Abstract
Globular somatic embryos can be induced from immature cotyledons of soybean (Glycine max L. Merr. cv Jack) placed on high levels of the auxin 2,4-dichlorophenoxyacetic acid (2,4-D). Somatic embryos develop from the adaxial side of the cotyledon, whereas the abaxial side evolves into a callus. Using a 9,280-cDNA clone array, we have compared steady-state RNA from the adaxial side from which embryos develop and from the abaxial callus at five time points over the course of the 4 weeks necessary for the development of globular embryos. In a second set of experiments, we have profiled the expression of each clone in the adaxial side during the same period. A total of 495 genes differentially expressed in at least one of these experiments were grouped according to the similarity of their expression profiles using a nonhierarchical clustering algorithm. Our results indicate that the appearance of somatic embryos is preceded by dedifferentiation of the cotyledon during the first 2 weeks on auxin. Changes in mRNA abundance of genes characteristic of oxidative stress and genes indicative of cell division in the adaxial side of the cotyledons suggest that the arrangement of the new cells into organized structures might depend on a genetically controlled balance between cell proliferation and cell death. Our data also suggest that the formation of somatic globular embryos is accompanied by the transcription of storage proteins and the synthesis of gibberellic acid.
Due to their ability to regenerate into full plants, somatic embryos are the tissue of choice for transformation by particle bombardment in several crop species including soybean (Glycine max L. Merr. cv Jack; Finer and McMullen, 1991). In soybean, somatic embryos are obtained by induction, or culturing, of immature cotyledons on high concentration of auxin. Globular somatic embryos originate from epidermal and subepidermal cells of the adaxial side of the cotyledon (Finer, 1988), whereas the abaxial side evolves into a callus. The adaxial side is the “flat” side of the cotyledons, closest to the axis of the embryo, whereas the abaxial side is the side of the cotyledon in contact with the endosperm and the seed coat. However, the response to tissue culture is highly genotype dependent (Meurer et al., 2001), and the ability to transform a wider range of cultivars could accelerate the production of transgenic plants.
Somatic and zygotic embryos follow the same general pattern of development (Zimmerman, 1993; Goldberg et al., 1994). However, large quantities of somatic embryos can be produced in vitro, making them more amenable to experimentation than their zygotic counterparts, which are protected by fruit structures and less accessible. Therefore, somatic embryos constitute a model system to study basic aspects of embryogenesis, as well as a tool for efficient transformation.
Little is known of the genes expressed in early globular stage embryos (Zimmerman, 1993). Choi evaluated that only 10% of the proteins visible on a two-dimensional gel are embryo specific (Choi and Sung, 1984). Differential screening of cDNA libraries with RNA from embryogenic and nonembryogenic calli led to the identification of few genes specific to somatic embryos in carrot (Daucus carota; Aleith and Richter, 1990) and in alfalfa (Medicago sativa; Giroux and Pauls, 1997), most of them with no homology to known genes.
Today, microarray technology is an obvious choice to identify global gene expression patterns during development. Its sensitivity and reliability has been demonstrated in the study of a variety of phenomena including fruit ripening (Aharoni et al., 2002), the hypersensitive response in response to pathogen (Schenk et al., 2000), and the response to wounding (Cheong et al., 2002). In particular, microarray data was shown to be highly consistent with results obtained by RNA gel multiple times (Desikan et al., 2001; Perez-Amador et al., 2001). Over 80 cDNA libraries have now been constructed as part of a “Public EST Project for Soybean” sponsored by the soybean grower organizations. More than 250,000 expressed sequence tags (ESTs) from these libraries have been entered into dbEST soybean (Shoemaker et al., 2002). As part of an National Science Foundation-sponsored “Soybean Functional Genomics Project,” low-redundancy sets of cDNAs (“unigenes”) have been selected, sequenced at the 3′ end, and used to build soybean microarrays.
We have sampled adaxial and abaxial sides of cotyledons separately, at 7-d intervals during the 4-week induction, and obtained RNA from the tissue. Expression in the adaxial side was compared with expression in the abaxial side collected at the same time point by hybridization of the corresponding labeled cDNAs to a soybean microarray representing 9,280 cDNA clones. In addition, transcript profiles of the genes expressed in the adaxial side were obtained by comparing each time point with the previous one. A total of 495 genes (5.3% of the genes on the array) that were differentially expressed in at least one of these experiments were clustered into 11 sets using a nonhierarchical method (k-means). Our results give a global picture of the molecular events unfolding in the cotyledons during their reprogramming.
RESULTS
Quality Evaluation of the Microarray Hybridizations
We used a 9,728-element microarray consisting of 9,216 single-spotted soybean cDNA clones (Gm-r1070 library) and 64 choice clones, each printed eight times. Each cDNA clone was chosen as a representative of a unigene. The unigene set represented on this array was created by contigging expressed sequenced tags (ESTs) generated by 5′ sequencing of cDNA clones from embryo, seed coat, flower, or pod libraries (Table I). A total of 9,216 clones from these libraries, corresponding to singletons or 5′-most members of a contig were reracked into a new library, Gm-r1070. The estimated redundancy is between 10% and 15%; therefore, the 9,216 cDNA clones represent approximately 8,000 unique genes. First pass sequencing of the 3′ ends of Gm-r1070 cDNA clones was performed. For clones of interest to our study (overexpressed or underexpressed in at least one experiment), the tentative contigs (TCs) to which the 3′ and 5′ ESTs belonged were identified in the Soybean Gene Index (The Institute for Genome Research [TIGR]; Quackenbush et al., 2000). Annotations given here are based on the matching of the TC consensus sequence, or the 3′ EST in the case of a singleton, by BLASTX (Altschul et al., 1997).
Table I.
Origin of the cDNA clones on the array
| Tissuea | Stage | No. of Unigenes Represented on the Array |
|---|---|---|
| Embryo | 20–50-mg Seed | 589 |
| Embryo | 100–300-mg Seed | 2,050 |
| Seed coats | 100–300-mg Seed | 1,770 |
| Flowers | Immature | 2,242 |
| Flowers | Mature | 1,696 |
| Whole pods | 1–2 cm | 869 |
| Total | – | 9,216 |
Tissue used for cDNA library.
We performed 22 hybridizations corresponding to nine experiments as described in “Materials and Methods.” Raw and normalized data from these hybridized slides were deposited in the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo; see “Materials and Methods” for accession nos.). Five experiments consisted of the comparison of adaxial and abaxial tissue at time points 0, 7, 14, 21, and 28 d. A time course in the adaxial side was obtained by comparing adaxial tissue at 14 and 7 d, 21 and 14 d, 28 and 21d, and 28 and 7 d. Due to the small amount of material collected at each time point for each replicate, we pooled the RNA obtained from seven different replicates (see “Materials and Methods”). Pooling RNA before labeling has the advantage of reducing the variation due to biological replication and sample handling (Churchill, 2002). To estimate this variation, we competitively hybridized cDNA from two 14-d adaxial samples collected at different times to a microarray (GEO accession no. GSM3384). The correlation coefficient between the normalized intensities of the two channels was of 0.982, and only four spots showed differences in intensities above 2-fold. The same experiment conducted with 21-d adaxial samples returned a correlation coefficient of 0.979 (GEO accession no. GSM3385), and five spots showed differences in intensities above 2-fold (see supplemental data at www.plantphysiol.org for scatter plots). These very high correlation coefficients give a strong indication that biological replication is not a significant source of variation.
For each experiment, two to three replicate hybridizations were performed using the pooled RNA. After normalization, the coefficient of variation (CV) across replicates of the intensity ratio of each spot was calculated for each experiment. The median CV of the ratios of all the spots on the array ranged from 7% to 16% (average of 11.1%), depending on the experiment. These low CVs reflect the repeatability of our technique and show the robustness of our methods. In addition, inferences were only made from genes showing a ratio above 2 (below 0.5) in at least two of the two or three replicate hybridizations performed for a single experiment. This conservative criterion further ensures that our data are the result of treatment effect and not of technical variation.
The 28-d versus 7-d comparison confirmed the high quality of our data. For each clone, the product of the ratios measured in the experiments 14 d versus 7 d, 21 d versus 14 d, and 28 d versus 21 d should be the same as the ratio measured in the experiment 28 d versus 7 d. The correlation coefficient between the calculated and the measured ratios of all the clones was 0.90. Therefore, competitive hybridization of 28- and 7-d cDNA supplied additional evidence of the strength of our results.
Differential Expression in the Adaxial and Abaxial Tissue
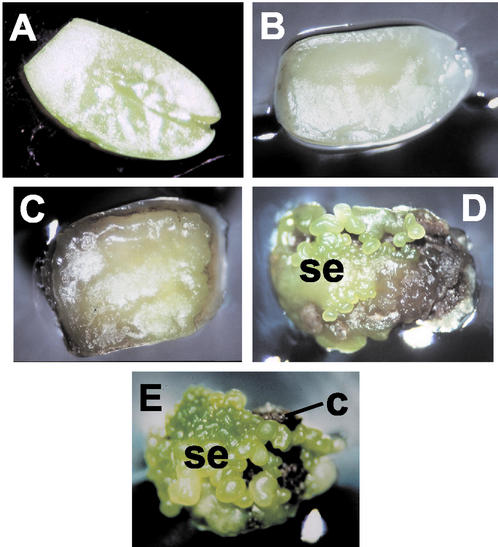
Our objective was to identify gene expression patterns during the development of somatic embryos. Immature soybean cotyledons have the ability to form somatic embryos when placed abaxial side down (Santarem et al., 1997) on Murashige and Skoog medium containing high levels (40 mg L−1) of 2,4-dichlorophenoxyacetic acid (2,4-D; Finer, 1988; Fig. 1A). We initiated such cultures and sampled the induced cotyledons every 7 d for 28 d. After 7 d on the medium, the cotyledons appeared lighter in color than at time of initiation (Fig. 1B). After 14 d, the adaxial sides of the cotyledons were swollen, and the abaxial sides started turning into a brown callus (Fig. 1C). At 21 d, some somatic embryos were visible (Fig. 1D) and at 28 d, the surfaces of the cotyledons were covered with globular embryos, whereas the abaxial sides were dark brown (Fig. 1E). At each time point, cotyledons from plates were sampled randomly, and the adaxial and abaxial sides were cut apart (Fig. 2).
Figure 1.
Induction of somatic embryos from immature cotyledons. Cotyledon, adaxial side facing up at: A, 0 d; B, 7 d; C, 14 d; D, 21 d; and E, 28 d. se, Somatic embryo; c, callus.
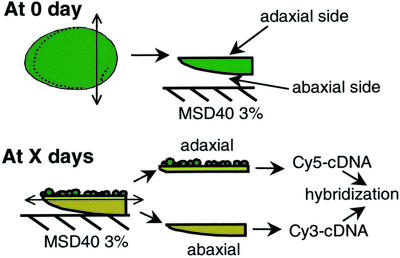
Figure 2.
Dissection of cotyledons at 0 d and at time of collection (X day). At 0 d, the chalazal end of the seed is cut. The cotyledons are pushed out of the seed coat and are placed on MSD40 3% (Murashige and Skoog basal medium, 40 mg L−1 2,4-D and 3% (w/v) Suc), abaxial side in contact with the medium. Adaxial and abaxial sides are separated at time of collection; their RNA is extracted and used to make fluorescent cDNA probes for microarray hybridization.
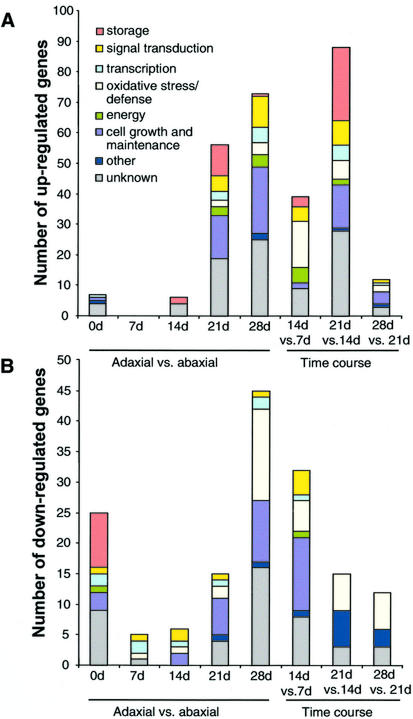
In a first series of experiments, adaxial and abaxial tissues sampled at the same time point were compared by competitive hybridization of labeled cDNA to a 9,280-clone microarray. This strategy was used to eliminate from our interpretation genes expressed in response to auxin but not directly involved in embryogenesis. For each time point, genes showing differential mRNA abundance (ratios above 2 or below 0.5) in at least two replicate slides were selected for further analysis. Surprisingly few genes fulfilled this condition: a total of 238 of 9,248 cDNA clones, or 2.6% of the cDNAs present on the array, were differentially expressed in at least one comparison of the five we performed. We classified the differentially regulated genes according to their probable functions, taking into account the redundancy on the array. The ratios of the clones found in the same TC in the TIGR soybean gene index were averaged and counted as one gene. The number of genes in each functional category at each time point is presented in Figure 3. Selected genes are presented in Table II, and the complete list of genes is available at www.plantphysiol.org (Supplemental Table S1).
Figure 3.
Number and functions of genes: A, overexpressed in each experiment performed; and B, underexpressed in each experiment performed. Redundancy was taken into account by counting clones belonging to the same TIGR soybean gene index TC as one gene.
Table II.
