apoptosis. Together, these independent lines of evidence suggest that important elements of the caspase-induced cell death cascade were present much earlier in evolution than is currently believed (Aravind et al., 2001). In contrast to other kingdoms of life, there is very little information available for protists; these organisms may provide the key to understanding the origin and evolution of apoptosis if we consider that the supposedly common ancestor (before the animal-plant divergence) was unicellular (Aravind et al., 1999).
The cell death phenomenon in D. tertiolecta confers no obvious ecological or evolutionary fitness. This alga cannot use the dissolved organic compounds released from lysis for its own growth, and the organism does not reproduce sexually; hence, suggestions that cell death has evolved for “altruistic” functions (Frohlich and Madeo, 2000; Lewis, 2000) or for cellular differentiation cannot be invoked in this organism. It is possible that the caspase-like activity that is present in normal growth serves some housekeeping functions in protein turnover (Zeuner et al., 1999), the value of which offsets the liability in terms of cell mortality. However, the cell death event triggered by darkness is surely maladaptive. Therefore, we must ask: How did D. tertiolecta acquire these caspase-like proteases, and what purpose(s) do they serve?
Over the past decade, it has become increasingly clear that the ocean contains an extraordinary number of viruses, between 105 and 107 mL−1 (Fuhrman, 1999; Wommack and Colwell, 2000). However, marine viral genomes are poorly characterized; viral attack on both prokaryotic and eukaryotic marine microbes appears to be relatively common and frequently leads to cell lysis (Bratbak et al., 1993; Suttle, 1994). We hypothesize that with the evolution of eukaryotes, viral proteolytic genes became incorporated into marine heterotrophs and autotrophs, and gene transfers were further facilitated laterally via viral infection. Interestingly, a reverse transcriptase gene of retroviral origin is contained and maintained within a nitrogen assimilation operon in the marine cyanobacterium Trichodesmium sp. IMS 101 (A. Post, personal communication). The amino acid motif QAC*G is found only in the active site of “true” caspases or “paracaspases,” and we have also found it on D. tertiolecta (T. Shi, unpublished data). However, based on EMBL database searches, homologous sequences are present in viruses: QACQG is in both the GAG protein from the simian immunodeficiency virus and bacteriophage phi-105 within the ORF10; QQACQG occurs in a hemagglutinin from the measles virus (MV), and QACGG occurs in the genome of human papillomavirus type 67. Defenses against expression of these genes might have included posttranslational modification and/or transposition such that some viral proteases may have been incorporated in the nuclear genome of their target cell as transposed elements. Under stress conditions, the silencing systems fail, and proteases are activated by stresses including energy deprivation and reactive oxygen species (Levine et al., 1994). Interestingly, viruses have evolved mechanisms to stop apoptosis to propagate infection cycles, whereas eukaryotic cells have devised ways to suppress anti-apoptotic viral proteins (see Bump et al., 1995; Uren et al., 1998; Barber, 2001).
Our results clearly establish a PCD pathway that is induced by a specific stress condition (i.e. darkness) in a eukaryotic obligate photoautotroph from an ancient lineage. We propose that the proteases themselves were originally inherited from a common ancestor through viral infection and were widely (but not necessarily universally) appropriated and maintained throughout the evolution of eukaryotes. This hypothesis explains why, belonging to different lineages within the kingdom of Eukaryotes, a unicellular chlorophyte alga such as D. tertiolecta, higher plants, and metazoans share conserved apoptotic machinery. Aravind et al. (1999) emphasized that the apoptosis machinery is based on several ancient domains that have not been “invented” for this function, but rather have been recruited from proteins that in unicellular organisms perform regulatory functions. Although the caspase-like proteins in D. tertiolecta may serve some housekeeping purposes, they appear to be activated during apoptosis. Some of these genes probably were laterally transferred from bacteria to the common ancestor by viral infection, and, in higher plants, mediate the HR in plant-pathogen interactions, floral and organ abortion, senescence, aerenchyma formation under hypoxia, somatic embryogenesis, and differentiation of tracheary elements. PCD triggered by darkness in D. tertiolecta might be the relic of what is now the common response to biotic and abiotic stress for higher plants.
MATERIALS AND METHODS
Culture Conditions
Dunaliella tertiolecta (CCAP strain 19/6) was grown in semicontinuous batch cultures in artificial seawater medium (Goldman and McCarthy, 1978) enriched with f/2 nutrients (Guillard and Ryther, 1962) at 26°C under continuous white light at 200 μmol quanta m−2 s−1. When cultures reached mid log-phase, they were placed in complete darkness while maintaining gentle stirring and bubbling with filtered air.
TEM
Cells were harvested by centrifugation (15 min at 7,000g) and fixed in cacodylate buffer containing 4% (w/v) glutaraldehyde and 8.6% (w/v) Suc. Pellets were washed in a series of cacodylate buffers with descending Suc concentration and postfixed in osmium tetroxide for 2 h. After dehydration in an ascending series of ethanol (70% to 100% [w/v]), samples were embedded in 4% (w/v) agar resin, sectioned (60-nm thickness) with a Reichert ultramicrotome, stained with uranyl acetate and lead citrate, and observed under a 100 CX transmission electron microscope (JEOL, Tokyo).
