Abstract
The cyanobacterium Synechocystis sp. strain PCC6803 possesses three modes of inorganic carbon (Ci) uptake that are inducible under Ci stress and that dramatically enhance the efficiency of the CO2-concentrating mechanism (CCM). The effects of Ci limitation on the mRNA transcript abundance of these inducible uptake systems and on the physiological expression of the CCM were investigated in detail in this cyanobacterium. Transcript abundance was assessed with semiquantitative and real-time reverse transcriptase-polymerase chain reaction techniques. Cells aerated with CO2-free air for 30 min in the light, but not in the dark, depleted the total [Ci] to near zero levels. Under these conditions, the full physiological expression of the CCM was apparent within 2 h. Transcripts for the three inducible Ci uptake systems, ndhF3, sbtA, and cmpA, showed near-maximal abundance at 15 min under Ci limitation. The transcriptional regulators, cmpR and ndhR, were more moderately expressed, whereas the rbcLXS and ccmK-N operons and ndhF4/ndhD4/chpX and ccaA genes were insensitive to the low-Ci treatment. The combined requirement of low Ci and light for the expression of several CCM-related transcripts was examined using real-time reverse transcriptase-polymerase chain reaction. CmpA, ndhF3, and sbtA were strongly expressed in the light, but not in the dark, under low-Ci conditions. We could find no evidence for induction of these or other CCM-related genes by a high-light treatment under high-CO2 conditions. This provided evidence that high-light stress alone could not trigger the expression of CCM-related transcripts in Synechocystis sp. PCC6803. Potential signals triggering induction of the high-affinity state of the CCM are discussed.
Cyanobacteria possess a CO2-concentrating mechanism (CCM) that is maximally expressed under inorganic carbon (Ci) limitation. A constitutive level of CCM activity is apparent in cyanobacteria grown under high levels of Ci (Price et al., 1998). However, full induction of the CCM occurs when cells are aerated with low levels of CO2 (20–350 ppm CO2). The CCM functions to elevate the intracellular concentration of CO2 in the vicinity of Rubisco to compensate for the poor kinetic efficiency of this enzyme (Kaplan et al., 1980; Price et al., 1998, 2002; Kaplan and Reinhold, 1999). The accumulation of HCO3− against its electrochemical gradient underpins the operation of the CCM. Four systems for active Ci uptake in the model organisms Synechocystis PCC6803 and Synechococcus PCC7942 have recently been described (Price et al., 2002; Shibata et al., 2002b).
In cyanobacteria, NADPH dehydrogenase (NDH-1) complexes are crucial for dark respiration and cyclic electron transport. However, two specialized NDH-1 complexes, both containing several unique subunits, have been implicated in the active uptake of CO2 in Synechocystis and Synechococcus spp. (Klughammer et al., 1999; Shibata et al., 2001; Maeda et al., 2002; Price et al., 2002). Klughammer et al. (1999) first identified the ndhF3-ndhD3-orf427 (chpY) gene cluster in Synechococcus PCC7002 as being necessary for inducible, high-affinity CO2 uptake; (please note that chpY [Maeda et al., 2002] is also known as cupA [Shibata et al., 2001]). The same operon also has been shown to be essential for inducible, high-affinity CO2 uptake in Synechocystis sp. PCC6803 (Ohkawa et al., 2000a, 2000b; Shibata et al., 2001) and Synechococcus sp. PCC7942 (Maeda et al., 2002). A cbbR homolog, designated ndhR, appears to regulate the transcription of the ndhF3-ndhD3-chpY operon under low Ci in Synechocystis PCC6803 (Figge et al., 2001). Recent studies have also identified a constitutive, lower affinity system for CO2 uptake (Ohkawa et al., 2000a, 2000b; Maeda et al., 2002; Shibata et al., 2002a, 2002b). This system appears to employ NdhF4, NdhD4, and ChpY as unique subunits of a specialized NDH-1 complex that displays a high flux rate for CO2 uptake (Maeda et al., 2002). The chpX and chpY genes (CO2 hydration proteins; Maeda et al., 2002) also are known as cupB and cupA, respectively (Shibata et al., 2001); hereafter, we refer to these genes using the chp terminology.
In relation to HCO3− transport, the cmpABCD operon has been shown to encode an ABC-type, high-affinity HCO3− transporter in Synechococcus PCC7942 (Omata et al., 1999). The cmp operon in Synechocystis has also been shown to be low CO2 inducible (Omata et al., 2001), although the transporter is apparently ineffectual (Shibata et al., 2002a). A transcriptional regulator of the LysR family, cmpR, has been shown to regulate the low CO2 responsiveness of this operon in Synechocystis (Omata et al., 2001). In terms of a second mode of HCO3− uptake, a novel HCO3− transporter, encoded by sbtA, has recently been identified in Synechocystis (Shibata et al., 2002a). The sbtA gene was shown to be low CO2 inducible and dependent upon millimolar levels of Na+ for maximal activity (Shibata et al., 2002a).
Surprisingly little work has been targeted at characterizing the coordinate regulation of CCM genes in cyanobacteria. To address this deficiency, the present work was carried out to extend and further characterize the response of Synechocystis PCC6803 to Ci limitation, with special emphasis placed on the time-resolved kinetics of the changes in transcript abundance and physiological responses of Ci transport upon transfer of cells from high- to low-Ci conditions.