A selection of genes differentially regulated in the adaxial versus abaxial experiments.
| Clone Identification | Genbank Accession No.a | Ratio Averageb | Functionc | k-Means Setd | Annotatione |
|---|---|---|---|---|---|
| 0-d Adaxial vs. abaxial up-regulated | |||||
| Gm-b10BB-2 | BE806777 | 2.98 | oth | 10 | Phe ammonia lyase (PAL; Glycine max) |
| Gm-r1070-3714 | AI938893 | 2.09 | cgm | 10 | Trans-caffeoyl-CoA 3-O-methyltransferase(Medicago truncatula) |
| 0-d Adaxial vs. abaxial down-regulated | |||||
| Gm-r1070-4440 | BE820268 | 0.40 | to | 7, 8f | YABBY2 (Arabidopsis) |
| Gm-r1070-5305 | BE820875 | 0.32 | to | 11 | Filamentous flower protein (FIL)/YABBY1(Arabidopsis) |
| Gm-r1070-8983 | BE824085 | 0.44 | sp | 4 | Lipoxygenase 1 (Glycine max) |
| Gm-r1070-7666 | BE823049 | 0.44 | sp | 5, 1f | Beta-amylase (Glycine max) |
| Gm-r1070-1843 | BE658231 | 0.31 | sp | 8, 11f | Lectin precursor (agglutinin; Glycine max) |
| Gm-r1070-399 | BE657497 | 0.29 | sp | 4 | Bowman-Birk proteinase isoinhibitor C-II(Glycine max) |
| Gm-r1070-7656 | BE822991 | 0.23 | sp | 3 | Bowman-Birk proteinase isoinhibitor D-II(Glycine max) |
| 7-d Adaxial vs. abaxial down-regulated | |||||
| Gm-r1070-4440 | BE820268 | 0.55 | to | 7 | YABBY2 (Arabidopsis) |
| Gm-r1070-2656 | BE658799 | 0.45 | to | 8 | YABBY2 (Arabidopsis) |
| 14-d Adaxial vs. abaxial up-regulated | |||||
| Gm-r1070-9099 | BE824364 | 5.74 | sp | 3 | Albumin 1 precursor/ leginsulin (Glycine max) |
| Gm-r1070-120 | BE657237 | 1.83 | sp | 4 | Trypsin inhibitor A (Kunitz) precursor (Glycine max) |
| 14-d Adaxial vs. abaxial down-regulated | |||||
| Gm-r1070-4440 | BE820268 | 0.52 | to | 7 | YABBY2 (Arabidopsis) |
| Gm-r1070-149 | BE657203 | 0.53 | sig | 9 | Hypothetical calcium-binding protein(Arabidopsis) |
| Gm-r1070-8829 | BE824150 | 0.51 | sig | 2 | 1-Aminocyclopropane-1-carboxylate oxidase(Arabidopsis) |
| Gm-r1070-1711 | BE658306 | 0.55 | ox | 2 | Matrix metalloproteinase (MMP2; Glycine max) |
| 21-d Adaxial vs. abaxial up-regulated | |||||
| Gm-r1070-7726 | BE823315 | 3.13 | u | 11 | GA-regulated protein (Arabidopsis) |
| Gm-r1070-3826 | BE821412 | 2.41 | u | 3, 4f | ADR12-2 (Glycine max) |
| Gm-r1070-9099 | BE824364 | 4.14 | sp | 3 | Albumin 1 precursor/leginsulin (Glycine max) |
| Gm-r1070-7467 | AW459971 | 3.64 | sp | 5 | Glycinin A5A4B3 chain (Glycine max) |
| Gm-r1070-1653 | BE658147 | 2.74 | sp | 5 | Glycinin G1 precursor (Glycine max) |
| Gm-r1070-8607 | BE823766 | 2.69 | sp | 4 | Lipoxygenase 1 (Glycine max) |
| Gm-r1070-1184 | BE658067 | 2.48 | sp | 4 | Lipoxygenase 2 (Glycine max) |
| Gm-r1070-7343 | BE822998 | 2.44 | sp | 3 | Lipoxygenase 3 (Glycine max) |
| Gm-r1070-1596 | AI748023 | 2.20 | sp | 5 | Glycinin A3B4 subunit (Glycine soja) |
| Gm-r1070-6293 | BE821976 | 2.14 | sp | 3 | Napin-type 2S albumin (Glycine max) |
| Gm-r1070-111 | AI735902 | 1.94 | sp | 4 | Kunitz trypsin inhibitor 3 (Glycine max) |
| Gm-r1070-7723 | BE823259 | 2.52 | sig | 11 | Ent-kaurenoic acid hydroxylase (Arabidopsis) |
| Gm-r1070-7332 | AW460085 | 2.44 | sig | 5 | 14-3-3-Like protein (Glycine max) |
| Gm-r1070-8358 | BE823656 | 2.29 | sig | 11 | Ent-kaurene oxidase (Cucurbita maxima) |
| Gm-r1070-7777 | BE823087 | 2.24 | sig | 11 | Zinc finger protein like, Ser/Thr protein kinase like (Arabidopsis) |
| Gm-r1070-8002 | BE823535 | 2.01 | sig | 1 | Brassinosteroid biosynthetic protein (Pisum sativum) |
| Gm-r1070-970 | AW101691 | 1.88 | cgm | 1 | Histone H2A (Pisum sativum) |
| Gm-r1070-3909 | BE659557 | 1.95 | cgm | 1 | Plastid ribosomal protein L11 (Oryza sativa) |
| Gm-r1070-5132 | BE821002 | 2.52 | cgm | 7 | Histone H3 (Triticum aestivum) |
| Gm-r1070-7579 | BE823362 | 2.48 | cgm | 11 | Acetyltransferase homolog (Petunia hybrida) |
| Gm-r1070-8697 | BE823786 | 2.47 | cgm | 11 | Probable tyrosyl t-RNA synthase (Nicotiana tabacum) |
| Gm-r1070-3966 | BE659772 | 2.28 | cgm | 1 | Histone H4 (Zea mays) |
| Gm-r1070-7482 | AW460113 | 2.12 | cgm | 11 | Phophatidyl-Ser decarboxylase (Arabidopsis) |
| Gm-r1070-8547 | BE823943 | 2.09 | cgm | 5 | Transporter-like protein (Arabidopsis) |
| Gm-r1070-5465 | AW457965 | 2.07 | cgm | 7 | Putative DnaJ protein (Arabidopsis) |
| Gm-r1070-4091 | AW099385 | 2.05 | cgm | 1 | Acyl carrier protein (Olea europaea) |
| Gm-r1070-7571 | BE823365 | 2.02 | cgm | 11 | Histone H2A.F/Z (Arabidopsis) |
| 28-d Adaxial vs. abaxial up-regulated | |||||
| Gm-r1070-697 | BE657627 | 2.10 | u | 1 | GH1/IAA9 (Nicotiana tabacum) |
| Gm-r1070-7835 | BE823122 | 2.45 | to | 1 | Monopteros/ IAA24 (Arabidopsis) |
| Gm-r1070-7636 | BE822993 | 2.08 | sp | 11 | Kunitz-type trypsin inhibitor KTI2 precursor(Glycine max) |
| Gm-r1070-7723 | BE823259 | 3.65 | sig | 11 | Ent-kaurenoic acid hydroxylase (Arabidopsis) |
| Gm-r1070-7777 | BE823087 | 3.06 | sig | 11 | Zinc finger protein like, Ser/Thr protein kinase like (Arabidopsis) |
| Gm-r1070-819 | AW100772 | 2.90 | sig | 11 | Receptor-like kinase 2 (Glycine max) |
| Gm-r1070-6498 | BE822097 | 2.83 | sig | 1 | Receptor-like kinase 1 (Glycine max) |
| Gm-r1070-8358 | BE823656 | 2.68 | sig | 11 | Ent-kaurene oxidase (Cucurbita maxima) |
| Gm-r1070-8002 | BE823535 | 2.49 | sig | 1 | Brassinosteroid biosynthetic protein (Pisum sativum) |
| Gm-r1070-7250 | BE822570 | 2.30 | sig | 1 | Putative protein kinase (Arabidopsis) |
| Gm-r1070-8991 | BE824089 | 2.16 | sig | 11 | GA 20-oxidase (Phaseolus vulgaris) |
| Gm-r1070-6650 | BE822629 | 2.08 | sig | 1 | Peptidylprolyl isomerase FKBP62/ ROF1(Arabidopsis) |
| Gm-r1070-7836 | AW472167 | 2.01 | sig | 11 | Protein kinase C inhibitor like (Zea mays) |
| Gm-r1070-7709 | BE823267 | 1.99 | ox | 11 | Glutathione reductase chloroplast precursor(Glycine max) |
| Gm-r1070-6457 | BE822166 | 1.85 | ox | 1 | Probable amine oxidase (Arabidopsis) |
| Gm-r1070-4769 | BE820482 | 1.78 | ox | 1 | Putative phosphatase 2A inhibitor (Arabidopsis) |
| Gm-r1070-3997 | BE659580 | 2.62 | en | 11 | Chlorophyll a/b-binding protein (cab-3) of light-harvesting complex (LHC) II type I prec.(Glycine max) |
| Gm-r1070-7043 | BE822600 | 2.30 | en | 11 | Rubisco small subunit (RBCS1; Glycine max) |
| Gm-r1070-8717 | BE824039 | 2.18 | en | 11 | Chlorophyll a/b-binding protein type I prec.(Lycopersicon esculentum) |
| Gm-r1070-7920 | BE823502 | 1.89 | en | 11 | PSII type I chlorophyll a/b-binding protein(Glycine max) |
| Gm-r1070-8697 | BE823786 | 2.82 | cgm | 11 | Tyrosyl-tRNA synthase (Nicotiana tabacum) |
| Gm-r1070-7482 | AW460113 | 2.76 | cgm | 11 | Phosphatidyl-Ser decarboxylase (Arabidopsis) |
| Gm-r1070-3805 | BE821263 | 2.58 | cgm | 7 | Seed maturation protein (PM30; Glycine max) |
| Gm-r1070-7579 | BE823362 | 2.52 | cgm | 11 | Acetyltransferase (Olea europaea) |
| Gm-r1070-7571 | BE823365 | 2.48 | cgm | 11 | Histone H2A.F/Z (Arabidopsis) |
| Gm-r1070-5312 | BE821057 | 2.38 | cgm | 11 | ATP-dependent Clp protease proteolytic subunit (Lotus japonicus) |
| Gm-r1070-4091 | AW099385 | 2.35 | cgm | 1 | Acyl carrier protein (Olea europaea) |
| Gm-r1070-3909 | BE659557 | 2.35 | cgm | 1 | Plastid ribosomal protein L11 (Oryza sativa) |
| Gm-r1070-123 | BE657421 | 2.34 | cgm | 11 | Argonaute-like (zwille and pinhead) protein(Arabidopsis) |
| Gm-r1070-6877 | BE822339 | 2.25 | cgm | 11 | Ribosomal protein S2 (Lotus japonicus) |
| Gm-r1070-3713 | AI966777 | 2.22 | cgm | 1 | Putative xyloglucan endotransglycosylase(XET; Arabidopsis) |
| Gm-r1070-9084 | BE824267 | 2.18 | cgm | 11 | Putative epimerase/dehydratase (Oryza sativa) |
| Gm-r1070-7508 | BE822956 | 2.05 | cgm | 11 | SMT3 ubiquitin-like protein (Arabidopsis) |
| Gm-r1070-7940 | BE823195 | 2.05 | cgm | 1 | Histone H3 (Triticum aestivum) |
| Gm-r1070-8211 | AW508473 | 2.03 | cgm | 1 | Carbamoyl-phosphate synthetase small subunit (Nicotiana tabacum) |
| Gm-r1070-7659 | AW471559 | 2.02 | cgm | 1 | Alpha tubulin (Gossypium hirsutum) |
| Gm-r1070-87 | BE657412 | 2.01 | cgm | 1 | Histone H4 (Zea mays) |
| Gm-r1070-8438 | AW507810 | 1.99 | cgm | 1 | Putative chaperonin gamma chain (Arabidopsis) |
| Gm-r1070-8592 | BE824194 | 1.91 | cgm | 1 | 40S ribosomal protein S11 (Glycine max) |
| 28-d Adaxial vs. abaxial down-regulated | |||||
| Gm-r1070-6246 | BE822044 | 0.49 | u | 9 | Putative ethylene-responsive protein (Oryza sativa) |
| Gm-r1070-3118 | AW432447 | 0.44 | u | 6 | Putative ripening-related protein (Vitis vinifera) |
| Gm-r1070-8829 | BE824150 | 0.33 | sig | 2 | 1-Aminocyclopropane-1-carboxylate (ACC)oxidase (Arabidopsis) |
| Gm-r1070-2699 | BE658955 | 0.64 | ox | 9 | Glutaredoxin, putative (Arabidopsis) |
| Gm-r1070-5439 | BE821082 | 0.53 | ox | 6 | In2-1 protein (Glycine max) |
| Gm-r1070-7210 | BE822567 | 0.49 | ox | 9 | Cationic seed coat peroxidase precursor (Glycine max) |
| Gm-r1070-2114 | BE658477 | 0.49 | ox | 9 | 5′-Adenylylsulfate reductase (Arabidopsis) |
| Gm-r1070-9101 | BE824363 | 0.47 | ox | 4 | Putative Pro-rich cell wall protein (Asparagus officinalis) |
| Gm-r1070-2185 | BE658565 | 0.44 | ox | 2 | B′ regulatory subunit of PP2A (Arabidopsis) |
| Gm-r1070-4669 | BE820459 | 0.42 | ox | 2 | Extensin class 1 (hydroxy-Pro-rich protein;(Vigna unguiculata) |
| Gm-r1070-6150 | BE822027 | 0.40 | ox | 9 | Repetitive Pro-rich cell wall protein 1 precursor (Glycine max) |
| Gm-r1070-2684 | BE658937 | 0.40 | ox | 2 | Inhibitor of trypsin and Hageman factor(Cucurbita maxima) |
| Gm-r1070-1616 | AI938416 | 0.37 | ox | 2 | PR1a precursor (Glycine max) |
| Gm-r1070-1721 | BE658311 | 0.35 | ox | 2 | Amino acid-specific endopeptidase inhibitor(Momordica charantia) |
| Gm-r1070-2742 | BE659038 | 0.27 | ox | 2 | Trypsin inhibitor p20 (Glycine max) |
The full list is available at www.plantphysiol.org.
