Abstract
Single amino acid substitutions at either of two crucial positions in acetolactate synthase (ALS) result in a chlorsulfuron-insensitive form of this enzyme and, as a consequence, a herbicide-resistant phenotype. Here, we describe the successful in vivo targeting of endogenous tobacco (Nicotiana tabacum) ALS genes using chimeric RNA/DNA and all-DNA oligonucleotides at two different locations. Similar number of conversion events with two different chimeras indicates the absence of restricting influence of genomic target sequence on the gene repair in tobacco. Chlorsulfuron-resistant plants were regenerated from calli after mesophyll protoplast electroporation or leaf tissue particle bombardment with these specifically constructed chimeras. Sequence analysis and enzyme assays proved the resulting alterations to ALS at both DNA and protein levels. Furthermore, foliar application of chlorsulfuron confirmed the development of resistant phenotypes. Lines with proline-196-alanine, threonine, glutamine, or serine substitutions or with tryptophan-573-leucine substitutions were highly resistant at both cellular and whole plant levels, whereas lines with proline-196-leucine substitutions were less resistant. The stability of these modifications was demonstrated by the continuous growth of calli on chlorsulfuron-containing medium and by the transmission of herbicide resistance to progeny in a Mendelian manner. Ability of haploid state to promote chimera-mediated conversions is discussed.
The glut of sequence information now available for many organisms demands the application of reliable reverse genetics techniques to associate genes with their functions. Several reverse genetics strategies have been developed and are currently used in plants with different degrees of success. Some of them, like T-DNA or transposon insertional mutagenesis, fast neutron deletion mutagenesis, and the targeting of induced local lesions in genomes, are based on random processes (integration or mutation) and the subsequent application of various methods to identify desirable mutants. In many non-plant model species, homologous recombination is used to target genes for mutagenesis. Although gene targeting has been achieved in plants (for ectopic and endogenous genes), it is still far from routine. Low frequency of homologous recombination and inaccurate integration are basic barriers that must be overcome to make gene targeting work in higher plants (Morton and Hooykaas, 1995; Mengiste and Paszkowski, 1999).
A new strategy, chimera-directed gene alteration, was developed to correct or induce site-specific point mutations in mammalian cells and has been applied recently to animals and yeast (Yoon et al., 1996; Alexeev et al., 2000; Bartlett et al., 2000). This gene targeting strategy is based on the action of chimeric RNA/DNA oligonucleotides (chimeras; Yoon et al., 1996). Chimeric oligonucleotides are self-complementary and produce double-hairpin structures. In this duplex conformation, two complementary strands can be distinguished: a chimeric strand consisting of an interposed DNA fragment (5 bp) and two stretches of RNA (10 bp) flanking this region, and another strand that is composed only of DNA. The specific structure of this vector (two thymidine hairpins, a “GC” clamp at the 3′ end, and 2′-O-methylated RNA residues) makes it stable within cells. To target a gene, a chimera must be identical to the gene with the exception of a single nucleotide. This produces a mismatched base that is presumably recognized by endogenous repair machinery. The precise mechanism of this conversion is unknown, but it is believed that it is based on a two-step process: homologous pairing that results in the formation of a double d-loop structure and subsequent endogenous repair activity. From recent studies, it appears that the chimeric strand provides stability to the intermediate joint molecule, whereas the DNA strand is used by endogenous repair machinery as a template for correction (Gamper et al., 2000a). The possibility of using a chimeric oligonucleotide to promote site-specific alterations was shown for the first time in episomal DNA (Yoon et al., 1996). Later, it was successfully targeted to chromosomal sequence in mammalian cells and animal models (Kren et al., 1997, 1999; Alexeev and Yoon, 1998; Alexeev et al., 2000; Bartlett et al., 2000). These investigations in mammalian cells revealed several findings about the chimera strategy: (a) the efficiency is high and exceeds that found in experiments with homologous recombination, (b) it provides a high specificity of nucleotide conversion or nucleotide incorporation, and (c) corresponding DNA/DNA duplexes remain inactive under the same conditions.
In the plant kingdom, the production of chimera-mediated point and frame shift mutations in genomic (acetolactate synthase [ALS] gene) and in artificial (nonfunctional green fluorescent protein gene) targets, respectively, was described in somatic cells of tobacco (Nicotiana tabacum) and maize (Zea mays; Beetham et al., 1999; Zhu et al., 1999). However, in these studies, whole plants were regenerated only from maize callus. Segregation of herbicide-resistant and nonresistant phenotypes exhibited the expected Mendelian ratios (Zhu et al., 2000), which indicated that chimera-dependent changes could be stably transmitted through meiosis as well as mitosis. Unexpectedly, the application of chimera-mediated gene repair techniques in plants results in low efficiencies in comparison with animal cells. The appearance of semitargeted mutations as a shift of conversion from the expected nucleotide in the 5′ position or mutations other than those intended in the expected nucleotide also occur in plants. Studies in cell-free extract system by Rice et al. (2000) confirmed these peculiarities of chimera-dependent conversion in plants.
ALS catalyzes the initial step common to the biosynthesis of the branched chain amino acids Leu, Ile, and Val. Sulfonylureas are one class of herbicides that specifically inhibit this enzyme. In resistant lines, the herbicide insensitive form of ALS is present (Chaleff and Mauvais, 1984). The allotetraploid species tobacco has two genetically unlinked loci, SuRA and SuRB, for ALS. Mutations in either SuRA (Pro-196-Glu and Pro-196-Thr) or SuRB (Pro-196-Ser) locus result in single amino acid replacements at position 196 and a herbicide-resistant phenotype (Lee et al., 1988; Harms et al., 1992; Beetham et al., 1999). It was also shown that the S4-Hra mutant of tobacco bearing two linked mutations within locus SuRB had amino acid substitutions at positions 196 (Pro-196-Ala) and 573 (Trp-573-Leu), and it was more resistant to chlorsulfuron. Genetic linkage between these mutations did not allow the recovery of the independent contribution of each mutation for these highly resistant phenotypes (Creason and Chaleff, 1988). Lee et al. (1990) were able to target the wild-type ALS gene of tobacco via homologous recombination. To produce an amino acid substitution from Trp to Leu that would render a chlorsulfuron-resistant phenotype, a targeting vector carrying a 3′ end fragment of a mutant gene from S4-Hra was used. Although targeting was achieved, it was accompanied by random integration of the targeting vector into the genome, and the phenotype conferred by Hra mutation remained unknown. Interestingly, the same type of amino acid change (Trp to Leu) in the conserved region near the C terminus of ALS (as a result of 1-bp substitution, G to T) resulted in sulfonylurea resistance in a Xanthium sp. and Brassica napus (Bernasconi et al., 1995; Hattori et al., 1995). Moreover, in cocklebur (Xanthium), all possible mutations affecting the Trp at this position were investigated using site-directed mutagenesis, and only Trp-Leu substitution yielded an active, herbicide-insensitive form of ALS (Bernasconi et al., 1995).