TUNEL Staining
nDNA fragmentation was identified in situ by TUNEL labeling (Gavrieli et al., 1992). Cells were fixed with 0.1% (w/v) glutaraldehyde and centrifuged (5 min at 14,000g). Cells were permeabilized (0.1% [w/v] Triton X-100 for 15 min), washed with phosphate-buffered saline, and labeled following the manufacturer's instructions (Apoptag Direct Kit, Oncor, Gaithersburg, MD). Samples were then resuspended in phosphate-buffered saline, filtered (50 mm Hg) onto 0.8-μm, 25-mm black polycarbonate filters (catalogue no. 13062, Osmotics, Minnetonka, MN), and observed using a Laborlux D epifluorescence microscope (excitation 490 nm, emission of 525 nm, Leitz, Leica Microsystems (UK) Ltd, Bucks Milton Keynes, UK). Positive controls consisted of cells pretreated with 10 μg mL−1 DNAse I (nickase); for negative controls, distilled water was substituted for the terminal deoxynucleotidyl transferase.
Activity Assays
Caspase-like activities were measured using commercial kits (R & D Systems, Minneapolis). Cells were harvested by centrifugation (as above), resuspended in lysis buffer, and sonicated (25-Jencons Vibracell, Jencons (Scientific) Ltd, Bedfordshire, UK) on ice. Extracts were mixed with 50 μm 7-amino-4-fluoromethyl coumarin-labeled substrates for caspases 1 (WEHD), 3 (DEVD), 6 (VEID), 8 (IETD), and 9 (LEHD), and fluorescence was measured for 4 h at 26°C (excitation 400 nm, emission 505 nm) in a SOFTmax Pro plate reader. Parallel reactions were performed with increasing concentrations of the irreversible caspase inhibitors Boc-D-FMK (Calbiochem, San Diego), Ac-VAD-FMK (Calbiochem), and Ac-YEVD-CMK (synthesized and provided by Prof. B. Walker, Queen's University of Belfast, UK); homogenates were pre-incubated with inhibitors for 60 min before running reactions. Caspase activities were standardized to protein concentration measured using the bicinchorinic acid method (Smith et al., 1985; Pierce Chemical Co., Rockford, IL) with bovine serum albumin as a standard.
Western Blots
Samples for immunochemical analysis were loaded on an equal protein basis, separated on 15% (w/v) polyacrylamide gels and western blotted onto polyvinylidene difluoride membranes (Laemmli, 1970). Blots were probed with polyclonal antibodies raised against caspases 1 (catalogue nos. RDI-RTICEP106abr and RDI-CPP32abR, Research Diagnostics Inc., Flanders, NJ), 2, 7, 8, 10 (R & D Systems, catalogue nos. AF826, AF823, AF832, AF831, and AF834), and 3, 6, 9 (catalogue nos. 235412, pc365, and 218794, respectively, Calbiochem), detected using a chemiluminescence system (SuperSignal, Pierce), and the intensity of cross-reactions were quantified with Image-Pro Plus software (Media Cybernetics).
ACKNOWLEDGMENTS
We thank Eric Lam and Kay Bidle for discussions and Gerry Brennan and George McCartney for assistance with the TEM.
Footnotes
This work was supported by the European Union (Individual Marie Curie Postdoctoral Fellowship to M.S.), by the Natural Environment Research Council (UK; grants to J.A.B.), and by the U.S. National Institutes of Health (to P.G.F.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.017129.