EST from the 3′ or 5′ end of the cDNA clone b average over two or three replicates c cgm, cell growth and maintenance; en, energy; oth, other; ox, oxidative stress/defense; sig, signaling; sp, seed protein; to, transcription; u, unknown d refers to the sets presented on Figure 5.
BLASTX hit of the 3′ end EST or TIGR contig.
Gene represented by clones clustering in different sets.
At 0 d, transcripts corresponding to 32 genes on the array exhibited differences in abundance in the adaxial and abaxial sides of the 4- to 6-mm cotyledon (Fig. 3). Higher mRNA levels of genes encoding seed storage proteins in the abaxial side were largely responsible for this polarity (Fig. 3B). Transcripts of the homologs of the transcription factors YABBY2 and FIL/YABBY1 were more abundant in the abaxial side as well. At 7 d, only five genes of the estimated 8,000 showed differential mRNA levels within the cotyledon (all of them overexpressed in the abaxial side), suggesting that the accumulation of transcripts of storage proteins genes stopped. Similarly, little difference was observed between the mRNA populations of the two sides of the cotyledons after 14 d on auxin (12 genes were differentially expressed). In the abaxial side, overexpressed homologs to an ACC oxidase, a calcium-binding protein and a metalloproteinase (Delorme et al., 2000; Swidzinski et al., 2002), suggest a response similar to senescence.
Somatic embryos started appearing after 14 d on auxin-containing medium. Consistent with this observation, the number of genes exhibiting different mRNA levels in the adaxial and abaxial sides increased to 71 genes in the 21-d samples. Of the 54 genes up-regulated in the adaxial side at 21 d, 14 genes are involved in cell maintenance and growth (Fig. 3A). They include histones (H3, H4, and H2A), homologs to tyrosyl-tRNA, DnaJ, and plastid ribosomal proteins (Table II), and suggest a higher level of cellular activity in the adaxial side than in the abaxial side. Relative transcript abundance of homologs to ent-kaurenoic hydroxylase, ent-kaurene oxidase, a GA-regulated protein, and an enzyme of the brassinosteroid biosynthesis pathway suggest higher levels of GAs and brassinosteroid in the developing embryos than in the subtending callus. Genes for seed proteins such as glycinins, lipoxygenases, Kunitz trypsin inhibitors, and 2S albumin were also up-regulated in the adaxial side. The functional distribution of the genes overexpressed in the adaxial side at 28 d was similar to that of genes overexpressed at 21 d (Fig. 3A). In fact, 16 genes were up-regulated at both time points including histones and homologs to GA and brassinosteroid synthesis genes. Important differences were the reduced number of up-regulated genes encoding seed proteins (only one, for Kunitz trypsin inhibitor) and the presence of several kinases overexpressed in the adaxial side at 28 d (Table II). In the abaxial side at 28 d (Fig. 3B), 15 of 48 up-regulated genes are possibly involved in the control of oxidative damage (homologs to a putative glutaredoxin and a 5′-adenylsulfate-reductase, peroxidases, and Pro-rich proteins) or induced during senescence (putative ripening-related protein, ACC oxidase, and ethylene-responsive protein). However, it should be noted that other genes, also acting in the protection of cells against oxidation or death, were up-regulated in the adaxial side at 28 d; for example, glutathione reductase, a putative phosphatase 2A inhibitor. Taken together, the comparison of adaxial and abaxial tissue by hybridization to microarrays showed that differences in steady-state mRNA between the adaxial and abaxial sides of cotyledons cultured on MSD40 emerge between 14 and 21 d, which corresponds to the time of physical appearance of embryos.
Time Course of Genes Expressed in the Adaxial Side
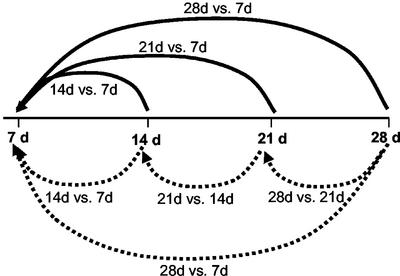
To refine our analysis, we quantified the evolution of each transcript involved in embryo development over time. We performed a time course experiment in which RNA from the adaxial sides of tissue at a specific time point was compared with RNA from the same tissue at the previous time point on a microarray. Because our first results indicated that genetic events related to somatic embryogenesis per se occur after 14 d of culture of the cotyledons on auxin, 14-d samples were compared with 7-d samples, 21-d samples were compared with 14-d samples, and 28-d samples were compared with 21-d samples (Fig. 4, dotted lines). A total of 226 clones (2.4% of the 9,280 clones on the array) were differentially regulated in at least one experiment. These genes were classified according to their probable functions, and their distribution is shown in Figure 3. A selection of these genes is also presented in Table III. The complete list is available at http://www.plantphysiol.org (Supplemental Table S2).
Figure 4.
Experimental design of the time course experiment. The dotted lines represent the expression ratios obtained experimentally. The solid lines represent the ratios expressed in reference to 7 d, which are plotted on Figure 5. For each clone, the calculated ratio 21 versus 7 d is the product of the ratios 21 versus 14 d and 14 versus 7 d, and the calculated ratio 28 versus 7 d is the average of the measured 28 versus 7 d and of the product of the ratios 28 versus 21 d, 21 versus 14 d, and 14 versus 7 d.
Table III.
A selection of genes differentially regulated in the time course experiments.
| Clone Identification | Genbank Accession No.a | Ratio Averageb | Functionc | k-Means setd | Annotatione |
|---|---|---|---|---|---|
| 14-d Adaxial vs. 7-d adaxial up-regulated | |||||
| Gm-r1070-9099 | BE824364 | 6.23 | sp | 3 | Albumin 1 precursor/leginsulin (Glycine max) |
| Gm-r1070-941 | BE657741 | 2.18 | sp | 3 | Insulin-like growth factor S11 precursor(Glycine max) |
| Gm-r1070-7926 | BE823202 | 2.01 | sp | 3 | Alpha′-subunit of beta-conglycinin (Glycine max) |
| Gm-r1070-8829 | BE824150 | 4.30 | sig | 2 | ACC oxidase (Arabidopsis) |
| Gm-r1070-8543 | BE823989 | 2.30 | sig | 9 | Receptor-like kinase (Arabidopsis) |
| Gm-r1070-11 | AI735898 | 2.28 | sig | 4 | FKBP12-interacting protein (FIP37; Arabidopsis) |
| Gm-r1070-1932 | BE658454 | 2.26 | sig | 9 | Calcium/proton exchanger (Arabidopsis) |
| Gm-r1070-5129 | BE820838 | 2.15 | sig | 9 | Contains similarity to receptor-like proteinkinase 5 (Arabidopsis) |
| Gm-r1070-1616 | AI938416 | 11.41 | ox | 2 | PR1a precursor (Glycine max) |
| Gm-r1070-2742 | BE659038 | 3.49 | ox | 2 | Trypsin inhibitor p20 (Glycine max) |
| Gm-r1070-4109 | BE659729 | 2.99 | ox | 2 | Wound-induced protein (Arabidopsis) |
| Gm-r1070-1711 | BE658306 | 2.74 | ox | 2 | MMP2 (Glycine max) |
| Gm-r1070-1721 | BE658311 | 2.59 | ox | 2 | Amino acid-specific endopeptidase inhibitor (Momordica charantia) |
| Gm-r1070-8498 | BE823815 | 2.59 | ox | 3, 7f | Cationic peroxidase 2 (Glycine max) |
| Gm-r1070-2280 | AW133370 | 2.53 | ox | 2 | Thaumatin-like protein 1 (Pyrus pyrifolia) |
| Gm-r1070-1671 | BE658299 | 2.53 | ox | 2 | Alpha-amylase/subtilisin inhibitor (Oryza sativa) |
| Gm-r1070-249 | BE657221 | 2.52 | ox | 3 | Cys-rich protein (Vigna radiata) |
| Gm-r1070-1990 | BE658457 | 2.49 | ox | 2 | Amino acid-specific endopeptidase inhibitor (Momordica charantia) |
| Gm-r1070-2185 | BE658565 | 2.47 | ox | 2 | B′-regulatory subunit of PP2A (Arabidopsis) |
| Gm-r1070-2779 | BE659112 | 2.36 | ox | 2 | Alliinase precursor (Allium cepa) |
| Gm-r1070-1963 | BE658517 | 2.34 | ox | 2 | Endonuclease III homolog (Arabidopsis) |
| Gm-r1070-4139 | AW099251 | 2.11 | ox | 8 | Aspartyl aminopeptidase-like protein(Arabidopsis) |
| Gm-r1070-2395 | AW156731 | 3.27 | en | 2 | Phosphoenolpyruvate carboxykinase (Flaveria trinervia) |
| Gm-r1070-2535 | BE658836 | 2.40 | en | 9 | ATP-dependent phosphoenolpyruvate carboxykinase (Cucumis sativus) |
| Gm-r1070-1216 | BE820007 | 2.36 | en | 8 | Chlorophyll a/b-binding protein CP24 (Vigna radiata) |
| Gm-b10BB-41 | AI495218 | 2.17 | en | 8 | Rubisco (small chain 1 precursor; Glycine max) |
| Gm-r1070-7385 | BE823015 | 2.13 | en | 8 | Protoporphyrinogen oxidase (Nostoc sp. PCC7120) |
| Gm-r1070-1411 | BE657885 | 2.31 | cgm | 2 | Glucosyltransferase-like protein (Arabidopsis) |
| Gm-r1070-5369 | BE820955 | 2.09 | cgm | 9 | Putative alcohol dehydrogenase (Arabidopsis) |
| 14-d Adaxial vs. 7-d adaxial down-regulated | |||||
| Gm-r1070-8454 | BE823813 | 0.41 | u | 1 | GH1 protein (fragment; Glycine max) |
| Gm-r1070-7835 | BE823122 | 0.48 | to | 1 | Monopteros/ IAA24 (Arabidopsis) |
| Gm-r1070-8651 | BE823776 | 0.50 | cgm | 1 | Probable chaperonin-containing TCP-1 gamma chain (Arabidopsis) |
| Gm-r1070-8476 | BE824165 | 0.49 | cgm | 1 | Actin (Vigna radiata) |
| Gm-r1070-1814 | AW508084 | 0.46 | cgm | 1 | Elongation factor 1-alpha 1 (Lilium longiflorum) |
| Gm-r1070-7490 | BE822955 | 0.46 | cgm | 1 | Pinhead (Arabidopsis) |
| Gm-r1070-8771 | AW508084 | 0.46 | cgm | 1 | Putative polygalacturonase (Arabidopsis) |
| Gm-r1070-5432 | AW459885 | 0.46 | cgm | 1 | Glutamyl-tRNA reductase precursor (Glycine max) |
| Gm-r1070-6175 | AW186469 | 0.45 | cgm | 1 | Coat protein gamma-COP homolog (Arabidopsis) |
| Gm-r1070-6723 | BE822303 | 0.45 | cgm | 1 | Probable nucleolar GTP-binding protein 1(Arabidopsis) |
| Gm-b10BB-1 | AW318030 | 0.42 | cgm | 1 | EF1-a (elongation factor 1-alpha; Glycine max) |
| Gm-r1070-5868 | BE821694 | 0.35 | cgm | 1 | Cell division cycle protein 48 homolog(Glycine max) |
| Gm-r1070-8752 | BE823964 | 0.34 | cgm | 1 | Vacuolar-processing enzyme precursor(Glycine max) |
| 21-d Adaxial vs 0.14-d adaxial up-regulated | |||||
| Gm-r1070-806 | BE657863 | 3.94 | u | 3, 4, 5f | ADR12-2 (Glycine max) |
| Gm-r1070-7439 | BE823019 | 2.49 | to | 5 | DNA-dependent RNA polymerase II(Arabidopsis) |
| Gm-r1070-9099 | BE824364 | 8.13 | sp | 3 | Albumin precursor/leginsulin (Glycine max) |
| Gm-r1070-8983 | BE824085 | 5.12 | sp | 4 | Lipoxygenase 1 (Glycine max) |
| Gm-r1070-6293 | BE821976 | 4.50 | sp | 3 | Napin-type 2S albumin 1 precursor (Glycine max) |
| Gm-r1070-8909 | BE824331 | 4.47 | sp | 4 | Beta-subunit of beta conglycinin (Glycine max) |
| Gm-r1070-484 | BE819850 | 4.25 | sp | 3 | Bowman-Birk-type proteinase inhibitorprecursor (Glycine max) |
| Gm-r1070-1184 | BE658067 | 4.17 | sp | 4, 5f | Lipoxygenase 2 (Glycine max) |
| Gm-r1070-111 | AI735902 | 3.71 | sp | 4 | Kunitz trypsin inhibitor 3 (Glycine max) |
| Gm-r1070-7343 | BE822998 | 3.58 | sp | 3, 4, 5f | Lipoxygenase 3 (Glycine max) |
| Gm-r1070-8014 | BE823524 | 3.45 | sp | 4 | Beta-conglycinin alpha-subunit (Glycine max) |
| Gm-r1070-9021 | BE824328 | 3.42 | sp | 4, 3f | Alpha′-subunit of beta-conglycinin (Glycine max) |
| Gm-r1070-941 | BE657741 | 3.41 | sp | 3 | Insulin-like growth factor S11 precursor(Glycine max) |