In our study, we used an oligonucleotide-mediated strategy to create single point mutations at different positions within ALS genomic sequences of tobacco. Sequence analysis confirmed that application of oligonucleotides with various targeting sequences resulted in the production of predicted alterations. The same frequency of chimera-mediated conversions at different target sites suggests the absence of influence of genomic target sequence on the gene repair in tobacco. The assay of ALS activity in the leaves of resistant lines in the presence of chlorsulfuron demonstrated the appearance of an herbicide-insensitive form of the enzyme. Our data also suggest a correlation between the appearance of nonspecific chimera-dependent alterations and the ability of selection systems applied to detect them.
RESULTS
Production of Site-Specific Modifications to the ALS Gene by Chimera-Mediated Mutagenesis
In the allotetraploid species tobacco, there are two highly conserved ALS genes (SuRA and SuRB) that are expressed in all tissues (Keeler et al., 1993). Two chimeric RNA/DNA oligonucleotides were designed to obtain separate targeted single-nucleotide conversion at two different positions within either of these genes (Fig. 1, A and B). The chimeras used in our study had the same structure as those that directed repair mutations in animals (Kren et al., 1999; Alexeev et al., 2000; Bartlett et al., 2000). The targeting region of these chimeras consisted of a 25-bp region homologous to the target DNA with the exception of 1 centralized bp. ChALS-588 was designed to produce single nucleotide substitution from C to A at nucleotide position 588, and ChALS-1719 was designed to convert G to T at nucleotide position 1,719. These nucleotide conversions led to amino acid changes Pro-Gln at position 196 and Trp-Leu at 573, respectively. Both predicted amino acid substitutions, Pro-196-Glu and Trp-573-Leu, must result in a herbicide-insensitive form of ALS. Therefore, the desired conversion of the targeted base could be recovered by growing transformed cell colonies on chlorsulfuron-containing medium (Fig. 2A). Chimeric RNA/DNA oligonucleotides (ChALS-588 and ChALS-1719) were introduced into tobacco cells independently. In each experiment, several types of controls were used: untreated cells or protoplasts, treated without oligonucleotides, treated with nonspecific chimeric oligonucleotide, and treated with oligonucleotide composed of DNA only (ChALS-587). Electroporation of protoplasts and bombardment of cell colonies were used to deliver the chimeric oligonucleotide ChALS-1719. Four colonies were recovered on selection medium with 140 nm chlorsulfuron after particle bombardment of 3 × 106 cells and five colonies after electroporation of 6 × 106 protoplasts. Then, we examined the capacity of the haploid state to promote chimera-mediated conversions. For this purpose, haploid lines of tobacco were generated by anther culture on double-layer medium H (Nitsch and Nitsch, 1969) with activated charcoal. The reduction in chromosome number was verified by caryological analysis (data not shown). The chimeric oligonucleotide ChALS-588 was delivered into protoplasts by electroporation. After treatment of 7 × 106 protoplasts of wild type and 6 × 106 protoplasts of haploid lines six and four, respectively, green-resistant colonies were found on medium containing 56 nm chlorsulfuron. In all control experiments (except cells treated with DNA oligonucleotide), 39 × 106 cells were used and two resistant colonies (Mu-1 and Mu-2) were obtained (but one colony was unable to grow when the selective agent in the medium was increased up to 140 nm chlorsulfuron). In experiments with a DNA-only oligonucleotide (ChALS-587), a total of 15 × 106 cells were treated, and two resistant colonies were recovered (rchl-17 and rchl-42).
Figure 1.
Sequences of targeting oligonucleotides and strategy of gene correction by chimeric oligonucleotides. A, Oligonucleotide sequences. B, Schematic presentation of chimera-mediated gene conversion. The endogenous DNA repair system mediates conversion of G to T at position 1,719, gives rise to substitution of Trp to Leu, and leads to appearance of chlorsulfuron-resistant phenotype. Lowercase letters indicate 2′-O-methyl RNA residues, and uppercase letters indicate DNA bases. The mismatched base is underlined. The chimeric ChALS-588 and the DNA-composed oligonucleotide ChALS-587 are designed to make conversions at the codon for Pro-196, whereas chimera ChALS-1719 is designed to alter the codon for Trp-573. ChALS(−) is a nonspecific chimera.
Figure 2.
Resistance to chlorsulfuron at cellular and whole plant levels. A, Green-resistant colony on selective medium. B, Plants of wild-type (WT) and resistant (R) line rchl-6.6 growing on medium containing 560 nm herbicide. C and D, Response of resistant and wild-type plants to foliar application of chlorsulfuron.
Analysis of Targeted Sequences from the Resistant Lines
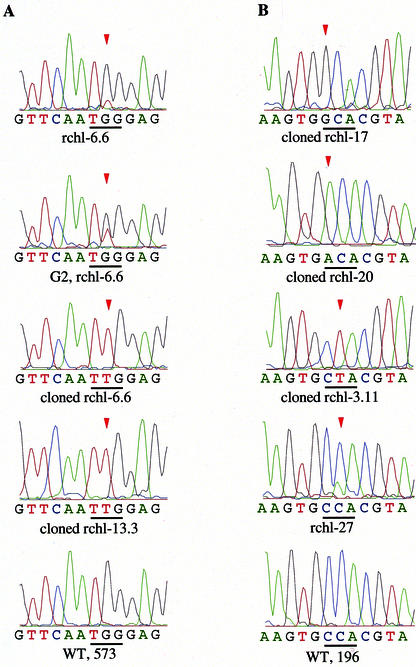
To confirm that resistant lines appeared due to chimera-dependent specific conversion, total DNA was isolated, and fragments including the target site near the 5′ or 3′ ends of the ALS gene were amplified by PCR. Direct sequencing of these fragments revealed a small new peak at the targeted codon on sequence chromatograms. The PCR products from wild type had unchanged sequence patterns (Fig. 3). The observed weak peak could be explained as artifacts or real conversion events that occurred only in one of several copies of ALS genes. To clarify this, we produced haploid plants from our resistant lines and selected them by growing on chlorsulfuron-containing medium. The idea was to decrease the number of unchanged copies of the ALS gene. Sequence analysis of the targeted region of resistant haploid lines revealed the presence of increased signal corresponding to the same nucleotide that was observed on chromatograms in the original T0 plants. Furthermore, PCR products of targeted regions of T0 resistant plants were ligated into the pPCR-Script Amp SK(+) vector and individually sequenced. In all cases, sequence analysis demonstrated the alteration of one nucleotide at the codon corresponding to Pro-196 or Trp-573. Because a change in nucleotides at different positions within the ALS gene could provide chlorsulfuron resistance, we performed sequence analysis of PCR fragments generated from the 5′ and 3′ ends of ALSs from all analyzed resistant lines to ensure that resistance was a result of chimera-mediated conversion. In the case of the chimeric oligonucleotide ChALS-1719, the observed change was limited to a single nucleotide substitution (G to T) at position 1,719; the codon for Pro-196 was unchanged. Application of the chimeric oligonucleotide ChALS-588 resulted in changes to the codon Pro-196 as predicted; no changes were found at the second possible region. Interestingly, in experiments targeting the Trp-573 codon, only predicted base replacements (G→T) were found. However, the second chimeric oligonucleotide (ChALS-588) targeting the Pro-196 codon also resulted in semitargeted conversions. Sequence analysis of two lines (rchl-17 and rchl-42) obtained using DNA-only construct (ChALS-587) resulted in single nucleotide conversions (C→G and C→A, respectively) at predicted positions only (Fig. 3; Table I).