LITERATURE CITED
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Aravind L, Dixit VM, Koonin EV. The domains of death: evolution of the apoptotic machinery. Trends Biochem Sci. 1999;22:47–53. doi: 10.1016/s0968-0004(98)01341-3. [DOI] [PubMed] [Google Scholar]
- Aravind L, Dixit VM, Koonin EV. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291:1279–1286. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- Barber GN. Host defense, viruses and apoptosis. Cell Death Diff. 2001;8:113–126. doi: 10.1038/sj.cdd.4400823. [DOI] [PubMed] [Google Scholar]
- Berges JA, Falkowski PG. Physiological stress and cell death in marine phytoplankton: induction of proteases in response to nitrogen or light limitation. Limnol Oceanogr. 1998;43:129–135. [Google Scholar]
- Bhattacharya D, Medlin L. Algal phylogeny and the origin of land plants. Plant Physiol. 1998;116:9–15. [Google Scholar]
- Bratbak G, Egge JK, Heldal M. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar Ecol Prog Ser. 1993;93:39–48. [Google Scholar]
- Bump NJ, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Franklin S, Ghayur T, Li P, Licari P et al. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YB, Yu CD, Guo BC, Ding M, Guo SZ, Zeng D, Wen LP. Cloning and functional expression of CPP32 cDNA in E. coli. Prog Biochem Biophys. 1998;25:151–154. [Google Scholar]
- Delorme VGR, McCabe PF, Kim DJ, Leaver CJ. A matrix metalloproteinase gene is expressed t the boundary of senescence and programmed cell death in cucumber. Plant Physiol. 2000;123:917–927. doi: 10.1104/pp.123.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo O, Lam E. Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr Biol. 1998;20:1129–1132. doi: 10.1016/s0960-9822(98)70469-5. [DOI] [PubMed] [Google Scholar]
- Dickman MB, Park YK, Oltersdorf T, Li W, Clemente T, French R. Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc Natl Acad Sci USA. 2001;98:6957–6962. doi: 10.1073/pnas.091108998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich KU, Madeo F. Apoptosis in yeast: a monocellular organism exhibits altruistic behaviour. FEBS Lett. 2000;473:6–9. doi: 10.1016/s0014-5793(00)01474-5. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell-death in situ via specific labelling of nuclear-DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman JC, McCarthy JJ. Steady state growth and ammonium uptake of a fast growing marine diatom. Limnol Oceanogr. 1978;23:695–703. [Google Scholar]
- Greenberg JT. Programmed cell death: a way of life for plants. Proc Natl Acad Sci USA. 1996;93:12094–12097. doi: 10.1073/pnas.93.22.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard RRL, Ryther JH. Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt and Detonula confervacea (Cleve) Gran. Can J Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- Heldal M, Bratbak G. Production and decay of viruses in aquatic environments. Mar Ecol Prog Ser. 1991;72:205–212. [Google Scholar]
- Jabs T. Reactive oxygen intermediates as mediators of programmed cell death in plants and animals. Biochem Pharmacol. 1999;57:231–245. doi: 10.1016/s0006-2952(98)00227-5. [DOI] [PubMed] [Google Scholar]
- Korthout HAAJ, Berecki G, Bruin W, van Duijn B, Wang M. The presence and subcellular localization of caspase 3-like proteinases in plant cells. FEBS Lett. 2000;475:139–144. doi: 10.1016/s0014-5793(00)01643-4. [DOI] [PubMed] [Google Scholar]
- Lacomme C, Cruz SS. Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc Natl Acad Sci USA. 1999;96:7956–7961. doi: 10.1073/pnas.96.14.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lam E, del Pozo O. Caspase-like protease involvement in the control of plant cell death. Plant Mol Biol. 2000;44:417–428. doi: 10.1023/a:1026509012695. [DOI] [PubMed] [Google Scholar]
- Lam E, Kato N, Lawton M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature. 2001;411:848–853. doi: 10.1038/35081184. [DOI] [PubMed] [Google Scholar]
- Leist M, Nicotera P. The shape of cell death. Biochem Biophys Res Commun. 1997;236:1–9. doi: 10.1006/bbrc.1997.6890. [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- Lewis K. Programmed cell death in bacteria. Microbiol Mol Biol Rev. 2000;64:503–514. doi: 10.1128/mmbr.64.3.503-514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps JH. Fossil Protists. Oxford: Blackwell; 1993. [Google Scholar]
- Madeo F, Herker E, Maldener C, Wissing S, Lächelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S et al. A caspases-related protease regulates apoptosis in yeast. Mol Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- Noble RT, Fuhrman JA. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microbiol Ecol. 1998;14:113–118. [Google Scholar]
- Pennell RI, Lamb C. Programmed plant cell in plants. Plant Cell. 1997;9:1157–1168. doi: 10.1105/tpc.9.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen G, Nagase H. Inhibition of proteolytic enzymes. In: Beyon RJ, Bond JS, editors. Proteolytic Enzymes: A Practical Approach. Oxford: IRL Press; 1989. pp. 83–104. [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell. 1999;11:431–444. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. The significance of viruses to mortality in aquatic microbial communities. Microbiol Ecol. 1994;28:237–243. doi: 10.1007/BF00166813. [DOI] [PubMed] [Google Scholar]
- Thornberry NA. Caspases: a decade of death research. Cell Death Diff. 1999;6:1023–1027. doi: 10.1038/sj.cdd.4400607. [DOI] [PubMed] [Google Scholar]
- Uren AG, Walsh MC, Van dam K, Westerhoff HV. Conservation of baculovirus inhibitor of apoptosis repeat proteins (BIRPs) in viruses, nematodes, vertebrates and yeasts. Trends Biochem Sci. 1998;23:159–162. doi: 10.1016/s0968-0004(98)01198-0. [DOI] [PubMed] [Google Scholar]
- Uren AG, O'Rourke K, Aravind L, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- Vardi A, Berman-Frank I, Rozenberg T, Hadas O, Kaplan A, Levine A. Programmed cell death of the dinoflagellate Peridinium gatunense is mediated by CO2 limitation and oxidative stress. Curr Biol. 1999;9:1061–1064. doi: 10.1016/s0960-9822(99)80459-x. [DOI] [PubMed] [Google Scholar]
- Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuner A, Eramo A, Peschle C, De Maria R. Caspase activation without death. Cell Death Diff. 1999;6:1075–1080. doi: 10.1038/sj.cdd.4400596. [DOI] [PubMed] [Google Scholar]