| Gm-r1070-7467 | AW459971 | 3.36 | sp | 5, 7f | Glycinin A5A4B3 subunits precursor (amino acids −23 to 539; Glycine max) |
| Gm-r1070-1653 | BE658147 | 3.21 | sp | 5 | Glycinin G1 precursor (glycinin A1A and BX subunit; Glycine max) |
| Gm-r1070-1840 | BE658199 | 3.05 | sp | 4, 5, 8f | Beta-conglycinin alpha-chain precursor(Glycine max) |
| Gm-r1070-265 | AW397006 | 2.99 | sp | 3 | Bowman-Birk proteinase isoinhibitor D-II(Glycine max) |
| Gm-r1070-1596 | AI748023 | 2.83 | sp | 5, 4f | Glycinin A3B4 subunit (Glycine soja) |
| Gm-r1070-90 | BE657402 | 2.71 | sp | 5 | Glycinin G3 precursor (Glycine max) |
| Gm-r1070-1597 | BE658140 | 2.61 | sp | 4 | 2S albumin precursor (Glycine max) |
| Gm-r1070-5245 | BE820890 | 2.57 | sp | 4 | Lipoxygenase 7 (Glycine max) |
| Gm-r1070-120 | BE657237 | 2.55 | sp | 4 | Trypsin inhibitor A (Kunitz) precursor (Glycine max) |
| Gm-r1070-1818 | AI938452 | 2.39 | sp | 5 | Putative beta-amylase (Cicer arietinum) |
| Gm-r1070-7552 | BE822977 | 2.39 | sp | 5 | p24 Oleosin isoform b (p91; Glycine max) |
| Gm-r1070-9195 | BE824378 | 3.58 | ox | 5 | ATP-dependent protease LA, putative(Methanococcus jannaschii) |
| Gm-r1070-8951 | BE824071 | 3.46 | ox | 5 | Putative Ser peptidase (Oryza sativa) |
| Gm-r1070-249 | BE657221 | 2.84 | ox | 3 | Cys-rich protein (Vigna radiata) |
| Gm-r1070-9092 | BE824254 | 2.55 | ox | 5 | Cys peroxiredoxin (Xerophyta viscosa) |
| Gm-r1070-9071 | BE824348 | 2.36 | ox | 5 | Cys proteinase (Glycine max) |
| Gm-r1070-8498 | BE823815 | 2.23 | ox | 3 | Cationic peroxidase 2 (Glycine max) |
| Gm-r1070-7468 | BE823425 | 3.05 | cgm | 5 | Formate dehydrogenase (Arabidopsis) |
| Gm-r1070-1750 | BE658179 | 3.00 | cgm | 4 | Suc-binding protein precursor (SBP; Glycine max) |
| Gm-r1070-1783 | AI941280 | 2.97 | cgm | 4 | S-adenosyl-Met synthetase (Phaseolus lunatus) |
| Gm-r1070-8705 | BE823791 | 2.81 | cgm | 4 | Seed maturation protein PM34 (Glycine max) |
| Gm-r1070-1205 | AW570313 | 2.62 | cgm | 3 | Ribonucleoside-diphosphate reductase small chain (Nicotiana tabacum) |
| Gm-r1070-7536 | BE823431 | 2.57 | cgm | 4 | PII protein (Medicago truncatula) |
| Gm-r1070-8231 | BE823589 | 2.52 | cgm | 5 | Actin-related protein, putative (Arabidopsis) |
| Gm-r1070-8986 | BE824223 | 2.51 | cgm | 5 | Vacuolar-sorting receptor protein BP-80(Glycine max) |
| Gm-r1070-5257 | BE820863 | 2.42 | cgm | 11 | Putative XET (Arabidopsis) |
| Gm-r1070-5132 | BE821002 | 2.24 | cgm | 7 | Histone H3 (Medicago sativa) |
| Gm-r1070-729 | AW397196 | 2.22 | cgm | 5 | Putative carbamoyl phosphate synthetase small subunit (Oryza sativa) |
| Gm-r1070-9037 | BE824099 | 2.20 | cgm | 5 | Cytosine methyltransferase (Arabidopsis) |
| Gm-r1070-165 | BE657201 | 2.17 | cgm | 5 | Ribosomal protein L2 (Glycine max) |
| Gm-r1070-8406 | BE823658 | 2.08 | cgm | 5 | Similar to ATP binding associated with celldifferentiation (Arabidopsis) |
| 21-d Adaxial vs. 0.14-d adaxial down-regulated | |||||
| Gm-r1070-2675 | BE658859 | 0.46 | ox | 6 | Cytochrome P450 82A4 (Glycine max) |
| Gm-r1070-4841 | BE820550 | 0.42 | ox | 6 | Glutathione S-transferase (GST) 10 (Glycine max) |
| Gm-r1070-5621 | AW459775 | 0.40 | ox | 6 | Probable 2,4-D-inducible glutathionetransferase (Glycine max) |
| Gm-r1070-3058 | BE659268 | 0.36 | ox | 6 | NtPRp27 (Nicotiana tabacum) |
| Gm-r1070-1963 | BE658517 | 0.48 | ox | 2 | Endonuclease III homolog (Arabidopsis) |
| Gm-r1070-2779 | BE659112 | 0.45 | ox | 2 | Alliinase precursor (Allium cepa) |
| Gm-b10BB-4 | AW707011 | 0.46 | oth | 6 | CHS2 (chalcone synthase gene 2; Glycine max) |
| Gm-b10BB-7 | AI441937 | 0.43 | oth | 6 | CHS1 (chalcone synthese gene 1; Glycine max) |
| Gm-b10BB-10 | BG157194 | 0.47 | oth | 6 | CI (chalchone isomerase; Glycine max) |
| Gm-b10BB-6 | AI437793 | 0.41 | oth | 6 | CHS7 (Glycine max) |
| Gm-b10BB-2 | BE806777 | 0.45 | oth | 10 | PAL (Glycine max) |
| Gm-b10BB-18 | AW569417 | 0.37 | oth | 6 | F3′5′H (flavonoid-3′,5′-hydroxylase; Glycine max) |
| 28-d Adaxial vs. 21-d adaxial up-regulated | |||||
| Gm-r1070-3826 | BE821412 | 2.69 | u | 3, 4f | ADR12-2 (Glycine max) |
| Gm-r1070-8705 | BE823791 | 2.03 | cgm | 4 | Seed maturation protein PM34 (Glycine max) |
| Gm-r1070-1205 | AW570313 | 2.29 | cgm | 3 | Ribonucleoside-diphosphate reductase small subunit (Nicotiana tabacum) |
| Gm-r1070-536 | BE819864 | 2.13 | cgm | 8 | Seed maturation protein PM41 (Glycine max) |
| 28-d Adaxial vs. 21-d adaxial down-regulated | |||||
| Gm-r1070-324 | BE657471 | 0.47 | ox | 6 | GST 11 (Glycine max) |
| Gm-r1070-8532 | BE824189 | 0.42 | ox | 10 | GST 8 (Glycine max) |
| Gm-r1070-1721 | BE658311 | 0.37 | ox | 2, 9f | Amino acid-specific endopeptidase inhibitor (Momordica charantia) |
| Gm-r1070-7210 | AW278706 | 0.30 | ox | 9 | Cationic seed coat peroxidase precursor(Glycine max) |
| Gm-r1070-1811 | AI938533 | 0.32 | ox | 9 | Peroxidase 1A (Medicago truncatula) |
| G0m-b10BB-37 | AW309104 | 0.42 | ox | 9 | PRP (Pro-rich protein SBPRP1; Glycine max) |
| Gm-b10BB-33 | AW278000 | 0.34 | oth | 6 | IFR1 (isoflavone reductase 1; Glycine max) |
| Gm-b10BB-19 | BE805102 | 0.56 | oth | 6 | F3′5′H (Glycine max) |
| Gm-b10BB-6 | AI437793 | 0.42 | oth | 6 | CHS7 (Glycine max) |
The full list is available at http://www.plantphysiol.org.
EST from the 3′ or 5′ end of the cDNA clone.
Average of two replicates.
cgm, Cell growth and maintenance; en, energy; oth, other; ox, oxidative stress/defense; sig, signaling; sp, seed protein; to, transcription; u, unknown.
Refers to the sets presented on Figure 5.
BLASTX hit of the 3′ end EST or TIGR contig.
Gene represented by clones clustering in different sets.
Substantive changes occurred in the adaxial side between 7 and 14 d because 76 genes were differentially regulated (Fig. 3). Fifteen stress-related genes, such as a cationic peroxidase, and homologs to a PR1a precursor, thaumatin, and endonuclease III, showed increased mRNA abundance between 7 and 14 d. Together with the few differences noted previously between the adaxial and the abaxial side at 7 and 14 d (Fig. 3, A and B), it suggests that the stress occurs throughout the cotyledon. Three genes for storage proteins were up-regulated. One of them, leginsulin, exhibited a particularly dramatic increase (ratio 6.23). Five genes associated with photosynthesis showed increased mRNA levels. Decrease in steady-state RNA levels of homologs of actin and cell division cycle protein 48 suggests that cell division might be slowing down between 7 and 14 d. The down-regulation of homologs of glutaminyl-tRNA reductase, elongation factor 1 alpha, and Pinhead, which belongs to a family of translation initiation factors, also supports the hypothesis of a slow-down in translation (Fig. 3B).
By 21 d, a sharp increase in mRNA abundance of seed proteins (mainly glycinin, conglycinin, and lipoxygenase) was observed. The up-regulation of homologs of histone 3, ribonucleoside-diphosphate reductase, and carbamoyl phosphate synthase suggests an increase in DNA replication, and that of RNA polymerase suggests an increase in transcription. Fewer genes involved in defense or oxidation than in the 14-d versus 7-d comparison were up-regulated. Two GST genes were down regulated, one of which is inducible by 2,4-d. Expression of genes involved in the biosynthesis of phytoalexins such as PAL, CHS, CI, and F3′5′H also decreased. Little changes were observed between 21 and 28 d in the adaxial side. The most remarkable differences were the induction of seed maturation genes (PM34 and PM41) and the further decrease in transcripts for GSTs and genes of the flavonoid pathway (CHS7, IFR1, and F3′5′H).
DISCUSSION
Induction of Somatic Embryos Occurs in Two Distinct Phases
In many tissue culture systems such as the ones developed for carrot or Norway spruce (Picea abies), the addition of auxin to the medium leads to the formation of a pro-embryogenic callus that only differentiates into embryos upon removal of auxin (Zimmerman, 1993; Filonova et al., 2000). The results of the adaxial versus abaxial comparison we performed showed a two-step change in the induced cotyledons. The first step occurs between 0 and 14 d and is characterized by a reduction in the differences in the mRNA populations between the adaxial and abaxial side. The near homogeneity in steady-state RNA populations across the cotyledon at 14 d suggests a dedifferentiation of the cotyledons during this first step. The second step starts after 14 d and is characterized by the development of globular embryos on the adaxial side and the degeneration of the abaxial side and coincides with the differential expression of a large number of genes between the two sides of the explant. This suggest that in soybean, the beginning of embryogenesis (up to the globular phase) can occur on auxin, but as in other systems the expression of the genes associated with embryogeesis might be delayed until partial depletion of auxin in the medium (after 14 d). The hypothesis of a depletion of the medium in auxin overtime is supported by the decrease in mRNA abundance between 7 and 14 d of GH1 and Monopteros, two genes participating in the auxin response (Guilfoyle et al., 1993; Ulmasov et al., 1997) and by the increase in steady-state RNA of the auxin down-regulated gene ADR12-2 (Datta et al., 1993) between 14 and 21 d and 21 and 28 d (Table III).
A Determinant of Abaxial Cell Fate May Influence Polarity in the Induction of Somatic Embryos
In this study, we placed the cotyledon adaxial side up on auxin medium. The orientation of the explant is critical for successful induction of somatic embryos in several species including alfalfa (Chen et al., 1987) and soybean (Santarem et al., 1997) and is consistent with the fact that shoot apical meristems form from cells with adaxial cell fate (McConnell and Barton, 1998). The explanation for this requirement most likely lies in the polar expression of one or several factors in plant lateral organs. At 0 d, homologs of the transcription factors YABBY2 and FIL/YABBY1 showed higher expression in the abaxial side of cotyledons (ratios of 0.42 and 0.33, respectively) than in the adaxial side. The YABBY family has been shown to specify abaxial cell fate in Arabidopsis leaves, cotyledons, and ovules (Sawa et al., 1999; Siegfried et al., 1999), probably in conjunction with other proteins. In our study, the polarity of YABBY2 mRNA persisted up to 14 d after the beginning of the 2,4-d treatment, whereas that of other indicators such as seed proteins did not. This observation supports the hypothesis that YABBY is a determinant of abaxial cell fate in soybean as in Arabidopsis and that its low abundance in the adaxial cells allows the formation of shoot apical meristems and somatic embryos from these cells.