Figure 3.
Confirmation of oligonucleotide-mediated acetolactate syntase gene conversion by sequence analysis. Nucleotide sequences from control wild type (WT) and independent resistant lines rchl-6.6, rchl-17, rchl-20, and rchl-3.11 are shown. G2 rchl-6.6 is sequence of haploid from line rchl-6.6. Plate A, Nucleotide substitutions at the codon for Trp-573. Plate B, Nucleotide substitutions at the codon for Pro-196. The targeted codon is underlined and conversion events are indicated with arrows.
Table I.
Summary of chimera-directed alterations of ALS gene
| Target (Position of Nucleotide) | Line | Predicted Changes
|
Observed Changes
|
||
|---|---|---|---|---|---|
| Nucleotide | Amino acid | Nucleotide | Amino acid | ||
| ALS-588 | rchl-27 | CCA→CAA | Pro-196-Gln | CCA→CAA | Pro-196-Gln |
| rchl-3.11 | CCA→CTA | Pro-196-Leu | |||
| rchl-20.1 | CCA→ACA | Pro-196-Thr | |||
| rchl-5.5 | CCA→TCA | Pro-196-Ser | |||
| ALS-1719 | rchl-6.6a | TGG→TTG | Trp-573-Leu | TGG→TTG | Trp-573-Leu |
| ALS-587 | rchl-17 | ||||
| rchl-4.2 | CCA→GCA | Pro-196-Ala | CCA→GCA | Pro-196-Ala | |
| CCA→ACA | Pro-196-Thr | ||||
All lines have the same substitution.
Resistance on Cellular and Whole-Plant Levels
Callus cells remained chlorsulfuron resistant during several passages, demonstrating stable transmission of the modified gene in somatic cells. To induce organogenesis, resistant colonies were transferred to regeneration medium (Murashige and Skoog, 1962; 1 mg L−1 6-benzyl-aminopurine, and 0.1 mg L−1 α-naphthaleneacetic acid) with 140 nm herbicide. Regenerated plantlets were cultured on Murashige and Skoog medium containing 420 nm chlorsulfuron. Wild type could not grow at such herbicide concentrations, but all resistant lines were able to produce roots and elongate shoots (Fig. 2B). The inhibition of root production was detected only for line rchl-3.11. T0 plants were transferred into the greenhouse, and 3-week-old plants were sprayed with chlorsulfuron. Different concentrations (84, 140, and 280 μm) of herbicide were applied. Two weeks after treatment, wild-type plants were discolored, widely necrotic, and sometimes the whole leaf blade was dry (Fig. 2D). A concentration of 84 μm was sufficient to distinguish resistant and nonresistant phenotypes (Fig. 2C). Resistant lines were significantly different from the wild type in fresh weight. Plants of resistant lines had green leaves, and no necroses were detected except for plants of line rchl-3.11. Some leaves of these plants had small necrotic spots (Table II).
Table II.
Assessment of wild-type and independent resistant lines after herbicide application and segregation of chlorsulfuron resistance in T1 progeny
| Line | Response on Application of 84 μm Chlorsulfuron
|
T1 Progeny Seedlings No. of Plants
|
||||
|---|---|---|---|---|---|---|
| Plant phenotype | Leaf color | Presence of necroses | Fresh weighta | Resistant | Sensitive | |
| g | ||||||
| WT (Pro-196 and Trp-573) | Sensitive | Bleached | +++ | 197.02 ± 8.36 a | 0 | 330 |
| rchl-3.11 (Leu-196) | Susceptible | Green | + | 214.6 ± 12.87 a | 149 | 61 |
| χ2 = 1.835 | ||||||
| rchl-20b (Thr-196) | Resistant | Green | – | 316.25 ± 9.27 c | 400 | 0 |
| rchl-17 (Ala-196) | Resistant | Green | – | 262.02 ± 5.31 b | 113 | 37 |
| χ2 = 0.009 | ||||||
| rchl-5.5 (Ser-196) | Resistant | Green | – | 254.12 ± 6.23 b | 167 | 53 |
| χ2 = 0.097 | ||||||
| rchl-6.6 (Leu-573) | Resistant | Green | – | 242.17 ± 15.92 b | 184 | 68 |
| χ2 = 0.529 | ||||||
| rchl-2.5 (Leu-573) | Resistant | Green | – | 237.02 ± 14.01 b | 175 | 65 |
| χ2 = 0.556 | ||||||
Values presented as a mean ± se from measurements of five independent plants. Within-column means without common letters are significantly different at P < 0.05.
Resistant dihaploid line. +, Indicates that some leaves had small necrotic spots; +++, indicates wide necroses up to dryness of the whole leaf blade. χ2 values were calculated on the basis of expected ratio of three resistant to one sensitive plant.
It is worth noting that during regeneration of resistant colonies that were selected in the experiments with haploid plants, chromosome doubling took place (data not shown). Hence, regenerated dihaploid plants were homozygous for the altered ALS gene that led to higher viability after herbicide treatment (Table II).