RESULTS
Changes in Culture [Ci] during Aeration with CO2-Free Air
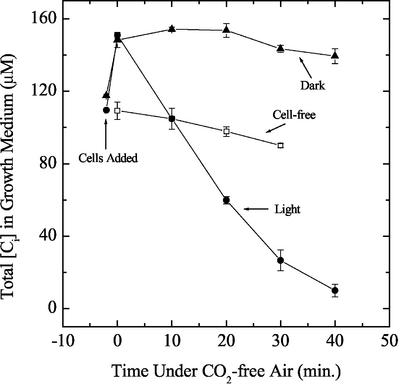
The protocol used for CCM induction in this study was relatively severe in that the cells were rapidly switched from high-CO2 conditions to Ci-deficient conditions by centrifugation and resuspension in a medium low in Ci (110–130 μm Ci). This was followed by aeration with CO2-free air for varying periods of up to 5 h. It should be noted that control cultures were subjected to the same centrifugation-resuspension regime and then returned to media equilibrated with elevated CO2. The cells were kept at 25°C to 30°C during all manipulations. To more fully define the conditions for induction, it was deemed necessary to measure the [Ci] in the extracellular growth medium during exposure of cell cultures to CO2-free air. Typical changes in the extracellular medium in illuminated and darkened cultures aerated with CO2-free air are shown in Figure 1. In the light, Ci in cell cultures was depleted much more quickly than growth medium alone and reached near-zero levels within 40 min of exposure to CO2-free air. In darkened cell cultures, the decline in [Ci] was more gradual. Clearly, cellular photosynthetic activity was necessary for reduction of the Ci concentration to low levels.
Figure 1.
The effect of aeration with CO2-free air on total [Ci] in illuminated (black circles) and darkened (black triangles) cultures of Synechocystis PCC6803. Culture conditions and [Ci] measurement are as described in “Materials and Methods.” For incubations done in the dark, the culture tube was wrapped in aluminum foil. The light intensity used for the “+ Light” treatment was 60 μmol photons m−2 s−1. The mean ± sd of three replicate measurements is shown.
Increases in Affinity for Ci in Photosynthesis during Exposure to CO2-Free Air
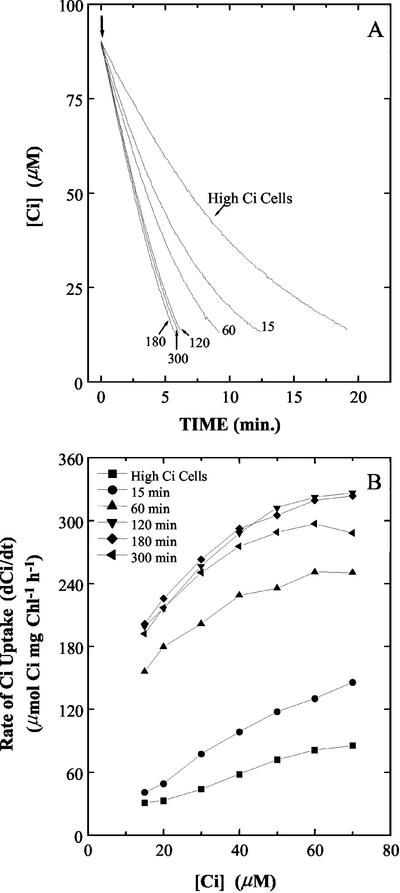
To determine the rate at which cellular affinity for Ci changed, aliquots of cells were harvested after various periods of exposure to CO2-free air in a 5-h time course experiment and then resuspended in the cuvette of the mass spectrometer. Affinity for Ci was determined from drawdown experiments in which exogenously added Ci was depleted from 90 to around 10 μm Ci (Fig. 2). High-Ci cells depleted the Ci in the assay medium in approximately 20 min. Cells exposed to CO2-free air for 15 min and 1 h depleted the Ci in the subsequent assay in approximately 13 and 10 min, respectively (Fig. 2A). Cells aerated with CO2-free air for 2, 3, and 5 h all depleted the Ci in the assay medium in approximately 6 min, indicating that 2 h of this treatment was sufficient for maximal expression of the CCM (Fig. 2A). The rates of Ci uptake (dCi/dt), calculated from the drawdown traces shown in Figure 2A and normalized for chlorophyll a concentration, are shown in Figure 2B.
Figure 2.
A, Photosynthetic depletion of Ci in the cuvette of the mass spectrometer by Synechocystis PCC6803 cells exposed to CO2-free air for 15, 60, 120, 180, and 300 min. Other conditions were as detailed in “Materials and Methods.” The cell suspensions were re-illuminated (↓) and allowed to deplete the Ci in the medium until near the CO2-compensation point. A representative set of traces for three independent experiments is shown. B, Rates of Ci uptake below 70 μm calculated from the Ci drawdown traces plotted in A by estimating the dCi/dt at the indicated external Ci levels.