Clustering Shows That an Increase in Transcripts for Oxidative Stress Response Precedes the Appearance of Embryos on the Adaxial Side
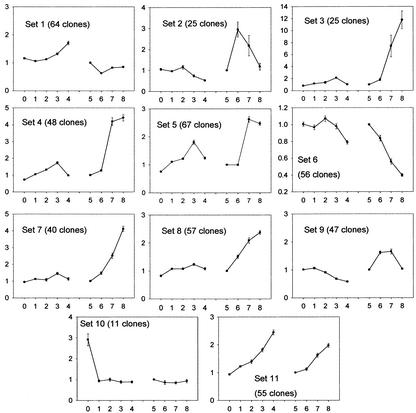
To obtain a global perspective on their expression, we clustered the differentially expressed genes into 11 sets according to their profiles in the adaxial versus abaxial and in the time course experiments using a nonhierarchical clustering method, the k-means technique. The underlying assumption of the cluster analysis is that genes sharing similarities in their profiles are likely to be involved in the accomplishment of a common function. Our time course analysis was so far restricted to genes showing a minimum of a 2-fold change within a week. To also include genes showing a slower trend in the course of the 4 weeks of induction, we compared expression in the adaxial side at 28 d with expression in the adaxial side at 7 d (Fig. 4, dotted line). Including this last experiment, a total of 495 clones show differential expression in at least one of the nine experiments we conducted. Their ratios in each hybridization are available at http://www.plantphysiol.org (Indiv_profiles). To facilitate the interpretation, the time course data was transformed so that differences in expression at any time are graphed in reference to expression at 7 d (see “Materials and Methods” and Fig. 4, solid lines). The average profile of the genes in each set is shown on Figure 5. The complete list of clones in each set is available at www.plantphysiol.org (sets 1–11).
Figure 5.
Cluster analysis of 495 cDNA clones differentially expressed in at least one of nine experiments. The clones were classified based on the similarity of their expression profiles using the k-means technique. The following experiments are on the x axis. 0 through 4, Adaxial versus abaxial experiments at 0, 7, 14, 21, and 28 d, respectively. 5 through 8, Time course experiments: experiment 5, 7 d (equal to 1, see “Materials and Methods”); experiment 6, 14 versus 7 d; experiment 7, 21 versus 7 d; experiment 8, 28 versus 7 d. On the y axis are the average ratios of the clones in the set. Bars = se around the mean of the ratios. See “Materials and Methods” and Figure 4 for ratio calculations.
Although all the intermediates of the response to auxin are not known, it is established that 2,4-D and light can induce an oxidative burst in the target tissue by generating reactive oxygen species (ROS; Pfeiffer and Hoftberger, 2001). Consistent with these findings, we observed many genes, characteristic of a response to oxidative burst, up-regulated in the cotyledons during the first 14 d on 2,4-D (Fig. 3, 14 versus 7 d). Although the tissue was wounded during sample collection and an oxidative burst can also result from mechanical stress (Gus-Mayer et al., 1998), all samples at each time point were treated similarly: put on ice immediately after cutting and frozen in liquid nitrogen within 30 min. Because all tissues were similarly wounded at every time point, it is unlikely that differences in mRNA abundance of any genes was caused by wounding.
Most of the genes participating in the response to auxin or oxidative stress clustered in sets 6, 2, and 9. A large number of GST (glutathione-S-transferase) genes are found in set 6: GST7, GST8, GST10, GST11, GST16, GST19, and two probable glutathione-S-transferases (Table IV). GSTs catalyze the conjugation of a broad range of substrates to glutathione (McGonigle et al., 2000). GSTs are induced by the ROS hydrogen peroxide (H2O2; Levine et al., 1994). They detoxify xenobiotics, metabolize the by-products of membrane oxidation lipid hydroperoxides (Berhane et al., 1994), and may serve as carriers for plant hormones, including auxin (Edwards et al., 2000). Due to their presence in the same cluster, we can speculate on a GST role for five cytochrome P450s of unknown function and for the safener-induced In2-1 gene. Some GSTs might be targets of the transcription factors WRKY (Du and Chen, 2000), two homologs of which are also in this cluster. In addition, set 6 contains many genes of the flavonoid pathway (for example, CHS, CI, F3′5′H, IFS, and IFR) that synthesize phytoalexins and anthocyanins. As opposed to GSTs, these genes are not directly induced by H2O2 (Levine et al., 1994), but phytoalexins are produced during oxidative stress (Wojtaszek, 1997) and might be transported to the vacuole by GSTs (Marrs et al., 1995; Edwards et al., 2000). Expansin, pectinesterase, and glucanase, also in this set, are expressed in response to auxin and induce loosening of the cell wall. The mRNA abundance of genes in set 6 is highest at 7 d and decreases consistently in the adaxial side over the course of the experiment. This pattern, consistent with the decrease of auxin in the medium over time, which was mentioned previously, suggests that their expression profile is modulated by the concentration of 2,4-D in the medium.
Table IV.
k-Means set 6
| Clone Identification | GenBank Accession No.a | Functionb | Annotationc |
|---|---|---|---|
| Gm-r1070-1847 | BE658232 | cgm | Glc-6-phosphate 1-dehydrogenase (cytoplasmic; Medicago sativa) |
| Gm-r1070-2578 | BE658921 | cgm | Probable coatomer complex subunit 33791-27676 (Arabidopsis) |
| Gm-r1070-9132 | BE824285 | cgm | Glc-6-phosphate 1-dehydrogenase (cytoplasmic; Medicago sativa) |
| Gm-r1070-1797 | BE658222 | cgm | Beta-glucosidase (Arabidopsis) |
| Gm-b10BB-19 | BE805102 | oth | F3′5′H (Glycine max) |
| Gm-b10BB-33 | AW278000 | oth | IFR1 (Glycine max) |
| Gm-b10BB-29 | AW201205 | oth | IFS2 (isoflavone synthase 2; Glycine max) |
| Gm-b10BB-30 | BG044305 | oth | IFS1 (isoflavone synthase 1; Glycine max) |
| Gm-b10BB-31 | AW707047 | oth | CYP93A1 (dihydroxypterocarpan-6a-hydroxy;Glycine max) |
| Gm-b10BB-5 | L03352 | oth | CHS6 (homologous to CHS1–5; Glycine max) |
| Gm-b10BB-10 | BG157194 | oth | CI (Glycine max) |
| Gm-b10BB-18 | AW569417 | oth | F3′5′H (Glycine max) |
| Gm-b10BB-4 | AW707011 | oth | CHS2 (Glycine max) |
| Gm-b10BB-6 | AI437793 | oth | CHS7 (Glycine max) |
| Gm-b10BB-7 | AI441937 | oth | CHS1 (Glycine max) |
| Gm-r1070-2852 | BE659055 | ox | Putative NtPRp27-like protein (Nicotiana tabacum) |
| Gm-r1070-3579 | BE821421 | ox | GST 16 (Glycine max) |
| Gm-r1070-3642 | BE821453 | ox | Probable glutathione transferase (Glycine max) |
| Gm-r1070-4730 | BE820598 | ox | Endo-beta-1 4-glucanase (Fragaria ananassa) |
| Gm-r1070-4752 | BE820601 | ox | GST 7 (Glycine max) |
| Gm-r1070-5160 | BE820969 | ox | Probable disease resistance response protein(Arabidopsis) |
| Gm-r1070-6214 | AW186185 | ox | GST 19 (Glycine max) |
| Gm-r1070-8040 | BE823238 | ox | Probable glutathione transferase (Glycine max) |
| Gm-r1070-929 | BE819902 | ox | GST 8 (Glycine max) |
| Gm-r1070-3058 | BE659268 | ox | NtPRp27 (Nicotiana tabacum) |
| Gm-r1070-324 | BE657471 | ox | GST 11 (Glycine max) |
| Gm-r1070-4841 | BE820550 | ox | GST 10 (Glycine max) |
| Gm-r1070-5439 | BE821082 | ox | In2-1 protein (Glycine max) |
| Gm-r1070-5505 | BE821526 | ox | Expansin (Cicer arietinum) |
| Gm-r1070-5621 | AW459775 | ox | Probable glutathione transferase (Glycine max) |
| Gm-r1070-4989 | BE820570 | sig | Calcium-binding protein like (Arabidopsis) |
| Gm-r1070-6079 | AW185677 | sig | Auxin-responsive GH3 product (Glycine max) |
| Gm-r1070-1441 | BE657902 | to | Homeodomain-Leu zipper protein 56 (Glycine max) |
| Gm-r1070-1657 | BE658290 | to | WRKY4 (Petroselium crispum) |
| Gm-r1070-2262 | BE658511 | to | NtWRKY2 (Nicotiana tabacum) |
| Gm-r1070-2675 | BE658859 | u | Cytochrome P450 82A4 (Glycine max) |
| Gm-r1070-1577 | BE658278 | u | Cytochrome P450 (Pisum sativum) |
| Gm-r1070-2238 | BE658653 | u | None |
| Gm-r1070-2823 | BE659130 | u | None |
| Gm-r1070-2998 | BE659085 | u | None |
| Gm-r1070-3023 | BE659194 | u | None |
| Gm-r1070-3778 | BE821387 | u | Unknown protein |
| Gm-r1070-3918 | BE659767 | u | Cytochrome P450 82A4 (Glycine max) |
| Gm-r1070-4927 | AW396292 | u | None |
| Gm-r1070-4972 | BE820646 | u | Cytochrome P450 (Pyrus communis) |
| Gm-r1070-5056 | BE820941 | u | None |
| Gm-r1070-549 | BE657516 | u | Cytochrome P450 (Pisum sativum) |
| Gm-r1070-5643 | BE821162 | u | None |
| Gm-r1070-6965 | BF219577 | u | Unknown protein |
| Gm-r1070-7084 | BE822789 | u | Unknown protein |
| Gm-r1070-8628 | BE824219 | u | Hypothetical protein |
| Gm-r1070-2522 | BE658907 | u | Unknown protein |
| Gm-r1070-7753 | AW472132 | u | Unknown protein |
| Gm-r1070-2856 | BE659056 | u | None |
| Gm-r1070-3118 | AW432447 | u | Putative ripening-related protein (Vitis vinifera) |
| Gm-r1070-3957 | BE659569 | u | Putative ripening-related protein (Vitis vinifera) |
EST from the 3′ or 5′ end of the cDNA clone.
cgm, Cell growth and maintenance; en, energy; oth, other; ox, oxidative stress/defense; sig, signaling; to, transcription; u, unknown.
BLASTX hit of the 3′ end EST or TIGR contig.
Genes in sets 2 and 9 have similar mRNA abundance profiles as genes in set 6, but, in the adaxial side, the steady-state RNA levels of genes in sets 2 and 9 peaked later (14 and 21 d, respectively) than those of genes in set 6. Both sets 2 and 9 contain genes commonly induced during wounding or pathogen infection. For example, set 2 comprises homologs of genes encoding a PR1a precursor, a thaumatin, a wound-inducible protein, and ACC oxidase, which catalyzes the last step in ethylene synthesis (Ecker, 1995). Involved in defense and in set 9 are also homologs to receptor-like protein kinases (Cheong et al., 2002) that are under the control of the transcription factors WRKY (Du and Chen, 2000), and homologs to transcription factors containing an AP2 domain, a RING zinc finger, or a bHLH domain (Cheong et al., 2002). RING zinc finger proteins participate in the ubiquitination of targets destined to proteasomal degradation (Freemont, 2000). Unsurprisingly, a homolog to a subunit of the 26S proteasome also clusters in set 9. Genes involved in cell wall remodeling, which often occurs in response to stress, are found in set 2: MMPs, which facilitate extracellular matrix degradation in senescing tissue (Delorme et al., 2000) and during tumor invasion (Zhu et al., 2001), homologs of glucosyltransferases, extensin, which participate in the loosening of the cell wall, and a homolog of the carrot EDGP (extra dermal glycoprotein precursor), a protein found in the extracellular matrix of unorganized callus tissue (Satoh et al., 1992). Several genes in set 9 are active in the cell wall as well: peroxidases that scavenge ROS, Pro-rich proteins that fortify the cell wall in response to wounding, a reticuline oxidase-like gene (Wojtaszek, 1997), and a pectinesterase.
Several genes in set 9 are reportedly directly involved in the control of oxidative stress. They comprise homologs to a gene for adenosine 5′-adenylphosphosulfate reductase, which participates in sulfate assimilation (Kopriva et al., 2001), and to a putative glutaredoxin, both of which are involved in the synthesis of reducing thiol compounds; and a homolog to a regulatory subunit of protein phosphatase 2A, which blocks the induction of H2O2 (Levine et al., 1994). In set 2, a homolog to an Arabidopsis endonuclease III involved in the replacement of oxidized pyrimidines in DNA (Roldan-Arjona et al., 2000) could induce DNA fragmentation, a phenomenon characteristic of programmed cell death (PCD). PCD can be triggered by high levels of ROS (Tenhaken et al., 1995). It occurs at various times during plant development and is interpreted as a way for plants to recycle nutrients from unneeded structures. In particular, PCD occurs during the switch from pro-embryogenic masses to somatic embryos upon removal of plant growth regulators in Norway spruce as detected by DNA fragmentation (Filonova et al., 2000).
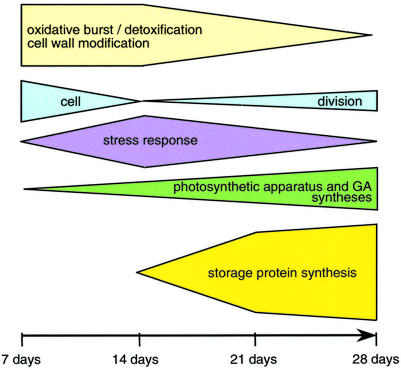
Our analysis show that many genes increasing in expression in the cotyledons during the first 2 weeks on auxin (sets 2, 6, and 9) are involved in detoxification, defense, or in the maintenance of the redox state in other contexts. It suggests that an oxidative burst, most probably caused by 2,4-D (Pfeiffer and Hoftberger, 2001), occurs during the development of somatic embryos and might lead to PCD of certain cells (Fig. 6). Interestingly, Santarem et al. (1997) showed that wounding of the cotyledons accelerates the appearance of somatic embryos in soybean. The oxidative stress is likely to trigger a specific signaling pathway. Ethylene could be involved because an ACC oxidase gene clusters in set 2. Salicylic acid might also participate because PR1, CHS, flavonol synthase, thaumatin, GST, ACC oxidase, peroxidase, an AP2 domain gene, and EDGP, homologs of which are present in sets 6, 2 or 9, were shown to be induced by salicylic acid in Arabidopsis (Schenk et al., 2000).