Biochemical Analysis
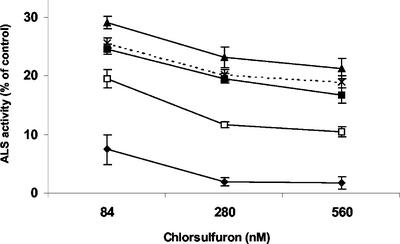
To prove that chimera-mediated conversions resulted in the appearance of a chlorsulfuron-insensitive form of ALS, enzyme activity was assayed in the presence and absence of chlorsulfuron. ALS activity in the leaf extracts from wild type was greatly inhibited under all tested concentrations. All resistant lines demonstrated lower sensitivity of ALS in extracts to the inhibition by chlorsulfuron. Line rchl-3.11 was intermediate in its response to increasing herbicide concentrations in comparison with other lines that demonstrated similarly high degrees of resistance to chlorsulfuron (Fig. 4).
Figure 4.
Inhibition of ALS activity by chlorsulfuron in leaf extracts. ALS activities are presented as percentages of the respective activity in the absence of herbicide. ▴, Line rchl-6.6; *, rchl-5.5; □, rchl-3.11; ▪, rchl-17; ♦, wild type. Each data point is the mean ± se from measurements of activity of five independent plants.
Transmission of the Altered Gene through Meiosis
Seeds from self-fertilized resistant lines (T0 plants) were collected and after germination were transferred to the medium containing 560 nm chlorsulfuron. T1 seedlings segregated for resistant and nonresistant phenotypes (Table II). On average, about one-third of the seedlings were sensitive to the herbicide. These results indicated that chlorsulfuron resistance was inherited as a single dominant Mendelian trait.
DISCUSSION
Homologous recombination-dependent gene targeting in plants is characterized by low frequency of integration and random integration of targeting vector elsewhere in the genome (Morton and Hooykaas, 1995; Mengiste and Paszkowski, 1999). In addition, some examples of mitotic and meiotic instability of targeted genes have been described (Paszkowski et al., 1988; Lee et al., 1990; Schaefer and Zryd, 1997). In contrast, oligonucleotide-directed gene targeting is not accompanied by random construct integration, and converted genes are regulated by their own promoters in their natural genetic background (Kren et al., 1997; Zhu et al., 2000). Altered genes are also stably maintained during mitosis (Alexeev and Yoon, 1998; Beetham et al., 1999; Igoucheva and Yoon, 2000) and transmitted in a Mendelian fashion to subsequent generations (Zhu et al., 2000).
In this study, we report the use of synthetic RNA/DNA oligonucleotides to produce distinct, precise modifications within the ALS gene of tobacco at two different target sites. Two chimeras were designed and delivered in independent experiments into wild-type cells of tobacco. Such an approach should allow us to distinguish between chimera-mediated conversions and random mutations. In each set of experiments in which different chimeric oligonucleotides were used, detection of conversion event at the targeted region and absence of any changes at the second possible site served as additional proof that the herbicide-resistant phenotype appeared as a result of chimera-directed gene targeting. On the other hand, this should answer the question about the possibility of a single mutation within the ALS gene at nucleotide 1,719 by itself to provide chlorsulfuron resistance, and if this is the case, about the degree of this resistance. A previously described double mutant of tobacco S4-Hra bearing two linked mutations in Pro-196-Ala and Trp-573-Leu possessed a highly chlorsulfuron-resistant phenotype (Creason and Chaleff, 1988; Lee et al., 1988). Because one mutation was produced in the genetic background of another, it was not possible to examine the contribution of each of them in the resulting phenotype. Our experiments with chimera ChALS-1719 clearly demonstrated that single amino acid substitution Trp-573-Leu alone can provide a chlorsulfuron-resistant phenotype. The fact that this mutation was generated on the wild-type background allowed its phenotypic expression to be revealed. As follows from results of enzyme activity assays, two resistant lines, rchl-17 (Ala-196 corresponding to S4 mutation) and rchl-6.6 (Leu-573 corresponding to Hra mutation), possessed similar level of ALS activity (Fig. 4) and responded in similar manners to herbicide treatment (Table II). These data suggest that the high level of chlorsulfuron resistance seen for the double mutant S4-Hra probably arose from an additive effect of two mutations.
Application of only DNA-composed oligonucleotide ALS-587 resulted in the generation of alterations at the intended nucleotide (Table I; Fig. 3). However the frequency of changes was low. There are two lines of evidence that a DNA duplex was able to provide repair activity in vivo. First, changes were found in the codon for Pro-196, which was chosen as a target, but a second possible place (codon Trp-573) alteration, which also could result in generation of herbicide resistance, remained unchanged. Second, even alterations in the codon Pro-196 at any nucleotide could lead to production of herbicide-resistant phenotype. However, only the targeted nucleotide was modified, which also indicated oligonucleotide-directed character of conversions. This is consistent with findings in the in vitro system (plant cell-free extracts), where it was shown that all-DNA oligonucleotides mediated targeted repair (Rice et al., 2000). In contrast, such activity has never been observed in experiments with mammalian cells (Yoon et al., 1996; Kren et al., 1997).
Using both chimeric oilgonucleotides resulted in the intended conversions of the ALS gene of tobacco; however, some semitargeted alterations also were detected (Table I). These results are contradictory to those obtained in a study by Beetham et al. (1999), where a chimera-mediated change of the codon for Pro-196 in the ALS gene of tobacco was achieved, but in all cases the only nucleotide located near the 5′ end of the targeted base was changed. In our opinion, detection of only semitargeted modifications in the same model system could reflect peculiarities of the selection system used (see below). The big pool of spontaneous mutations, which was observed in this work, and the inability to regenerate plants could be referred to using tobacco cell suspension culture. Reduction of morphogenic potential and appearance of mutations during maintenance of cell suspension cultures is well documented (Halperin, 1986; Sree Ramulu, 1986). Our results showed that mesophyll protoplast cells are competent in the chimera-mediated conversions. Using mesophyll protoplasts also allowed the reduction of the appearance of spontaneous mutations and the regeneration of plants to be achieved. This made possible further study of the inheritance of the modified gene.