An Initial Screen of Low-Ci-Responsive Genes
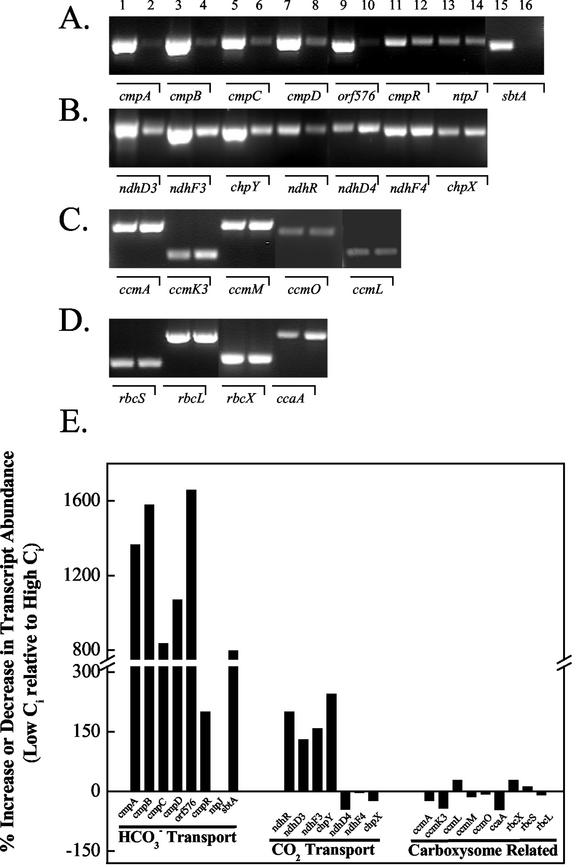
Changes in the expression patterns of several genes known, or suspected, to be important for CCM expression in Synechocystis were investigated using a semiquantitative RT-PCR technique to analyze mRNA transcripts after rapid transfer of high-Ci cells via centrifugation to media bubbled with CO2-free air. One hour of exposure to CO2-free air was chosen as a convenient time point for the analysis because it preceded the full physiological expression of the CCM (Fig. 2). It should be emphasized that the particular method of gene expression analysis employed here was used primarily to provide insight into which genes were the most responsive under the low-Ci induction protocol used. As such, the results are not to be interpreted as being rigorously quantitative. The results of the experiment are shown in Figure 3, A to E.
Figure 3.
Initial screen by semiquantitative reverse transcriptase (RT)-PCR of Ci-responsive genes from cells grown continuously on 3% (v/v) CO2 (even-numbered lanes) or exposed to 1 h of CO2-free air (odd-numbered lanes). RT-PCR conditions were as described in “Materials and Methods” with the exception that 30 cycles of PCR amplification were used. Equal amounts of RNA were used in the first strand synthesis reaction. A, cmpA-D (slr0040-slr0044) operon plus its transcriptional regulator cmpR (sll0030), ntpJ (slr1509) Na+-ATPase subunit J, and sbtA (slr1512)-Na+-dependent HCO3− transporter. B, ndhF3/D3/chpY operon (sll1732-sll1734) plus its transcriptional regulator ndhR (sll1594); ndhD4/F4/chpX (sll0026, sll0027, and slr1302). C, ccmA, ccmK3, ccmLMO (sll0934, slr1838, sll1030-sll1032, slr1032) operon involved in carboxysome assembly and function. D, rbcLXS (slr0009-slr0012) operon plus ccaA (slr1347) carboxysome-localized carbonic anhydrase (CA). E, Plot of the expression profiles from the RT-PCR reactions shown in A to D.
It was readily apparent that within 1 h of aeration with CO2-free air that the transcripts for genes encoding inducible systems of Ci transport (cmpA-D, ndhF3/D3/chpY, and sbtA) were highly up-regulated relative to high-Ci growth conditions (Fig. 3, A and B). A gene, ntpJ, which encodes a protein recently implicated as being necessary for energizing Na+-dependent HCO3− transport in Synechocystis sp. (Shibata et al., 2002a), was not differentially expressed under these conditions nor was the homolog of a gene identified as necessary for HCO3− transport in Synechococcus ictB (data not shown; Bonfil et al., 1998). Both the cmpA-D and the ndhF3/D3/chpY gene clusters appeared to be cotranscribed, suggesting that both clusters may act as operons, as suggested by Omata et al. (2001) and Ohkawa et al. (2000b), respectively. Slr0042 (orf576, Price et al., 1998) was cotranscribed with cmpABCD, indicating its participation as a member of this operon in Synechocystis. This gene is absent from the cmp operon in Synechococcus PCC7942 (Omata et al., 2002).
Interestingly, both of the transcriptional regulators for the cmpA-D and ndhF3/D3/chpY operons, namely cmpR and ndhR, respectively, were up-regulated by about the same amount (Fig. 3, A, B, and E). This is the first report, to our knowledge, of the effect of Ci limitation on the abundance of the cmpR transcript. The expression of the low-affinity CO2 transporter, encoded by the ndhF4/D4 and chpX genes, was essentially unaffected by Ci stress, indicating that these genes were constitutively expressed under these conditions. Somewhat surprisingly, the expression of the rbcLXS and ccmK-N operons, ccmO and ccaA, encoding carboxysomal CA, were relatively insensitive to severe Ci limitation and, as a consequence, these were not investigated any further (Fig. 3, D and E).