Figure 6.
Summary of the putative physiological changes occurring in the adaxial sides of immature cotyledons during the differentiation of somatic embryos. The timescale represents the number of days the cotyledons were on 40 mg L−1 2,4-D. The heights of the blocks reflect the intensity of the response at the transcript level.
Transcript Profiling Suggests That Cell Proliferation Peaks in the First 7 d
Histological data in soybean and conifers showed that somatic embryos form by division of epidermal and subepidermal cells of the cotyledon (Santarem et al., 1997; Salajova and Salaj, 2001). Genes indicative of cell division cluster in set 1. Transcript abundance of genes in set 1 peaks in the adaxial side at 7 d, decreases by one-half by 14 d, and is increasingly high in the adaxial side compared with the abaxial side overtime. Markers of the S-phase of the cell cycle such as histone H4 (Frank et al., 2000), histone H3 (Gown et al., 1996), and histone H2A (Tanimoto et al., 1993) suggest that genes in this set are involved in DNA replication. Their clustering with a cell division cycle 48 homolog (CDC48) suggests that DNA replication is concomitant with cytokinesis because CDC48 participates in the formation of spindle poles in proliferating tissue of plants (Feiler et al., 1995). Several other genes in set 1 have been linked to cell proliferation, such as homologs of actin, alpha-tubulin, the chaperonin TCP1 (Ursic et al., 1994), and FKBP62, which binds the cell proliferation inhibitor rapamycin (Vucich and Gasser, 1996). This set also includes homologs of genes participating in translation: translation factors 1-alpha and Pinhead (Lynn et al., 1999), glutaminyl-t-RNA reductase, 40S ribosomal protein, and plastid ribosomal protein S11. Although the variations in expression of genes in set 1 are small over the time course (their average ratios range from 0.5–1.6), the average profile of the genes in this set suggest that cell proliferation peaks at 7 d in both the adaxial and abaxial sides of cotyledons, thus coinciding with the potential oxidative burst mentioned previously, but becomes localized to the adaxial side after 14 d (Fig. 6).
Transcripts for GA Synthesis Increase Steadily in the Developing Embryos
Set 11 (Table V) includes homologs of genes participating in photosynthesis (several chlorophyll-binding proteins, a Rubisco small subunit, and a chloroplast ribonucleoprotein) and translation (homologs to the translation factor argonaute and ribosomal protein S2). Most noticeable in this set are homologs to GA3 biosynthesis genes: GA 20 oxidase (two clones), ent-kaurenoic acid hydroxylase (two clones), ent-kaurene oxidase, and GA-regulated genes (homologs to GAST1, LTCOR11, and a GA-regulated protein). Two clones annotated as cytochrome P450 and identified as specific to the pollen tubes of orchid (Nadeau et al., 1996) are present in this set and could also participate in the synthesis of GA. Transcripts of genes in set 11 show increasing polarity between the adaxial and abaxial side and increasing expression in the developing somatic embryos over time (Fig. 6). Interestingly, the polarity in expression for genes in this set is apparent as early as 7 d after the beginning of the treatment (average ratio of 1.27), suggesting that they may play a role early in the initiation of somatic embryos. Little is known of the effect of GA on embryo development. Conflicting reports exist on the effect of exogenous application of GA on somatic embryogenesis. In alfalfa, application of GA3 increased the number of somatic embryos, whereas inhibitors of GA prevented the formation of embryos (Rudus et al., 2002). The exact opposite effects were observed in Pelargonium × hortorum Bailey, using hypocotyls as explants (Hutchinson et al., 1997). In pea, a mutation in ent-kaurene oxidase caused lower rate of survival and reduced seed weight (Swain et al., 1997), and GA plays an essential role in axis elongation by promoting cell elongation (Hays et al., 2002). On 2,4-D medium, soybean somatic embryos are arrested in the globular stage, suggesting that GA is not sufficient to induce axis elongation. Two clones with homology to Arabidopsis XET are found in sets 11 and 1. XET enzymes catalyze the cleavage of xyloglucan, thereby inducing loosening of the cell wall. Expectedly, XETs are highly expressed in developing tissue, where they allow cell growth under turgor pressure (Fry et al., 1992). Given their clustering with genes involved in GA synthesis, it is possible that a soybean XET gene is up-regulated by GA as was demonstrated for the tomato LeXET2 expressed in mature tissues (Catala et al., 2001).
Table V.
k-Means set 11
| Clone ID | GenBank accessiona | Functionb | Annotationc |
|---|---|---|---|
| Gm-r1070-123 | BE657421 | cgm | Argonaute-like (zwille, pinhead) protein (Arabidopsis) |
| Gm-r1070-5257 | BE820863 | cgm | Putative xyloglucan endotransglycolase (Arabidopsis) |
| Gm-r1070-5312 | BE821057 | cgm | ATP-dependent Clp protease proteolytic subunit(Lotus japonicus) |
| Gm-r1070-6877 | BE822339 | cgm | Ribosomal protein S2 (Lotus japonicus) |
| Gm-r1070-7482 | AW460113 | cgm | Phosphatidyl-Ser decarboxylase (Arabidopsis) |
| Gm-r1070-7508 | BE822956 | cgm | SMT3 ubiquitin-like protein (Arabidopsis) |
| Gm-r1070-7571 | BE823365 | cgm | Histone H2A.F/Z (Arabidopsis) |
| Gm-r1070-7579 | BE823362 | cgm | Acetyltransferase (Olea europaea) |
| Gm-r1070-8697 | BE823786 | cgm | Tyrosyl-tRNA synthase (Nicotiana tabacum) |
| Gm-r1070-9084 | BE824267 | cgm | Putative epimerase/dehydratase (Oryza sativa) |
| Gm-r1070-5866 | BE821693 | cgm | Histone H2A.F/Z (Arabidopsis) |
| Gm-r1070-3997 | BE659580 | en | Chlorophyll a/b-binding protein of LHCII type I prec. (cab-3; Glycine max) |
| Gm-r1070-5038 | BE820966 | en | Chlorophyll a/b-binding protein of LHCII type I prec. (cab-3; Glycine max) |
| Gm-r1070-7043 | BE822600 | en | Rubisco small subunit RBCS1 (Glycine max) |
| Gm-r1070-7920 | BE823502 | en | PSII type I chlorophyll a/b-binding protein (Glycine max) |
| Gm-r1070-8717 | BE824039 | en | Chlorophyll a/b-binding protein type I prec. (Lycopersicon esculentum) |
| Gm-r1070-7893 | BE823134 | oth | 4-Coumarate-CoA ligase-like protein (Arabidopsis) |
| Gm-r1070-7709 | BE823267 | ox | Glutathione reductase chloroplast precursor (Glycine max) |
| Gm-r1070-42 | BE657386 | sig | Ent-kaurenoic acid hydroxylase (Arabidopsis) |
| Gm-r1070-7723 | BE823259 | sig | Ent-kaurenoic acid hydroxylase (Arabidopsis) |
| Gm-r1070-7797 | BE823089 | sig | GA 20-oxidase (Phaseolus vulgaris) |
| Gm-r1070-7836 | AW472167 | sig | Protein kinase C inhibitor-like (Zea mays) |
| Gm-r1070-819 | AW100772 | sig | Receptor-like kinase 2 (Glycine max) |
| Gm-r1070-8358 | BE823656 | sig | Ent-kaurene oxidase (Cucurbita maxima) |
| Gm-r1070-8991 | BE824089 | sig | GA 20-oxidase (Phaseolus vulgaris) |
| Gm-r1070-7777 | BE823087 | sig | Zinc finger protein like, Ser/Thr protein kinase like (Arabidopsis) |
| Gm-r1070-7851 | BE823255 | sig | Nodulin-like protein (Arabidopsis) |
| Gm-r1070-7636 | BE822993 | sp | Kunitz-type trypsin inhibitor KTI2 precursor (Glycine max) |
| Gm-r1070-5613 | BE821543 | sp | Beta-amylase (Glycine max) |
| Gm-r1070-8597 | BE823764 | sp | Lectin precursor (agglutinin; Glycine max) |
| Gm-b10BB-52 | AI736089 | sp | Lectin precursor (Glycine max) |
| Gm-r1070-7209 | BF219549 | to | Putative ribonucleoprotein chloroplast precursor (Nicotiana sylvestris) |
| Gm-r1070-5305 | BE820875 | to | Filamentous flower protein FIL/YABBY1 (Arabidopsis) |
| Gm-r1070-7252 | BE822573 | u | GA-regulated protein GAST1 like (Arabidopsis) |
| Gm-r1070-7726 | BE823315 | u | GA-regulated protein (Arabidopsis) |
| Gm-r1070-8165 | BE823572 | u | None |
| Gm-r1070-3678 | AI965366 | u | None |
| Gm-r1070-5655 | AW457938 | u | Hypothetical protein |
| Gm-r1070-7540 | BE822967 | u | Cytochrome P-450 (Phalaenopsis sp. SM9108) |
| Gm-r1070-7549 | BE822890 | u | None |
| Gm-r1070-7671 | BE823390 | u | None |
| Gm-r1070-7968 | BE823514 | u | Unknown protein |
| Gm-r1070-7995 | BE823295 | u | None |
| Gm-r1070-7575 | BE823368 | u | None |
| Gm-r1070-8016 | BE823531 | u | Putative protein |
| Gm-r1070-8166 | BE823586 | u | Putative protein |
| Gm-r1070-8566 | BE823825 | u | None |
| Gm-r1070-138 | AW397125 | u | Hypothetical protein |
| Gm-r1070-3314 | AW432306 | u | None |
| Gm-r1070-3964 | AW119711 | u | None |
| Gm-r1070-7822 | BE823333 | u | None |
| Gm-r1070-6094 | BE821924 | u | Putative senescence-associated protein (Pisum sativum) |
| Gm-r1070-7816 | BE823346 | u | Putative protein |
| Gm-r1070-8594 | BE823833 | u | Cytochrome P-450 (Phalaenopsis cv. SM9108) |
| Gm-r1070-8987 | BE824086 | u | LTCOR11 (Lavatera thuringiaca) |
EST from the 3′ or 5′ end of the cDNA clone.
cgm, Cell growth and maintenance; en, energy; oth, other; ox, oxidative stress/defense; sig, signaling; sp, seed protein; to, transcription; u, unknown.
BLASTX hit of the 3′ end EST or TIGR contig.
Storage Proteins Transcripts Accumulate in the Globular Somatic Embryos
At 0 d, transcripts for storage proteins (Bowman-Birk trypsin inhibitor and lectin) were more abundant in the abaxial side of the cotyledons (Fig. 3B, 0-d adaxial versus abaxial). This observation supports in situ hybridizations performed with Kunitz trypsin inhibitor, beta-conglycinin, lectin, and glycinin probes that show progression of the expression in a wave-like pattern from the abaxial to the adaxial side during the development of the cotyledon (Goldberg et al., 1989).
We showed that developing somatic embryos accumulated transcripts for storage proteins from 14 d on (Fig. 4A, 21-d adaxial versus abaxial and 21 versus 14 d; Fig. 6). The corresponding genes clustered in sets 3 to 5. Their steady-state mRNA levels increased in the adaxial side over time and are higher in the adaxial side than in the abaxial side at 21 d. It is important to note that although the same amounts of transcripts for seed proteins were found within equal-sized pools of RNA from the adaxial and abaxial side of the cotyledons at 28 d (as reflected by the microarrays), at this time point the amount of extractable RNA in the abaxial side was 2 to 3 times lower than in the adaxial side (data not shown). Sets 3 to 5 differed primarily by the amplitude of the transcription response to the 2,4-D treatment, which is summarized in the 28- versus 7-d average ratio: 12 in set 3, 4.5 in set 4, and 2.5 in set 5. Members of set 3, and leginsulin in particular, showed the highest increase in the adaxial side of all the genes on the array in the course of the experiment. Soybean leginsulin was found to have insulin-like binding properties and to stimulate the phosphorylation of its receptor, a 7S globulin, Bg7S (Watanabe et al., 1994). Insulin-like growth factors were characterized in pea (Higgins et al., 1986), lupine (Lupinus angustifolius; Ilgoutz et al., 1997), and also in maize (Garcia Flores et al., 2001), where it was shown to enhance maize seedling growth presumably by increasing phosphorylation of a 40S ribosomal subunit protein, rpS6 (Garcia Flores et al., 2001). Rp S6 is speculated to preferentially induce the translation of 5′-terminal oligopyrimidine mRNAs (Brown and Schreiber, 1996), most of which are ribosomal proteins and proteins involved in translation (Levy et al., 1991). In humans, insulin-like growth factors were shown to mediate epidermal growth in cervical cancer cells (Steller et al., 1995) and to decrease apoptosis (Parrizas et al., 1997). Therefore, leginsulin could play an important role in cell proliferation and in the mitigation of cell death in the developing embryos. However, the gene for the soybean leginsulin receptor, Bg7S, also represented on the array, is expressed uniformly throughout the cotyledons.
Alternatively, leginsulin, the Bowman-Birk protease inhibitor, and the Cys-rich protein present in set 3 are all sulfur-rich proteins. Bowman-Birk protease inhibitors and the pea PA1, which encodes leginsulin, are transcriptionally up-regulated by sulfur (Higgins et al., 1986; Biermann et al., 1998). The timing of their increase (14 d for leginsulin or 21 d for the others) in the adaxial side coincides with the decline in abundance of transcripts associated with oxidative stress, a time when the availability of reducing thiol compounds such as glutathione might become less important, and sulfur reserves can be replenished. The clustering of Bowman-Birk and leginsulin genes with ADR12-2, a small polypeptide characterized by its negative regulation by auxin (Datta et al., 1993), suggests that they might be down regulated by auxin or the ensuing oxidative burst and is consistent with our hypothesis.