In experiments with maize, a precise targeted correction mediated by a chimera was reported, although additional semitargeted conversions were also found (Zhu et al., 1999). Using a plant cell-free extract system, the diminished fidelity of chimera-directed gene repair (incorrect conversion at the predicted nucleotide position as well as a shift in the 5′ side from the targeted nucleotide) was confirmed (Rice et al., 2000). In vitro and in vivo experiments with plant semitargeted conversions were always detected at one position in 5′ direction to the targeted nucleotide. The shift of conversion in 3′ side was never observed. Because formation of a complement-stabilized D-loop is a key step in chimera-mediated gene repair, the appearance of these non-correct mutations could be explained by the existence of alternative pathways (high-fidelity repair and different mutagenic pathways) for processing this joint molecule (Gamper et al., 2000a). The correlation between appearance of unspecific base alterations and nonoptimal modifications of the backbone of the chimera (Gamper et al., 2000a) or single-stranded oligonucleotides (Gamper et al., 2000c) also supports this assumption. Interestingly, the frequency of conversion events for chimera-mediated repair in plants was 3 orders of magnitude less than was observed for mammalian cells (Zhu et al., 1999). In combination with observations that a DNA duplex can provide targeted gene conversions and that chimera-dependent nonspecific mutations can be generated, this finding indicates the possible existence of different repair pathways in animals and plants.
In our experiments, a second nucleotide at the codon corresponding to Pro-196 or Trp-573 was chosen as a target for chimera-mediated conversion. Such an approach should allow us to detect both types of previously revealed nonspecific alterations as a shift of conversion and incorrect conversion if those would take place and distinguish oligonucleotide-dependent alterations and random mutations. Surprisingly, chimera ChALS-1719 led to only targeted conversion, whereas application of ChALS-588 resulted in both targeted and semitargeted alterations (Table I; Fig. 3). In our opinion, this can be explained by the selection system applied. Recovery of conversion events in a big pool of treated cells was based on expression of the altered form of an ALS gene, the product of which renders tobacco cells resistant to the herbicide. Several single amino acids substitutions at position 196 of ALS, Pro-196-Glu, Pro-196-Ser, Pro-196-Ala, and Pro-196-Thr (Lee et al., 1988; Harms et al., 1992; Beetham et al., 1999) and Pro-196-Leu (this study, Table I) led to the appearance of chlorsulfuron-resistant phenotypes. Thus, any chimera-dependent nucleotide changes at this codon (no matter if the first or second nucleotide was changed) could be recovered. In contrast, within other conserved sites of the ALS protein involved in the herbicide binding, only one specific amino acid substitution, Trp-573-Leu, led to sulfonylurea resistance (Creason and Chaleff, 1988; Bernasconi et al., 1995; Hattori at al., 1995). Therefore, even though other types of alteration took place in the experiments with ChALS-1719, these would not have been recovered because they were not accompanied by chlorsulfuron resistance. The results obtained with all-DNA construct (ChALS-587) also support such conclusions. A shift of repair conversion in the 5′ side from the predicted nucleotide was described (Beetham et al., 1999; Zhu et al., 1999; Rice et al., 2000). Thus, theoretically we could expect such an effect, but only conversions at the targeted nucleotide 587 were obtained (Table I). The substitution of C for G or A can be understood because any nucleotide substitutions for the first nucleotide of the Pro-196 codon could be identified using our selection system (which is based on recovery of chlorsulfuron-resistant cells). In contrast, even if the third nucleotide (which is at a 5′ side of the targeted nucleotide) of the adjacent codon of Val-195 is changed to another nucleotide, it would result in a silent mutation, and such an event would remain undetectable because it would not render resistance to the herbicide.
In previous studies, the appearance of chimera-directed conversions were proved on the DNA level by RFLP and sequence analyses. Together with the latter, we also used enzyme activity assay to verify conversions at the protein level. Variable levels of ALS resistance to chlorsulfuron among different mutant lines were revealed (Table II; Fig. 4). Thus, the line rchl-3.11 containing substitution Pro-196-Leu had approximately 2-fold less resistance to chlorsulfuron than other lines. This was accompanied by stronger inhibition of callus, seedling growth, and delay of root production on selective medium. The observed differences suggest that different types of amino acid substitution confer unequal levels of chlorsulfuron resistance, and, under strong selective conditions, cells with defined type of alterations could not survive. This finding explains why in the study of Beetham et al. (1999), after application of a chimeraplast designed to modify codon for Pro-196 (CCA to CTA), only semitargeted alterations were detected. In that report, regeneration of herbicide-resistant plants was not achieved and conversion events were identified as calli that were able to grow under strong chlorsulfuron selection. Our data clearly show that under such conditions, cells bearing Leu (CTA) at the position 196 in the amino acid sequence of ALS are susceptible (Table II; Fig. 4). Thus, the selection system applied in those experiments could be one of the reasons why only conversions adjacent to the targeted nucleotide were observed. Zhu et al. (1999) demonstrated that chemical selection influenced the frequency of chimera-mediated alterations. In maize, the frequency of green fluorescent protein-expressing cells that appeared because of conversion events was higher in comparison with experiments where chemical selection was employed. These data and our results demonstrate that the selection system applied may influence the frequency of chimera-mediated targeting events and the ability to detect semitargeted chimera-dependent conversions.
Studies with yeast, bacteria, mammalian, and plant cells did not reveal obvious restriction to DNA sequences that can be targeted by chimeric oligonucleotides (Graham and Dickson, 2002; Kren and Steer, 2002, and refs. therein). However, when the APOAI gene in human (Homo sapiens) HepG2 cells was targeted at two locations, the conversions were obtained only at one target site (apoAIParis). A limited correction at second site (apoAIMilano) was achieved in CHO cells. This finding pointed out the possible limitation of chimera-mediated correction by differences between the two target sequences (Graham et al., 2001). In our experiments, application of ALS-588 and ALS-1769 chimeraplasts, mediating conversion at two different sites inside the ALS gene, resulted in comparable frequency of conversion events. Together with the fact that our chimeras had the equal purity and the same level of GC content, this suggests that the influence of genomic target sequence on the gene repair in tobacco was not detected.
Gene targeting of plants using homologous recombination is currently limited by low efficiency and inaccurate integration. Only moss (Physcomitrella patens) demonstrates a high rate of homologous recombination. It has been speculated that the efficiency of homologous integration might correlate with the haploid state (Schaefer and Zryd, 1997). The mechanisms of chimera-directed gene repair and homologous recombination are different (Gamper et al., 2000a; Chen et al., 2001). However, experiments with chicken (Gallus gallus) B cells and embryonic mouse (Mus musculus) fibroblasts lacking an active p53 protein in which high frequency of homologous recombination was correlated with high frequency of chimera-mediated conversion demonstrated that cell recombination activity is important for chimera-dependent repair (Igoucheva et al., 1999; Alexeev and Yoon, 2000). The observation that the RecA protein can catalyze strand exchange between chimera and DNA duplex also suggests that recombination is a necessary step in chimera-dependent correction (Gamper et al., 2000a, 2000b). In view of these results, we assessed the role of the haploid state in increasing frequency of chimera-mediated conversion. For this purpose, haploid plants of tobacco were produced, and chimera ChALS-588 was electroporated into protoplasts. However, the number of recovered chlorsulfuron-resistant colonies did not differ significantly from those derived from wild-type protoplasts (see results). This observation is consistent with studies on the haploid green alga Volvox carteri and haploids of tobacco in which it has been shown that a haploid nature per se was not enough for successful targeting of arylsulfatase gene (Hallmann et al., 1997) or disruption of the nitrate reductase gene (Lebel, 1994). Although our results suggest that the haploid state by itself could not elevate the frequency of targeted conversion, using haploids in chimera-targeting experiments provides some new insights. First, it gives opportunity to operate with recessive traits, and second, it makes possible one-step production of homozygous plants with respect to the modified gene.