Time Course of the Response of Inducible Transcripts under Low-Ci Stress
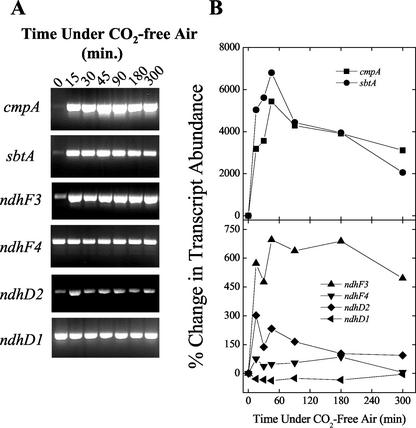
One specific goal was to investigate the kinetics of low-Ci-responsive gene expression in relation to the rapid physiological changes in the affinity during Ci limitation (Fig. 2), particularly the expression profiles of transcripts encoding components of inducible Ci transporters highlighted in Figure 3. Emphasis was placed on obtaining high temporal resolution during the first 90 min of exposure to CO2-free air. The sbtA, cmpA, and ndhF3 transcripts were rapidly induced, accumulating to approximately 75%, 55%, and 80% of the maximum, respectively, in as little as 15 min under Ci limitation (Fig. 4, A and B). The maximum levels of sbtA and cmpA transcripts occurred at 45 min under low-Ci conditions and declined steadily afterward (Fig. 4B). The maximum accumulation of ndhF3 transcript also occurred at 45 min but, in contrast to sbtA and cmpA, the transcript abundance was largely sustained for approximately 3 h of low-Ci stress, after which it also began to decline (Fig. 4B). The pattern of sbtA and cmpA expression and the relative amounts of these transcripts tracked one another quite closely. The maximum relative percentage increase in sbtA and cmpA was approximately 10-fold greater than that of ndhF3. Somewhat unexpectedly, the ndhF4 transcript did show a mild increase in transcript abundance, but this increase was small relative to ndhF3 (Fig. 4B). Interestingly, ndhD2, a gene encoding a subunit of an NDH-1 complex, which possibly functions in respiration and cyclic electron flow around PSI, was also up-regulated under these conditions, although not as strongly as the genes encoding subunits of Ci transporters (Fig. 4B). The ndhD1 gene was taken to be constitutively expressed under these conditions based on its invariance with respect to total RNA (Fig. 4B) and on literature showing no involvement of the gene in CO2 uptake, plus a lack of responsiveness to Ci limitation when assessed by northern analysis (Ohkawa et al., 1998, 2000a). Thus, ndhD1 was included in this analysis as a reference gene against which transcript quantitations were normalized.
Figure 4.
A, Semiquantitative RT-PCR amplification of RNA extracted from cells during induction of the CCM. Amplification conditions and quantitation of band intensities were as described in “Materials and Methods.” No bands were detected in the minus RT controls (data not shown). B, Expression profiles from the RT-PCR reactions shown in A. The reference gene, ndhD1, was used to correct for errors in the estimation of RNA concentrations and for differences in RT reaction efficiency.
Effect of Treatment with CO2-Free Air on Photosynthesis, CCM Induction, and Growth
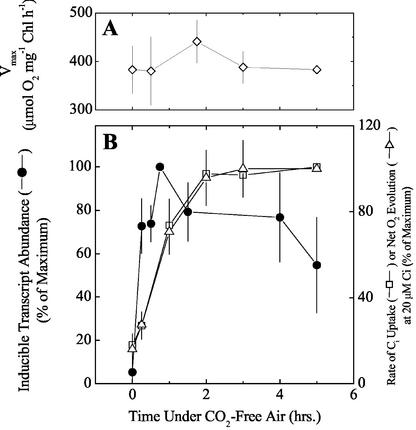
The response to low-CO2 stress of various physiological parameters related to photosynthesis, CCM induction, and growth are shown together in Figure 5. The maximum rate of photosynthesis, measured as net O2 evolution, was unaffected over 5 h of aeration with CO2-free air (Fig. 5A). This indicates that the rates of photo-inhibition and repair were balanced under severe Ci limitation. It should be noted that under severe Ci stress, there was an approximate 10% decrease in cell density over 5 or even 18 h of treatment (results not shown); however, this does not preclude expression of Ci-responsive genes. The results in Figure 4 showed that the responses of cmpA, sbtA, and ndhF3 transcripts were qualitatively similar on exposure to CO2-free air, especially during the 1st h. In view of this, we considered it acceptable to express the response of these most strongly induced transcripts collectively as a pooled average and to compare this expression with the physiological changes in affinity for Ci over the same time period of low-Ci exposure.
Figure 5.
Physiological responses of cells of Synechocystis PCC6803 to aeration for up to 6 h with CO2-free air. A, Vmax of net O2 evolution. The mean ± sd of three separate experiments are shown. B, Pooled average increase in ndhF3, sbtA, and cmpA transcripts replotted from the RT-PCR experiment shown in Figure 4 (black circles) and the rates of Ci uptake (white squares) or net O2 evolution (white triangles) supported by 20 μm Ci in the assay medium replotted from the drawdown experiments shown in Figure 2B.
The increases in affinity for Ci, indicated by the rate of Ci uptake and net O2 evolution supported by 20 μm Ci, and the corresponding collective response of inducible transcripts to CO2-free air, are shown in Figure 5B. The data were derived from data shown in Figures 2 and 4, respectively. When compared graphically, it was evident that the initial rate of transcript induction was more rapid than the physiological response. Most of the collective increase in transcript abundance was complete by 15 min of exposure to CO2-free air, whereas full physiological expression of the CCM was evident by 2 h under these conditions, indicating a strong relationship between gene transcription and subsequent translation into functional products. There was no evidence of a rapid induction response (Sültemeyer et al., 1998) in the first 15 to 30 min of induction. This was consistent with studies by Benschop et al. (2003) showing a lack of fast induction in Synechocystis cells grown under the same conditions.
The Effect of High-Light Stress on Low-Ci-Responsive Transcripts
Initial experiments were aimed at testing if cells grown at a low light intensity (40 μmol photons m−2 s−1) and 3% (v/v) CO2 would show any induction of low-Ci-responsive genes when shifted to high light (200 or 400 μmol photons m−2 s−1) when CO2 was held at the 3% (v/v) level. The highly responsive transcripts, cmpA, ndhF3, or sbtA, were unaltered by high-light treatment (results not shown). However, in a recent DNA microarray study in Synechocystis, the expression of several low-Ci-inducible genes involved in the CCM was reported as strongly up-regulated within 15 min of treatment with high light (Hihara et al., 2001). We investigated this result in more detail using real-time RT-PCR.