Set 4 includes conglycinin genes, a Met-rich 2S albumin, but also lipoxygenase 1, 2, and 3. Lipoxygenases constitute 1% to 2% of the protein content of soybean seeds (Loiseau et al., 2001). These iron-containing enzymes, in their oxidative state, catalyze the hydroxiperoxidation of fatty acids (Maccarrone et al., 2001). Their action can lead to membrane degradation in oxidative conditions, and can result in the synthesis of jasmonic acid, which is involved in senescence or necrosis (Creelman and Mullet, 1997). However the expression profile of lipoxygenases indicate that they are transcribed later than the genes involved in the putative oxidative burst (mostly in sets 2, 6, and 9) and are therefore not likely to play a major role in the oxidative process. Their clustering with the storage protein conglycinin suggests that they have a storage rather than a signaling function. Set 5 is more diverse and comprises genes of the glycinin and lipoxygenase families and genes associated with carbohydrate metabolism such as homologs to a hexokinase, a transcriptional activator of sugar kinases, and beta amylase.
Our data show that most genes encoding storage proteins cluster in sets 3, 4, and 5, indicating that their expression increases very early in the globular phase of embryo development. Although small amounts of storage compounds can be detected in globular embryos of broad bean (Vicia faba; Panitz et al., 1999), zygotic embryos start accumulating transcripts for storage proteins at the maturation stage (Walling et al., 1986) and generally after cell divisions end. Therefore, the transcription of genes encoding storage proteins in globular embryos is unexpected and may represent an important difference between somatic and zygotic embryos.
Summary
Using a 9,280-cDNA clone array, we have identified 495 cDNA clones showing modulation of expression in response to 2,4-D treatment during the development of somatic embryos. Clustering the clones by similarity of expression profile over the course of the study allowed us to determine the timing of the molecular events taking place during embryogenesis. Of course, mRNA abundance data alone does not ensure that a physiological event is actually occurring because control of expression can be exerted at multiple levels. However, transcript profiles do give a strong point of reference and are particularly valuable for systems that have not been characterized extensively at the molecular level, such as somatic embryogenesis.
We have shown that 2,4-D induces the dedifferentiation of the cotyledon within 7 d and that differential expression in the adaxial and abaxial side of the cotyledons is apparent only after more than 14 d of treatment, when auxin levels have probably decreased in the medium. Transcripts participating in cell proliferation suggest that cell division is induced early (within 7 d) in both adaxial and abaxial sides of the cotyledons and persists at a slower rate until 28 d in the adaxial side. A possible oxidative burst concomitant with cell division reaches a peak at 14 d and gradually becomes more important in the abaxial side. Finally, we show strong indications that GA3 is produced in the adaxial side from 7 d on and that transcripts for storage proteins accumulate in the developing somatic embryos after 14 d on 2,4-D.
MATERIALS AND METHODS
Tissue Collection
Soybean (Glycine max L. Merrill cv Jack) plants were grown in the greenhouse. Pods containing 4- to 6-mm seeds were surface sterilized. Seeds were removed from the pods. The chalazal end of the seed was cut, thus separating the axis from the cotyledon, and the cotyledons were pushed out of the seed coat. Cotyledons were plated on MSD40 3% (Murashige and Skoog basal medium, 40 mg L−1 2,4-D, 3% (w/v) Suc; Finer, 1988), at a density of 25 cotyledons per plate, abaxial side in contact with the medium, and placed at 24°C in 3 to 7 μE of light. Cotyledons were collected randomly from each plate at 0, 7, 14, 21, and 28 d (Fig. 1). The adaxial side, consisting of one-fourth to one-half of the cotyledon thickness or when the somatic embryos were visible, the green tissue was carefully cut from the abaxial side of the cotyledons with a sterile scalpel (Fig. 2) so as to avoid any brown tissue. Adaxial and abaxial sides were frozen in liquid nitrogen and freeze dried. Seven replicate experiments were conducted, each comprising between 65 and 454 cotyledons.
Isolation and Pooling of RNA
RNA was extracted from each biological replicate's adaxial and abaxial sides for each time point as described previously (Sambrook et al., 1989). Equal microgram amounts of total RNA from tissue sampled at identical time points were pooled across biological replicates. Each pool was purified with RNeasy Mini columns (Qiagen, Valencia, CA) according to the manufacturer's instructions. The purified RNA was concentrated with YM-30 Microcon columns (Millipore, Bedford, MA) and used for the adaxial/abaxial comparison. More RNA was added to each pool for the time course experiment. Once again, the proportions were kept equal between the replicates, except for the 21-d adaxial tissue pool, in which replicate 5 is underrepresented, and the 7-d adaxial tissue pool, in which replicates 6 and 7 are overrepresented.
Preparation of Labeled Probes
For each probe, 35 to 60 μg of total purified RNA was reverse transcribed in the presence of Cy3- or Cy5-dUTP (Hegde et al., 2000). The same amount of total RNA was used for probes hybridized to the same array. In brief, the RNA and 5 μg of oligo(dT) (Operon, Qiagen) were denatured in a 10-μL volume at 70°C for 10 min and cooled on ice before the following 30-μL reaction was set up: 10 μL of the RNA and oligo(dT) mixture; 1× first strand reaction buffer; 10 mm dithiothreitol; 0.5 mm dATP, dCTP, and dGTP; 100 μm Cy3- or Cy5-dUTP (Amersham-Pharmacia Biotech, Uppsala); and 400 units of SuperscriptII (Invitrogen, Carlsbad, CA). The reaction was incubated 120 min at 42°C, with the addition of 200 units of SuperscriptII after the first 60 min, and treated with RNAse A and H (0.25 μg and 0.5 units, respectively) for 30 min at 37°C. The resulting Cy3 and Cy5 probes were paired according to the intended experiment, and unincorporated nucleotides were removed using a PCR cleaning kit (Qiagen). Cleaned probes were concentrated in a SpeedVac (Savant Instrument, Holbrook, NY).
Microarray Hybridization and Analysis
ESTs from embryo, seed coat, flower, and pod libraries were contigged to identify unigenes. Clones representative of 9,216 unigenes were reracked to build the library Gm-r1070 (Table I), and their 3′ ends were sequenced. Purified PCR products of the library Gm-r1070 were single spotted on amine slides (Telechem International, Sunnyvale, CA) using a PixSys 8200 arrayer (Cartesian, Irvine, CA). An additional 64 choice clones were each printed eight times on the array. Details of the unigene selection and of the microarray construction will be provided elsewhere. All cDNA clones are available to the public from the American Type Culture Collection (http://www.atcc.org). The accession number of the microarray platform in the Gene Expression Omnibus is GPL229 (http://www.ncbi.nlm.nih.gov/geo).
For each slide, the labeled cDNA and 15 μg of poly(A+) were denatured at 95°C for 3 min. An equal amount of prewarmed 2× hybridization buffer was added to the mixture, and the probe was deposited on the coverslip (Grace Biolab) or pipetted between the prehybridized slide (slides were incubated in 5× SSC, 0.1% [w/v] SDS, and 1% (w/v) bovine serum albumin at 42°C for 45–60 min) and the coverslip (LifterSlip, Erie Scientific Company, Portsmouth, NH). The slide was placed in a hybridization chamber (Corning, NY) overnight at 42°C and washed three times in 1× SSC and 0.2% (w/v) SDS, 0.2× SSC and 0.2% (w/v) SDS, and 0.1× SSC successively. The slides were scanned with a ScanArray 3000 or ScanArray Express (Perkin Elmer Life Sciences, Boston), the spots were found, and their fluorescence was quantitated by GenePix Pro 3.0 (Axon Instruments, Union City, CA). Local background was subtracted from each spot intensity. Spots showing signal intensity below the 95th percentile of the background distribution in the Cy3 or Cy5 channel were filtered out. The ratio of Cy5 mean to Cy3 mean (r) was computed and used to adjust the Cy3 values to Cy3 X sqrt(r) and the Cy5 values to Cy5/sqrt(r). A between-replicate correction was made using an ANOVA model, which equalized average grid intensity between replicates for Cy3 and Cy5 separately. The ratio of the resulting adjusted intensities of Cy5 to Cy3 was computed for each spot. The CV (sd/mean) across replicates was calculated for each spot to evaluate repeatability of the hybridizations.
Experimental Design
In a first set of experiments, labeled cDNA from the adaxial and abaxial sides of same-stage cotyledons were competitively hybridized to the microarray. The initial time point (0 d) comparison was done in duplicate. All the other comparisons were done in triplicate. To eliminate potential dye bias, the dyes were swapped. The correlation between intensities in replicate slides using same tissue/dye combinations were not higher than correlation between intensities in replicate slides with different tissue-dye combinations, and ranged from 0.86 to 0.96. Raw and normalized data from these hybridized slides was deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo). Accession numbers of these hybridizations are GSM3255, GSM3256, GSM3257, GSM3259, GSM3261, and GSM3263 to GSM3271.
A loop design (as shown in Fig. 4) was used for a second set of experiments. In these experiments, RNA from adaxial tissue at time point n was competitively hybridized to RNA from adaxial tissue at time point n − 1. The following hybridizations were performed: 14 versus 7 d (GSM3246 and GSM3247), 21 versus 14 d (GSM3248 and 3249), and 28 versus 21 d (GSM3250 and GSM3251; Fig. 4, dotted lines). Each was repeated once with a dye swap. We also hybridized two slides with cDNA from 28 d versus cDNA from 7 d (GSM3252 and GSM3254). The normalized ratio of each gene in this control should be the same as the product of the following normalized ratios: 14 versus 7 d, 21 versus 14 d, and 28 versus 21 d. The correlation factor was calculated to be 0.90.
Genes showing a ratio above 2 (below 0.5) in at least two of two or three replicates of the same experiments were considered up- (down) regulated. For these genes, TCs to which the corresponding ESTs belonged were identified in the TIGR soybean gene index (version 7; Quackenbush et al., 2000), and their consensus sequence, or the 3′ EST if a singleton, was subjected to BLASTX for annotation (Altschul et al., 1997). The E-value threshold was set at 1.0 E-6.
Cluster Analysis
Normalized data from the two sets of experiments were analyzed with GeneSpring 4.1 (Silicon Genetics, Redwood City, CA). A total of 120 of 9,280 cDNA clones (1.3%) exhibited poor quality in 11 or more of the 22 hybridizations performed and were not used in the analysis. A total of 495 of 9,280 (5.3%) of the cDNA clones on the array exhibited a ratio above 2 or below 0.5 in at least two replicates of one or more experiment. These 495 cDNA clones were clustered according to their expression patterns across the nine experiments into 11 sets using the k-means unsupervised clustering technique (Gordon, 1999). In brief, this algorithm arbitrarily separated the genes (“vectors”) into 11 groups. The centroid of each group was calculated by averaging the coordinates attached to each gene. In one iteration, each clone was then reassigned to the centroid to which it is closest and the coordinates of the centroids were recalculated. The data converged after 30 iterations. To simplify the interpretation, we transformed the ratios of the time course experiments so that the expression level at any time point is relative to the 7-d time point. Thus, we arbitrarily set the 7-d value to 1 and graphed the following ratios: 14 versus 7 d, 21 versus 7d (obtained by multiplying the ratios 14 versus 7 d and 21 versus 14 d), and 28 versus 7 d (obtained by averaging the measured ratio 28 versus 7 d and the product of the ratios 14 versus 7 d, 21 versus 14 d, and 28 versus 21 d; Fig. 4, solid lines). In addition, the ratios of all the genes in one set were averaged so that they are represented by a single point at each time point, with bars representing the se around the mean of the ratios (Fig. 5).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Shauna Somerville for her advice on setting up the microarray technology. We thank our colleagues Drs. Steven Clough, Orlando Gonzalez, and Jigyasa Tuteja for their helpful comments on the manuscript.
Footnotes
This work was supported by the National Science Foundation Plant Genome Research Program as part of a “Functional Genomics Project for Soybean” (grant no. DBI–9872565) and by grants from the United Soybean Board.
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.019968.