CONCLUSION
In summary, we produced chlorsulfuron-resistant lines of tobacco by modifying an endogenous ALS gene at different positions using in vivo site-specific oligonucleotide-mediated conversion. The limitation of chimera-mediated gene conversion by differences in genomic target sequences was not observed. Using this technique allowed the generation of plants with separate new point mutations causing amino acid substitutions Pro-196-Leu and Trp-573-Leu, which are characterized by different phenotypic and biochemical responses to the application of chlorsulfuron. The selection system applied may influence the capacity to detect different types of chimera-mediated alterations. Stable transmission of chlorsulfuron resistance in several generations of somatic cells of tobacco and Mendelian inheritance in T1 progeny verified the permanent character of chimeroplast and all-DNA oligonucleotide-dependent conversions.
The low frequency of observed alterations will still discourage wide application of this strategy. However, we hope that further elucidation of mechanism of oligonucleotide-mediated targeted gene conversion would help to improve the design of structure of RNA/DNA or single-stranded oligonucleotides that would result in increase of frequency. This would allow the expansion of the applicability of this technique as a powerful tool for the modification or disruption of the genes to create desirable phenotypes or knockout mutants.
MATERIALS AND METHODS
Design of Targeting Oligonucleotides
The oligonucleotide sequences (Interactiva, Ulm, Germany) are shown in Figure 1. They were designed to target the ALS gene of tobacco (Nicotiana tabacum) at the desired position and, with the exception of ChALS(−), shared homology to this gene. Chimera ChALS-1719 was designed to create a single G→T substitution at nucleotide position 1,719, and ChALS-588 was designed to create an A→C conversion at position 588. Oligonucleotide ChALS-587 was constructed to produce a single C→G conversion at position 587 and was composed of DNA residues only. The ChALS(−) chimeric oligonucleotide had no homology to the ALS sequence and served as a nonspecific control.
Plant Material and Transformation Procedure
Axenic shoots of tobacco cv Samsun were cultivated on Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 20 g L−1 Suc at 25°C, with illumination of 250 μmol m−2s−1 and a 16-h photoperiod. Mesophyll protoplasts were isolated using enzyme medium containing 1% (w/v) Cellulase R10 (Sigma, St. Louis) and 0.5% (w/v) Driselase (Fluka) in a solution of 0.5 m Suc and 5 mm CaCl2. After overnight incubation of the aseptic leaves in this mixture at 25°C, the suspension was diluted by 0.5 m Suc, passed through nylon mesh (100 μm), and centrifuged at 50g for 5 min. Floating protoplasts were collected and washed twice with W5 solution (Menzel and Wolfe, 1984). Isolated protoplasts (5 × 105) were electroporated with the appropriate oligonucleotide (0.5 μg) using a Bio-Rad Gene Pulser II (Bio-Rad Laboratories, Hercules, CA) in medium M (0.3 mL) containing 8 mm HEPES, 4 mm CaCl2, 70 mm KCl, and 0.4 m mannitol (pH 7.2). Electroporation conditions were as follows: voltage, 250 V; capacitor, 240 μF; and distance between electrodes, 4 mm. Protoplasts were cultivated in dark conditions at 25°C in 10 mL of K3NM medium (Nagy and Maliga, 1976) containing 0.4 m mannitol as an osmotic stabilizer. After 7 d, they were transferred to light conditions. Selection was started in liquid culture 2 weeks after protoplast isolation. Then, cell colonies were transferred to solidified selective medium with 56 and 140 nm chlorsulfuron for ChALS-588 and ChALS-1719, respectively. In particle bombardment experiments, protoplasts were cultured as described above and colonies about 1 to 2 mm in diameter were used for bombardment. Oligonucleotides (0.5 μg) were precipitated onto gold particles (1 μm) and delivered to microcalli using a Bio-Rad PDS-1000 He device as described previously (Klein et al., 1988). A helium pressure of 1,100 psi was used to accelerate the particles, and the vacuum in the chamber was 28 inches of Hg. After bombardment, colonies were plated on Murashige and Skoog medium with 0.2 m mannitol and were transferred in 2 or 3 d onto selective medium with 140 nm chlorsulfuron and without mannitol. After 3 to 4 weeks of culture, resistant colonies were transferred to Murashige and Skoog regeneration medium containing 1 mg L−1 6-benzyl-aminopurine and 0.1 mg L−1 α-naphthaleneacetic acid.
Haploid Plant Production and Caryotype Analysis
Anthers of tobacco were cultured on H-medium (Nitsch and Nitsch, 1969) using a double-layer method (Johansson et al., 1982). Root tip cells from the aseptically grown, haploid plants produced using this method were caryotyped as described previously (Kochevenko et al., 1996).
PCR Amplification and Sequencing Analysis
PCR was performed with Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA) to ensure high fidelity. Reactions were carried out in 50-μL volumes containing 10 mm KCl, 20 mm Tris-HCl (pH 8.8), 2 mm MgSO4, 0.1% (v/v) Triton X-100, 200 μm of each dNTP, 250 ng of each primer, and 100 ng of genomic DNA. After an initial denaturation step at 95°C for 5 min, the reaction mixtures were subjected to 35 amplification cycles of 95°C for 1 min, 65°C for 1.3 min, and 72°C for 2 min with subsequent incubations for 10 min at 72°C. The sequences of the primers used were (5′ to 3′): 1fwd, GGGTTACGCACGCGCCACCGG; 1rev, GGCTGATCCCAGTCAGGTATC; 2fwd, CACCAGATGTGGGCTGCTCAA; and 2rev, GCAGCAGGTACGCCACAAGCC. Amplification products were separated by electrophoresis on 1% (w/v) agarose gels, stained with ethidium bromide, and visualized under UV light. The amplified fragments with expected size were recovered and sequenced directly or subcloned into the pPCR-Script Amp SK(+) vector (Stratagene) for subsequent DNA sequence analysis. DNA sequencing was done by AGOWA GmbH (Berlin).