The effect of a 15-fold increase in light intensity from low light to high light (20–300 μmol photons m−2 s−1) in cultures bubbled with 3% (v/v) CO2 (pH 8.0 growth media) on the expression of several genes related to the CCM (Table I), along with a second experiment performed at 1% (v/v) CO2 (pH 7.0 media; Table I) that was closest to the conditions in Hihara et al. (2001). Control cultures, transferred to CO2-free air, at low and high light, were also included as verification that the cells were able to show a genuine low-Ci response (results not shown). Low-Ci-responsive transcripts, cmpA, ndhF3, and sbtA, did not respond to a transition to high light when CO2 in the cultures was kept high (Table I); in fact, a slight decline in abundance was typically seen. Other CCM genes such as ccmM, rbcL, and ndhF4 were not significantly changed by a transition to high light. Two genes, sodB (superoxide dismutase B) and the high-light inducible psbA2, showed a small, but significant, up-regulation of 250% and 150% (P < 0.1) at pH 8.0, indicating the induction of genuine light stress. However, at pH 7.0, an induction of psbA2 was not seen. Collectively, these experiments provided clear evidence that genes encoding inducible Ci transporters, and other CCM genes, were not induced under high light when high CO2 levels were maintained. This suggests that a high-light stress per se was incapable of triggering CCM induction in Synechocystis PCC6803. Thus, we were unable to confirm the results of Hihara et al. (2001), namely the high-light-induced expression of CCM-related genes.
Table I.
Relative changes in transcript abundance in cells of Synechocyctis PCC6803 exposed to high light as determined by real-time RT-PCR
| Gene Name | Change in Transcript Abundance in High Light Relative to Low Light
|
|
|---|---|---|
| pH 8.0, 3% CO2 | pH 7.0, 1% CO2 | |
| % | ||
| cmpA | −121 ± 52 | 9 ± 54 |
| ndhF3 | −34 ± 58 | −30 ± 76 |
| sbtA | −92 ± 56 | −67 ± 62 |
| sodB | 260 ± 65 | n.d. |
| psbA2 | 158 ± 80 | −22 ± 91 |
| rbcL | −10 ± 47 | 139 ± 73 |
| ccmM | −5 ± 65 | 107 ± 103 |
| ndhF4 | 24 ± 72 | n.d. |
| ndhD2 | 15 ± 53 | n.d. |
Cultures of low-light-grown cells (20 μmol photons m−2 s−1) were split and exposed to 300 μmol photons m−2 s−1 for 15 min at 30°C, or maintained at low light (low-light control). Transcript changes in the high-light treatment were referenced to the low-light treatment. The [CO2] was maintained at 3% (v/v) or 1% (v/v) in air, as indicated. The means ± se of four replicate measurements are shown.
DISCUSSION
Studies were carried out to investigate in detail the response of Synechocystis PCC6803 to severe Ci stress and to establish the temporal relationship between the initiation of CCM induction at the gene expression and physiological levels. Particular emphasis was placed on studying the expression of genes known to encode inducible Ci transporters (or components thereof) in this species, namely cmpA (BCT1, a high-affinity HCO3− transporter), sbtA (a Na+-dependent HCO3− transporter), and ndhF3 (a component of an NDH-1 complex involved in high-affinity CO2 uptake). One of the key findings was that the most responsive low-Ci-induced transcripts, cmpA, sbtA, and ndhD3, responded much more quickly than previously thought. Likewise, the rate of increase in affinity for external Ci, based on gene expression and de novo protein synthesis (i.e. not because of the allosteric, fast induction response), is much more rapid under these conditions than previously observed. With respect to methodologies for quantitation of transcript abundance, we found that semiquantitative and real-time RT-PCR generally gave similar results; however, largely as expected, it was clear that semiquantitative RT-PCR tended to underestimate strong changes in transcript abundance (e.g. quantitation of cmpA transcript).
Gene Expression under Ci Limitation in Synechocystis PCC6803
Considered together, the results from Figures 2 to 5 clearly show that Synechocystis sp. PCC6803 can respond very quickly to low-Ci stress. The inducible transcripts cmpA, sbtA, and ndhF3 reached, or exceeded, 75% of the maximal response within 15 min of exposure to Ci limitation, whereas physiological adaptation was close to maximal within 2 h. This physiological induction seems to be predominantly of a genetic basis because evidence of a fast induction response (Sültemeyer et al., 1998) was not seen in Synechocystis sp. PCC6803 under comparable conditions (Benschop et al., 2003; Fig. 5). This study appears to be the first study, to our knowledge, to determine the temporal relationship between gene transcription and subsequent translation, inferred from physiological change, for cyanobacteria CCM genes.
Clearly, the transcripts encoding inducible Ci transporters were the most responsive to low-Ci conditions of the genes examined. Both the cmp and ndhF3/D3/chpY gene clusters appeared to respond as classic operons, supporting suggestions made by Omata et al. (2001) and Ohkawa et al. (2000b). One novel finding was that slr0042 (orf576, Price et al., 1998) was co-expressed with other members of the cmp operon in Synechocystis. This result has not been demonstrated in previous studies. This gene encodes a putative membrane-spanning porin protein that could potentially assist in trans-location of HCO3− across the outer membrane. In addition, we demonstrated that the ndhF4/D4 and chpX genes were insensitive to low-Ci stress in Synechocystis PCC6803, representing an independent confirmation of the data of Ohkawa et al. (1998).