LITERATURE CITED
- Aharoni A, Keizer LC, Van Den Broeck HC, Blanco-Portales R, Munoz-Blanco J, Bois G, Smit P, De Vos RC, O'Connell AP. Novel insight into vascular, stress, and auxin-dependent and -independent gene expression programs in strawberry, a non-climacteric fruit. Plant Physiol. 2002;129:1019–1031. doi: 10.1104/pp.003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleith F, Richter G. Gene expression during induction of somatic embryogenesis in carrot cell suspensions. Planta. 1990;183:17–24. doi: 10.1007/BF00197562. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berhane K, Widersten M, Engstrom A, Kozarich JW, Mannervik B. Detoxication of base propenals and other alpha, beta-unsaturated aldehyde products of radical reactions and lipid peroxidation by human glutathione transferases. Proc Natl Acad Sci USA. 1994;91:1480–1484. doi: 10.1073/pnas.91.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biermann BJ, Debanzie JS, Handelsman J, Thompson JF, Madison JT. Methionine and sulfate increase a Bowman-Birk-type protease inhibitor and its messenger RNA in soybeans. J Agric Food Chem. 1998;46:2858–2862. [Google Scholar]
- Brown EJ, Schreiber SL. A signaling pathway to translational control. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- Catala C, Rose JKC, York WS, Albersheim P, Darvill AG, Bennett AB. Characterization of a tomato xyloglucan endotransglycosylase gene that is down-regulated by auxin in etiolated hypocotyls. Plant Physiol. 2001;127:1180–1192. doi: 10.1104/pp.010481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen THH, Marowitch J, Thompson BG. Genotypic effects on somatic embryogenesis and plant regeneration from callus cultures of alfalfa. Plant Cell Tissue Organ Cult. 1987;8:73–81. [Google Scholar]
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Sung ZR. Two-dimensional gel analysis of carrot somatic embryonic proteins. Plant Mol Biol Rep. 1984;2:19–25. [Google Scholar]
- Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002;32:490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:355–381. doi: 10.1146/annurev.arplant.48.1.355. [DOI] [PubMed] [Google Scholar]
- Datta N, LaFayette PR, Kroner PA, Nagao RT, Key JL. Isolation and characterization of three families of auxin down-regulated cDNA clones. Plant Mol Biol. 1993;21:859–869. doi: 10.1007/BF00027117. [DOI] [PubMed] [Google Scholar]
- Delorme VGR, McCabe PF, Kim D-J, Leaver CJ. A matrix metalloproteinase gene is expressed at the boundary of senescence and programmed cell death in cucumber. Plant Physiol. 2000;123:917–928. doi: 10.1104/pp.123.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, A.-H.-Mackerness S, Hancock JT, Neill SJ. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001;127:159–172. doi: 10.1104/pp.127.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du LQ, Chen ZX. Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salicylic acid-induced WRKY DNA-binding proteins in Arabidopsis. Plant J. 2000;24:837–847. doi: 10.1046/j.1365-313x.2000.00923.x. [DOI] [PubMed] [Google Scholar]
- Ecker JR. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- Edwards R, Dixon DP, Walbot V. Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000;5:193–198. doi: 10.1016/s1360-1385(00)01601-0. [DOI] [PubMed] [Google Scholar]
- Feiler HS, Desprez T, Santoni V, Kronenberger J, Caboche M, Traas J. The higher plant Arabidopsis thaliana encodes a functional cdc48 homologue which is highly expressed in dividing and expanding cells. EMBO J. 1995;14:5626–5637. doi: 10.1002/j.1460-2075.1995.tb00250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonova LH, Bozhkov PV, Brukhin VB, Daniel G, Zhivotovsky B, von Arnold S. Two waves of programmed cell death occur during formation and development of somatic embryos in the gymnosperm, Norway spruce. J Cell Sci. 2000;113:4399–4411. doi: 10.1242/jcs.113.24.4399. [DOI] [PubMed] [Google Scholar]
- Finer JJ. Apical proliferation of embryonic tissue of soybean [Glycine max (L.) Merrill] Plant Cell Rep. 1988;7:238–241. doi: 10.1007/BF00272532. [DOI] [PubMed] [Google Scholar]
- Finer JJ, McMullen MD. Transformation of soybean via particle bombardment of embryogenic suspension culture tissue. In Vitro Cell Dev Biol. 1991;27:175–182. [Google Scholar]
- Frank M, Rupp H-M, Prinsen E, Motyka V, Van Onckelen H, Schmulling T. Hormone autotrophic growth and differentiation identifies mutant lines of Arabidopsis with altered cytokinin and auxin content or signaling. Plant Physiol. 2000;122:721–730. doi: 10.1104/pp.122.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont PS. Ubiquitination: RING for destruction? Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J. 1992;282:821–828. doi: 10.1042/bj2820821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Flores C, Aguilar R, Reyes de la Cruz H, Albores M, Sanchez de Jimenez E. A maize insulin-like growth factor signals to a transduction pathway that regulates protein synthesis in maize. Biochem J. 2001;358:95–100. doi: 10.1042/0264-6021:3580095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux RW, Pauls KP. Characterization of somatic embryogenesis-related cDNAs from alfalfa (Medicago sativa L.) Plant Mol Biol. 1997;33:393–404. doi: 10.1023/a:1005786826672. [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Barker SJ, Perez-Grau L. Regulation of gene expression during plant embryogenesis. Cell. 1989;56:149–160. doi: 10.1016/0092-8674(89)90888-x. [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Depaiva G, Yadegari R. Plant embryogenesis: zygote to seed. Science. 1994;266:605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- Gordon AD. Classification. Classification. Ed 2. Boca Raton, FL: Chapman & Hall/CRC; 1999. pp. 41–43. [Google Scholar]
- Gown AM, Jiang JJ, Matles H, Skelly M, Goodpaster T, Cass L, Reshatof M, Spaulding D, Coltrera MD. Validation of the S-phase specificity of histone (H3) in situ hybridization in normal and malignant cells. J Histochem Cytochem. 1996;44:221–226. doi: 10.1177/44.3.8648081. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G, Li Y, Ulmasov T, Liu ZB, Strabala T, Gee M. Auxin-regulated transcription. Aust J Plant Physiol. 1993;20:489–502. [Google Scholar]
- Gus-Mayer S, Naton B, Hahlbrock K, Schmelzer E. Local mechanical stimulation induces components of the pathogen defense response in parsley. Proc Natl Acad Sci USA. 1998;95:8398–8403. doi: 10.1073/pnas.95.14.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays DB, Yeung EC, Pharis RP. The role of gibberellins in embryo axis development. J Exp Bot. 2002;53:1747–1751. doi: 10.1093/jxb/erf017. [DOI] [PubMed] [Google Scholar]
- Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J. A concise guide to cDNA microarray analysis. Biotechniques. 2000;29:548–562. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
- Higgins TJ, Chandler PM, Randall PJ, Spencer D, Beach LR, Blagrove RJ, Kortt AA, Inglis AS. Gene structure, protein structure, and regulation of the synthesis of a sulfur-rich protein in pea seeds. J Biol Chem. 1986;261:11124–11130. [PubMed] [Google Scholar]
- Hutchinson MJ, Krishnaraj S, Saxena PK. Inhibitory effect of GA3 on the development of thidiazuron-induced somatic embryogenesis in geranium (Pelargonium × hortorum Bailey) hypocotyl cultures. Plant Cell Rep. 1997;16:435–438. doi: 10.1007/BF01146789. [DOI] [PubMed] [Google Scholar]
- Ilgoutz SC, Knittel N, Lin JM, Sterle S, Gayler KR. Transcription of genes for conglutin gamma and a leginsulin-like protein in narrow-leafed lupin. Plant Mol Biol. 1997;34:613–627. doi: 10.1023/a:1005868105651. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Buchert T, Fritz G, Suter M, Weber M, Benda R, Schaller J, Feller U, Schurmann P, Schunemann V et al. Plant adenosine 5′-phospho sulfate reductase is a novel iron-sulfur protein. J Biol Chem. 2001;276:42881–42886. doi: 10.1074/jbc.M107424200. [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- Levy S, Avni D, Hariharan N, Perry R, Meyuhas O. Oligopyrimidine tract at the 5′ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc Natl Acad Sci USA. 1991;88:3319–3323. doi: 10.1073/pnas.88.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau J, Vu BL, Macherel MH, Le Deunff Y. Seed lipoxygenases: occurrence and functions. Seed Sci Res. 2001;11:199–211. [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- Maccarrone M, Melino G, Finazzi-Agro A. Lipoxygenases and their involvement in programmed cell death. Cell Death Differ. 2001;8:776–784. doi: 10.1038/sj.cdd.4400908. [DOI] [PubMed] [Google Scholar]
- Marrs KA, Alfenito MR, Lloyd AM, Walbot V. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature. 1995;375:397–400. doi: 10.1038/375397a0. [DOI] [PubMed] [Google Scholar]
- McConnell J, Barton M. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- McGonigle B, Keeler SJ, Lau S-MC, Koeppe MK, O'Keefe DP. A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiol. 2000;124:1105–1120. doi: 10.1104/pp.124.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer CA, Dinkins RD, Redmond CT, McAllister KP, Tucker DT, Walker DR, Parrott WA, Trick HN, Essig JS, Frantz HM et al. Embryogenic response of multiple soybean. In Vitro Cell Dev Biol. 2001;37:62–67. [Google Scholar]
- Nadeau JA, Zhang XS, Li J, O'Neill SD. Ovule development: identification of stage-specific and tissue-specific cDNAs. Plant Cell. 1996;8:213–239. doi: 10.1105/tpc.8.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panitz R, Manteuffel R, Wobus U. Tobacco embryogenesis: storage-protein-accumulating cells of embryo, suspensor, and endosperm are able to undergo cytokinesis. Protoplasma. 1999;207:31–42. [Google Scholar]
- Parrizas M, Saltiel AR, LeRoith D. Insulin-like growth factor 1 inhibits apoptosis using the phosphatidylinositol 3′-kinase and mitogen-activated protein kinase pathways. J Biol Chem. 1997;272:154–161. doi: 10.1074/jbc.272.1.154. [DOI] [PubMed] [Google Scholar]
- Perez-Amador MA, Lidder P, Johnson MA, Landgraf J, Wisman E, Green PJ. New molecular phenotypes in the dst mutants of Arabidopsis revealed by DNA microarray analysis. Plant Cell. 2001;13:2703–2717. doi: 10.1105/tpc.010295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer W, Hoftberger M. Oxidative burst in Chenopodium rubrum suspension cells: induction by auxin and osmotic changes. Physiol Plant. 2001;111:144–150. [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J. The TIGR gene indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 2000;28:141–145. doi: 10.1093/nar/28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan-Arjona T, Garcia-Ortiz MV, Ruiz-Rubio M, Ariza RR. cDNA cloning, expression and functional characterization of an Arabidopsis thaliana homologue of the Escherichia coli DNA repair enzyme endonuclease III. Plant Mol Biol. 2000;44:43–52. doi: 10.1023/a:1006429114451. [DOI] [PubMed] [Google Scholar]
- Rudus I, Kepczynska E, Kepczynski J. Regulation of Medicago sativa L. somatic embryogenesis by gibberellins. Plant Growth Regul. 2002;36:91–95. [Google Scholar]
- Salajova T, Salaj J. Somatic embryogenesis and plantlet regeneration from cotyledon explants isolated from emblings and seedlings of hybrid firs. J Plant Physiol. 2001;158:747–755. [Google Scholar]
- Sambrook J, Fritsch EE, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santarem ER, Pelissier B, Finer JJ. Effect of explant orientation, pH, solidifying agent and wounding on initiation of soybean somatic embryos. In Vitro Cell Dev Biol. 1997;33:13–19. [Google Scholar]
- Satoh S, Sturm A, Fujii T, Chrispeels MJ. cDNA cloning of and extracellular dermal glycoprotein of carrot and its expression in response to wounding. Planta. 1992;188:432–438. doi: 10.1007/BF00192811. [DOI] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Kanaya E, Morita EH, Okada K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 1999;13:1079–1088. doi: 10.1101/gad.13.9.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R, Keim P, Vodkin L, Retzel E, Clifton SW, Waterston R, Smoller D, Coryell V, Khanna A, Erpelding J et al. A compilation of soybean ESTs: generation and analysis. Genome. 2002;45:329–338. doi: 10.1139/g01-150. [DOI] [PubMed] [Google Scholar]
- Siegfried K, Eshed Y, Baum S, Otsuga D, Drews G, Bowman J. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- Steller MA, Delgado CH, Zou Z. Insulin-like growth factor II mediates epidermal growth factor-induced mitogenesis in cervical cancer cells. Proc Natl Acad Sci USA. 1995;92:11970–11974. doi: 10.1073/pnas.92.26.11970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Reid JB, Kamiya Y. Gibberellins are required for embryo growth and seed development in pea. Plant J. 1997;12:1329–1338. [Google Scholar]
- Swidzinski JA, Sweetlove LJ, Leaver CJ. A custom microarray analysis of gene expression during programmed cell death in Arabidopsis thaliana. Plant J. 2002;30:431–446. doi: 10.1046/j.1365-313x.2002.01301.x. [DOI] [PubMed] [Google Scholar]
- Tanimoto EY, Rost TL, Comai L. DNA replication-dependent histone H2A mRNA expression in pea root tips. Plant Physiol. 1993;103:1291–1297. doi: 10.1104/pp.103.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenhaken R, Levine A, Brisson LF, Dixon RA, Lamb C. Function of the oxidative burst in hypersensitive disease resistance. Proc Natl Acad Sci USA. 1995;92:4158–4163. doi: 10.1073/pnas.92.10.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- Ursic D, Sedbrook JC, Himmel KL, Culbertson MR. The essential yeast Tcp1 protein affects actin and microtubules. Mol Biol Cell. 1994;5:1065–1080. doi: 10.1091/mbc.5.10.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucich VA, Gasser CS. Novel structure of a high molecular weight FK506 binding protein from Arabidopsis thaliana. Mol Gen Genet. 1996;252:510–517. doi: 10.1007/BF02172397. [DOI] [PubMed] [Google Scholar]
- Walling L, Drews GN, Goldberg RB. Transcriptional and post-translational regulation of soybean seed protein mRNA levels. Proc Natl Acad Sci USA. 1986;83:2123–2127. doi: 10.1073/pnas.83.7.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Barbashov SF, Komatsu S, Hemmings AM, Miyagi M, Tsunasawa S, Hirano H. A peptide that stimulates phosphorylation of the plant insulin-binding protein: isolation, primary structure and cDNA cloning. Eur J Biochem. 1994;224:167–172. doi: 10.1111/j.1432-1033.1994.tb20008.x. [DOI] [PubMed] [Google Scholar]
- Wojtaszek P. Oxidative burst: an early plant response to pathogen infection. Biochem J. 1997;322:681–692. doi: 10.1042/bj3220681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Spitz MR, Lei L, Mills GB, Wu X. A single nucleotide polymorphism in the matrix metalloproteinase-1 promoter enhances lung cancer susceptibility. Cancer Res. 2001;61:7825–7829. [PubMed] [Google Scholar]
- Zimmerman JL. Somatic embryogenesis: a model for early development in higher plants. Plant Cell. 1993;5:1411–1423. doi: 10.1105/tpc.5.10.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.