Assay of ALS Activity
Plants were grown in a greenhouse at 26°C, with illumination of 250 μmol m−2s−1, and under a 16-h photoperiod. Leaf samples were harvested from 4-week-old plants and immediately frozen. One gram of leaf material was homogenized in 8 mL of extraction buffer containing 100 mm potassium phosphate buffer (pH 7.5), 0.5 mm MgCl2, 1 mm sodium pyruvate, 0.5 mm thiamine pyrophosphate, 10 μm FAD, 10% (v/v) glycerol, and 1% (w/v) polyvinylpolypyrrolidone (PVPP). Extracts were prepared and ALS activity was assayed as described by Chaleff and Mauvais (1984). Inhibition of ALS activity was measured in the presence of 84, 280, and 560 nm chlorsulfuron. The protein content in the same extracts was measured by the method of Bradford (1976).
Growth Test for Herbicide Resistance
Herbicide-resistant phenotypes were studied under greenhouse conditions after foliar application of chlorsulfuron. Three-week-old plants were sprayed with a solution (10 mL per plant) containing 0.2% (v/v) Tween 20, 10% (v/v) acetone, and the herbicide (84, 140, or 280 μm of chlorsulfuron). Plant fresh weight was determined 2 weeks later.
Segregation Analysis
Regenerants were self-fertilized in the greenhouse. Obtained seeds were surface sterilized and after germination, seedlings were transferred to Murashige and Skoog medium containing 560 nm chlorsulfuron. Plants showing root and shoot growth were scored as resistant.
ACKNOWLEDGMENTS
We thank Anna Lytovchenko and Megan McKenzie for proofreading and suggestions regarding the manuscript.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.016857.
LITERATURE CITED
- Alexeev V, Igoucheva O, Domashenko A, Cotsarelis G, Yoon K. Localized in vivo genotypic and phenotypic correction of the albino mutation in skin by RNA-DNA oligonucleotide. Nat Biotechnol. 2000;18:43–47. doi: 10.1038/71901. [DOI] [PubMed] [Google Scholar]
- Alexeev V, Yoon K. Stable and inheritable changes in genotype and phenotype of albino melanocytes induced by an RNA-DNA oligonucleotide. Nat Biotechnol. 1998;16:1343–1346. doi: 10.1038/4322. [DOI] [PubMed] [Google Scholar]
- Alexeev V, Yoon K. Gene correction by RNA-DNA oligonucleotides. Pigment Cell Res. 2000;13:72–79. doi: 10.1034/j.1600-0749.2000.130205.x. [DOI] [PubMed] [Google Scholar]
- Bartlett RJ, Stockinger S, Denis MM, Bartlett WT, Inverardi L, Le TT, Man NT, Morris GE, Bogan DJ, Metcalf-Bogan J et al. In vivo targeted repair of a point mutation in the canine dystrophin gene by a chimeric RNA/DNA oligonucleotide. Nat Biotechnol. 2000;18:615–622. doi: 10.1038/76448. [DOI] [PubMed] [Google Scholar]
- Beetham PR, Kipp PB, Sawycky XL, Arntzen CJ, May GD. A tool for functional plant genomics: chimeric RNA/DNA oligonucleotides cause in vivo gene-specific mutations. Proc Natl Acad Sci USA. 1999;96:8774–8778. doi: 10.1073/pnas.96.15.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi P, Woodworth AR, Rosen BA, Subramanian MV, Siehl DL. A naturally-occurring point mutation confers broad range tolerance to herbicides that target acetolactate synthase. J Biol Chem. 1995;270:17381–17385. doi: 10.1074/jbc.270.29.17381. [DOI] [PubMed] [Google Scholar]
- Bradford MM. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chaleff RS, Mauvais CJ. Acetolactate synthase is the site of action of two sulfonylurea herbicides in higher plants. Science. 1984;224:1443–1445. doi: 10.1126/science.224.4656.1443. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Felsheim R, Wong P, Augustin LB, Metz R, Kren BT, Steer CJ. Mitochondria isolated from liver contain the essential factors required for RNA/DNA oligonucleotide-targeted gene repair. Biochem Biophys Res Commun. 2001;285:188–194. doi: 10.1006/bbrc.2001.5156. [DOI] [PubMed] [Google Scholar]
- Creason GL, Chaleff RS. A second mutation enhanced resistance of a tobacco mutant to sulfonylurea herbicides. Theor Appl Genet. 1988;76:177–182. doi: 10.1007/BF00257843. [DOI] [PubMed] [Google Scholar]
- Gamper HB, Cole-Strauss A, Metz R, Parekh H, Kumar R, Kmiec EB. A plausible mechanism for gene correction by chimeric oligonucleotides. Biochemistry. 2000a;39:5808–5816. doi: 10.1021/bi9921891. [DOI] [PubMed] [Google Scholar]
- Gamper HB, Hou YM, Kmiec EB. Evidence for a four-strand exchange catalyzed by the RecA protein. Biochemistry. 2000b;39:15272–15281. doi: 10.1021/bi001704o. [DOI] [PubMed] [Google Scholar]
- Gamper HB, Parekh H, Rice MC, Bruner M, Youkey H, Kmiec EB. The DNA strand of chimeric RNA/DNA oligonucleotides can direct gene repair/conversion activity in mammalian and plant cell-free extracts. Nucleic Acids Res. 2000c;28:4332–4339. doi: 10.1093/nar/28.21.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IR, Dickson G. Gene repair and mutagenesis mediated by chimeric RNA-DNA oligonucleotides: chimeraplasty for gene therapy and conversion of single nucleotide polymorphisms(SNPs) Biochim Biophys Acta. 2002;1587:1–6. doi: 10.1016/s0925-4439(02)00068-6. [DOI] [PubMed] [Google Scholar]
- Graham IR, Manzano A, Tagalakis AD, Mohri Z, Sperber G, Hill V, Beattie S, Schepelmann S, Dickson G, Owen JS. Gene repair validation. Nat Biotechnol. 2001;19:507–508. doi: 10.1038/89209. [DOI] [PubMed] [Google Scholar]
- Hallmann A, Rappel A, Sumper M. Gene replacement by homologous recombination in the multicellular green alga Volvox carteri. Proc Natl Acad Sci USA. 1997;94:7469–7474. doi: 10.1073/pnas.94.14.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin W. Attainment and retention of morphogenetic capacity in vitro. In: Vasil IK, editor. Cell Culture and Somatic Cell Genetics of Plants. Vol. 3. Orlando, FL: Academic Press, Inc.; 1986. pp. 3–37. [Google Scholar]