Interestingly, cmpR and ndhR were up-regulated by approximately the same amount under low Ci, suggesting that the regulatory mechanism controlling the expression of the cmp and ndhF3/D3/chpY operons is potentially similar under these conditions. Despite this, however, there is evidence that these operons are regulated differently. Insertional inactivation of cmpR abolished the low-Ci responsiveness of the cmp operon in Synechocystis PCC6803, indicating positive transcriptional regulation (Omata et al., 2001). On the other hand, in an ndhR null mutant, the ndhF3/D3/chpY operon was more highly expressed compared with wild-type cells under Ci limitation, suggesting that ndhR negatively regulates (represses) the expression of this operon (Figge et al., 2001). However, the finding that ndhR transcript rises under low-Ci stress is not consistent with NdhR being a simple negative regulator of ndhF3/D3/chpY transcription.
Significantly, the rbcLXS and ccmK-N operons, ccmO, and the gene encoding carboxysomal CA (ccaA) did not show significant up-regulation under severe Ci limitation (Fig. 3, C and D). In Synechocystis PCC6803, Omata et al. (2001) reported only a slight increase in rbcL transcript after a shift from 2% to 0.005% (v/v) CO2 in air. However, in the same study, substantive increases in rbcL and ccmK-O transcript were reported in Synechococcus PCC7942 under the same low-Ci treatment. It has also been reported that the number of carboxysomes per cell increased in Synechococcus sp. as the [Ci] in the medium decreased (Turpin et al., 1984; McKay et al., 1993). This is consistent with the data of Omata et al. (2001) mentioned above in relation to ccm expression. However, a recent study by So et al. (2002) reported for Synechocystis PCC6803 that the number of carboxysomes per cell, determined by electron microscopy, do not change after the transfer of cells from high-Ci conditions to Ci-limited conditions. This result is consistent with our observations that carboxysome-related gene transcripts did not show significant up-regulation under severe Ci limitation. Nevertheless, the expression of carboxysome and Rubisco genes under more moderate Ci limitation will be addressed in future studies in this lab.
Another interesting observation is that transcripts for ndhD3, ndhF3, and chpY were found to be detectable at low abundance in cells grown under high-CO2 conditions (Fig. 4). Whether this transcript is translated into active Ci uptake system is unknown at present and will require some careful analysis of particular mutants. This aspect is being further studied in Synechococcus PCC7942, where we have knockout mutants available and can carry out detailed transcript and physiological analysis.
Rapid Transcriptional and Physiological Response to Ci Limitation
There have been relatively few studies that have investigated the effect of several hours of low-Ci stress on physiological adaptation in cyanobacteria. In Synechococcus PCC7942, it was estimated that 4 to 6 h was required for full induction of the CCM after cells were transferred from high-Ci conditions to 20 to 30 ppm CO2 (Yu et al., 1994). A similar value of 4 h was measured in Anabaena variabilis after aeration with CO2-free air (Marcus et al., 1982). In this study, full induction in Synechocystis was estimated at 2 h, indicating that the increase in affinity for Ci can be more rapid than previously reported. The difference between this and other studies is apparently related to the method used to obtain high-affinity cells. In previous studies, investigators have simply switched the bubbling-gas stream from 2% to 3% (v/v) CO2 to air (350 ppm CO2) or 20 to 30 ppm CO2. This approach is inefficient and would have been unlikely to allow attainment of low-Ci conditions for at least the 1st h after changeover. Consistent with this are the results of Shibata et al. (2002a), who showed, by semiquantitative RT-PCR, a lag of 2 and 6 h before the appearance of sbtA and cmpA transcript, respectively, after switching directly from aeration with high Ci to normal air. In the present study, the cells were rapidly collected by centrifugation, and the media was exchanged for low-Ci media before aeration with CO2-free air began. Omata et al. (2001) used an induction protocol very similar to the one employed here and showed a similar temporal response of cmpC transcript to low Ci in Synechococcus sp., although the level of expression was lower than that reported here.
As mentioned above, Shibata et al. (2002a) reported a nonsynchronous expression of sbtA and cmpA when Synechocystis cells were transferred to bubbling with air. In the present study, the expression of cmpA and sbtA was coincident and strongly controlled by Ci limitation during illumination (Figs. 3–5). Here, the initial [Ci] in the culture media at the beginning of CO2-free aeration was approximately 6-fold lower than in air-equilibrated media (Fig. 1), and the induction protocol imposed a more severe Ci stress than the approach of Shibata et al. (2002a). However, the latter work does suggest that the cmpA-D operon might be expressed under a relatively more severe Ci stress than sbtA. Preliminary work with real-time RT-PCR has confirmed that in the presence of diminishing total [Ci] in the light, sbtA was maximally expressed ahead of cmpA in Synechocystis (P.J. McGinn, G.D. Price, and M.R. Badger, unpublished data).
It should be stressed that the severity of the induction protocol used in this study did not inhibit photosynthetic capacity in these cells or induce net photo-inhibition because there was no significant change in the maximum rate of photosynthesis measured under saturating Ci conditions (Fig. 5A). However, net growth was completely inhibited under these conditions (data not shown). Severe inhibition of growth rate in cyanobacteria is a common feature of aeration with very low concentrations of CO2 (Sültemeyer et al., 1997).
The Role of Light in the Expression of Low-Ci-Induced Transcripts
Circumstantial evidence had previously pointed to high light as being able to induce CCM-related genes in cyanobacteria (Reddy et al., 1989; Hihara et al., 2001). However, the CCM-related genes examined in the present study either showed no significant stimulation by high light under high [Ci] conditions or showed a slight down-regulation (Table I). The reason for the discrepancy with the microarray study of Hihara et al. (2001) is not clear at present. However, the results of the present study suggest that light stress is not a primary factor in the induction of the CCM in Synechocystis sp. PCC6803 and that a strict redox control mechanism is unlikely to be solely responsible for CCM induction. It should be noted that under severe Ci limitation, it is possible to demonstrate a secondary involvement of light stress in the induction of highly responsive CCM genes (P.J. McGinn, G.D. Price, and M.R. Badger, unpublished data). This will be reported in a future paper.