- Harms CT, Armour SL, Dimaio JJ, Middlesteadt LA, Murray D, Negrotto DV, Thompson-Taylor H, Weymann K, Montoya AL, Shillito RD et al. Herbicide resistance due to amplification of a mutant acetohydroxy acid synthase. Mol Gen Genet. 1992;233:427–435. doi: 10.1007/BF00265440. [DOI] [PubMed] [Google Scholar]
- Hattori J, Brown D, Mourad G, Labbe H, Ouellet T, Sunohara G, Rutledge R, King J, Miki B. An acetohydroxy acid synthase mutant reveals a single-site involved in multiple herbicide resistance. Mol Gen Genet. 1995;246:419–425. doi: 10.1007/BF00290445. [DOI] [PubMed] [Google Scholar]
- Igoucheva O, Peritz AE, Levy D, Yoon K. A sequence-specific gene correction by an RNA-DNA oligonucleotide in mammalian cells characterized by transfection and nuclear extract using a lacZ shuttle system. Gene Ther. 1999;6:1960–1971. doi: 10.1038/sj.gt.3301042. [DOI] [PubMed] [Google Scholar]
- Igoucheva O, Yoon K. Targeted single-base correction by RNA-DNA oligonucleotides. Hum Gene Ther. 2000;11:2307–2312. doi: 10.1089/104303400750035861. [DOI] [PubMed] [Google Scholar]
- Johansson L, Andersson B, Eriksson T. Improvement of anther culture technique activated charcoal bound in agar medium in combination with liquid-medium and elevated CO2 concentration. Physiol Plant. 1982;54:24–30. [Google Scholar]
- Keeler SJ, Sanders P, Smith JK, Mazur BJ. Regulation of tobacco acetolactate synthase gene-expression. Plant Physiol. 1993;102:1009–1018. doi: 10.1104/pp.102.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein TM, Gradziel T, Fromm ME, Sanford JC. Factors influencing gene delivery into Zea mays cells by high velocity microprojectiles. Biotechnology. 1988;6:559–563. [Google Scholar]
- Kochevenko AS, Ratushnyak YI, Gleba YY. Protoplast culture and somaclonal variability of species of series Juglandifolia. Plant Cell Tissue Organ Cult. 1996;44:103–110. [Google Scholar]
- Kren BT, Cole-Strauss A, Kmiec EB, Steer CJ. Targeted nucleotide exchange in the alkaline phosphatase gene of HuH-7 cells mediated by a chimeric RNA/DNA oligonucleotide. Hepatology. 1997;25:1462–1468. doi: 10.1002/hep.510250626. [DOI] [PubMed] [Google Scholar]
- Kren BT, Parashar B, Bandyopadhyay P, Chowdhury NR, Chowdhury JR, Steer CJ. Correction of the UDP-glucuronosyltransferase gene defect in the Gunn rat model of Crigler-Najjar syndrome type I with a chimeric oligonucleotide. Proc Natl Acad Sci USA. 1999;96:10349–10354. doi: 10.1073/pnas.96.18.10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kren BT, Steer CJ. The application of DNA repair vectors to gene therapy. Curr Opin Biotechnol. 2002;13:473–481. doi: 10.1016/s0958-1669(02)00370-1. [DOI] [PubMed] [Google Scholar]
- Lebel E. Gene targeting in plants. Induction of cellular recombination and transposons as targeting vector. PhD thesis. Basel: Basel University; 1994. [Google Scholar]
- Lee KY, Lund P, Lowe K, Dunsmuir P. Homologous recombination in plant cells after Agrobacterium-mediated transformation. Plant Cell. 1990;2:415–425. doi: 10.1105/tpc.2.5.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Townsend J, Tepperman J, Black M, Chui CF, Mazur B, Dunsmuir P, Bedbrook J. The molecular basis of sulfonylurea herbicide resistance in tobacco. EMBO J. 1988;7:1241–1248. doi: 10.1002/j.1460-2075.1988.tb02937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T, Paszkowski J. Prospects for the precise engineering of plant genomes by homologous recombination. Biol Chem. 1999;380:749–758. doi: 10.1515/BC.1999.095. [DOI] [PubMed] [Google Scholar]
- Menzel L, Wolfe K. High frequency of fusion induced in freely suspended protoplast mixture by polyethylene-glycol and dimethylsulfoxide at high pH. Plant Cell Rep. 1984;3:196–198. doi: 10.1007/BF00270199. [DOI] [PubMed] [Google Scholar]
- Morton R, Hooykaas P-JJ. Gene replacement. Mol Breed. 1995;1:123–132. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nagy JI, Maliga P. Callus induction and plant regeneration from mesophyll protoplasts of Nicotiana sylvestris. Z Pflanzenphysiol. 1976;78:453–455. [Google Scholar]
- Nitsch JP, Nitsch C. Haploid plants from pollen grains. Science. 1969;163:85–87. doi: 10.1126/science.163.3862.85. [DOI] [PubMed] [Google Scholar]
- Paszkowski J, Baur M, Bogucki A, Potrykus I. Gene targeting in plants. EMBO J. 1988;7:4021–4026. doi: 10.1002/j.1460-2075.1988.tb03295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice MC, May GD, Kipp PB, Parekh H, Kmiec EB. Genetic repair of mutations in plant cell-free extracts directed by specific chimeric oligonucleotides. Plant Physiol. 2000;123:427–437. doi: 10.1104/pp.123.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer DG, Zryd JP. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- Sree Ramulu K. Case histories of genetic variability in vitro: potato. In: Vasil IK, editor. Cell Culture and Somatic Cell Genetics of Plants. Vol. 3. Orlando, FL: Academic Press, Inc.; 1986. pp. 449–469. [Google Scholar]
- Yoon K, Cole-Strauss A, Kmiec EB. Targeted gene correction of episomal DNA in mammalian cells mediated by a chimeric RNA center dot DNA oligonucleotide. Proc Natl Acad Sci USA. 1996;93:2071–2076. doi: 10.1073/pnas.93.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Mettenburg K, Peterson DJ, Tagliani L, Baszczynski CL. Engineering herbicide-resistant maize using chimeric RNA/DNA oligonucleotides. Nat Biotechnol. 2000;18:555–558. doi: 10.1038/75435. [DOI] [PubMed] [Google Scholar]
- Zhu T, Peterson DJ, Tagliani L, St-Clair G, Baszczynski CL, Bowen B. Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc Natl Acad Sci USA. 1999;96:8768–8773. doi: 10.1073/pnas.96.15.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]