Potential Signals Inducing CCM Expression
There have been relatively few studies directed at identifying the signal that triggers the induction of the CCM in cyanobacteria. Mayo et al. (1986) suggested that cells of Synechococcus sp. responded to the [HCO3−] in the extracellular medium. Badger and Gallagher (1987) showed a clear, inverse relationship between induced CCM activity and total Ci concentration in the growth medium.
In the experiments presented here, transcripts coding for Ci transporters were highly expressed in illuminated cells under low-Ci conditions well below 140 μm Ci (Fig. 2; data not shown). Badger and Gallagher (1987) showed significant CCM expression when cells of Synechococcus PCC6301 were grown in the presence of 100 to 200 μm Ci in the light. An intermediate level of adaptation was reported by Mayo et al. (1989) at 200 to 300 μm Ci. In the present study, there was no significant up-regulation of inducible transcripts in the presence of 140 to 150 μm Ci in darkened cells, relative to control cells in the light (data not shown). This suggests that a diminishing concentration of Ci per se does not trigger expression of the CCM in cyanobacteria but that a “cooperative” effect of light and Ci limitation is required. Whether or not the cells respond to a particular Ci species in the light, or to a photorespiratory intermediate, remains an open question (Kaplan and Reinhold, 1999). These possibilities are presently being examined in more detail.
MATERIALS AND METHODS
Growth Conditions and Induction of Cells
Cells of the cyanobacterium Synechocystis PCC6803 (the Glc-tolerant “DuPont” strain) were grown at 30°C in BG-11 medium at pH 8 buffered with 20 mm TES-KOH or HEPES-KOH in air enriched to 3% (v/v) CO2 in the presence of 60 to 70 μmol photons m−2 s−1 (Yu et al., 1994). For induction experiments, cells from 50-mL cultures were collected by centrifugation (4,800g for 10 min) and resuspended in an equal volume of sterile growth medium and aerated for between 15 min and 5 h with CO2-free air, which was prepared by passing humidified air through a plexiglass cylinder (4 × 26 cm) packed with CO2-absorbing soda lime (Drägersorb, Dräger, Germany). CO2-free air was passed through each 50-mL culture at a flow rate of 0.15 L min−1 using a glass pipette with a 1-mm annulus. The initial [Ci] in the growth medium after autoclaving was typically 110 to 130 μm. Control cultures were collected by centrifugation and resuspended in media equilibrated with 3% (v/v) CO2 and then aerated with 3% (v/v) CO2 for between 15 min and 5 h under illumination.
For high-light experiments, cells were grown for 2 to 3 d under low-light conditions (20 μmol photons m−2 s−1) under either 3% (v/v) CO2 in air (pH 8.0) or 1% (v/v) CO2 (pH 7.0). This involved several cycles of dilution and regrowth. For experimental treatments with pH 8.0-grown cells, approximately 50 mL of low-light-grown cells were collected by centrifugation (4,800g for 10 min) and resuspended at an OD730 of between 0.1 and 0.2 in two separate 80-mL cultures in sterile BG-11 medium. One of the resuspended cultures, the control, was put back under the previous growth conditions of low light, whereas the other was exposed to 300 μmol photons m−2 s−1 for 15 min before RNA isolation. Both cultures were bubbled continuously with 3% (v/v) CO2 in air. For experiments with pH 7.0-grown cells, the low-light culture was diluted to OD730 of 0.4 (100 mL) and grown for a further 1 h at low light. Thereafter, the culture was split, with one-half receiving a high-light intensity of 300 μmol photons m−2 s−1 for 15 min, whereas the other one-half was kept at low light for 15 min as the control. Both cultures were bubbled with 1% (v/v) CO2 in air. RNA was then extracted (see below).
Primer Sequence Selection
Primer sequences were chosen using the Primer 3.0 software (www.genome.wi.mit.edu; Whitehead Institute, Cambridge, MA) and designed to produce 450- to 550-bp PCR products with a melting temperature of 60°C, at least one GC clamp, and 18 to 20 nucleotides in length. Primers were purchased from Life Technologies (Melbourne, Australia). All primer sequences used in this study are listed in Table II.
Table II.
Oligonucleotide sequences used in this study
| Gene Name | Cyanobase Open Reading Frame Identification | Forward Primer (5′ → 3′) | Reverse Primer (5′ → 3′) |
|---|---|---|---|
| cmpA | slr0040 | CGAGCGATTGGGTAGATAA | TGGATTAGCAGGGTCAAAGG |
| cmpB | slr0041 | CGGGTTGCTCAAGGTTATTC | CGCACCGATGTAAAACACTG |
| orf576 | slr0042 | CCCTCTATCCCACCGAAAAT | TAGAGTCGCCTTCCGGATTA |
| cmpC | slr0043 | TTAGGAGTGCGGGAAGATTG | TGGTGAAATTGCGATGAATG |
| cmpD | slr0044 | CTTGGTTAACTGCCCTGGAA | TGGGCAAAGCGGTTATAGAG |
| ndhF3 | slll732 | GTGAAGCTAAGCCGATGACC | AAAGGCGCCAATTAAAGTCA |
| ndhD3 | sll1733 | CGGTACCGAGCTGAGTGTTT | TTCATTGGTGGTGGTTTGTG |
| chpY | sll1734 | ACGGTATTTTTGCCATTGCT | ATTGCCGATTCTTAGCCAAA |
| ndhF4 | sll0026 | GTGCCCATGGTGAGTTTGAT | CTGGACCAAAACTGCCTGAT |
| ndhD4 | sll0027 | ATCATTTCTGCCCTGCTGTT | ACTTCCCCCAATTCCTTGAC |
| chpX | slr1302 | TGTGGTGCAACATATCCAAGA | GTCTCCTGTTCCTGGGGATT |
| ndhD1 | slr0331 | CATACCCTGATGCTGGATGA | GCCGTAAAGCAGTGACTTCC |
| rbcX | slr0010 | GCACATAGCTCAGGCAACAG | GGGGGAGAATCGTTGTTGTT |
| rbcL | slr0009 | CGGCATTTTGCTCCATATTC | AACTCAGGGGACCAACGAC |
| rbcS | slr0012 | CTTACCTGCCCCCCTTTAACC | AGTAACGGCCTTGGTTTTGG |
| ntpJ | slr1509 | CGGGCTTTAACACCATTGAT | CCAGCAAATTTTCCTCTGGA |
| ccaA | slr1347 | GAGCCATGAAAGGTCTGCTC | ACGGGAGCCTCGATAAATG |
| ictB | slr1515 | CCGTGGCGGTTATATTAGGT | AACAATGGCCACTAGCAACC |
| ndhD2 | slr1291 | AGTAATGCCCACGGTTTTTG | AAGTCCGTGGTGGGAATGT |
| ccmK3 | slr1838 | CCGTTGGCGTGATACAGAC | GAAGCGAGCGACTTTTTCTG |
| ccmA | sll0934 | AGAAATCATTGCGGAAGTGG | GGCCAACGTCCTACAGTTTG |
| ccmL | sll1030 | GGGACCGTGGTCAGTACATC | TCCCATCATCCCTCTTGTTG |
| ccmM | sll1031 | AAGGCCAAATCCTGGGATAC | TTTAGCCGTGGGATCTATGC |
| ccmO | slr1032 | ACAGCAACGGGGTTACAGTC | CTTCCACTATGGGCTTTTCC |
| psbA2 | slr1311 | CTCTGATGGTATGCCCTTGG | TTGCGTTCGTGCATTACTTC |
| sodB | slr1516 | GACTACACCGCTCTGGAACC | GGAAATCGGCAATGTAATCG |
| cmpR | sll0030 | ACAAATTTTGGAGCGTTTGG | TGGAAACTGCTTCATCATGG |
| ndhR | sll0998 | ATCCCGGCATTGAAGTATCC | TGGGTAATCACCGACAGTTG |
| sbtA | slr1512 | TAATCGGGTCAAAATCTGGC | CAAAATCGTCAATAGCGGCG |
Cyanobase Web site: http://www.kazusa.or.jp/cyano/Synechocystis/index.html.
RNA Isolation
Cells were collected by a brief centrifugation (4,800g for 3–5 min) in 50-mL centrifuge tubes and concentrated in approximately 1 mL of residual growth medium and transferred to a 1.5-mL microcentrifuge tube. The cells were recentrifuged for 30 s (10,000g), and the pellets were snap frozen in liquid N2 and stored at −80°C for later processing (storage was usually <1 week). Total RNA was isolated from harvested cells or frozen pellets using the phenol-guanidine-isothiocyanate-chloroform extraction protocol (TRIzol Reagent, Invitrogen, Carlsbad, CA). The RNA solution was digested with 4 units of DNAse (RQ1 RNAse-free DNAse, Promega , Madison, WI) according to the manufacturer's instructions before cDNA synthesis to avoid amplifying genomic sequences. The digest was extracted with an equal volume of phenol:chloroform (5:1 [w/v]), and the RNA was precipitated by centrifugation after a 40-min incubation at −20°C in the presence of 75 mm sodium acetate buffer (pH 5.2) and 75% (v/v) ethanol.
cDNA Synthesis for Real-Time and Semiquantitative RT-PCR
For real-time RT-PCR analysis, first strand synthesis was carried out using the Superscript First Strand system (Invitrogen). The 2× master mix, which contained cDNA synthesis buffer (40 mm Tris-HCl [pH 8.4] and 100 mm KCl), 12.5 mm MgCl2, 20 mm dithiothreitol, and 20 units of RNAse inhibitor, was prepared and stored on ice. Approximately 1 μg of total RNA was incubated at 65°C for 5 min in the presence of 20 pmol gene-specific reverse primer and 1 mm of each dNTP. Ten microliters of RNA/primer/dNTP mixture was added to an equal volume of 2× master mix and incubated at 42°C for 2 min in an Eppendorf thermal cycler. Fifty units of Superscript RT enzyme was added and the tubes incubated at 42°C for 50 min followed by 70°C for 15 min to deactivate the RT enzyme. Negative controls received 1 μL of water instead of RT enzyme. cDNA products were diluted 10-fold for use in real-time RT-PCR amplification. For semiquantitative RT-PCR, first strand synthesis was carried out essentially as described above with some modification. The 2× master mix contained buffer [100 mm Tris-Acetate (pH 8.4), 150 mm CH3COOH, 16 mm Mg(C2H3O2)2, 4H2O, 10 mm dithiothreitol, 2 mm of each dNTP, 80 units of RNAse inhibitor, and 30 units of Thermoscript RT enzyme (Invitrogen, Groningen, The Netherlands)]. Approximately 0.23 μg of total RNA and 6 to 10 pmol reverse primer were mixed and denatured at 65°C for 5 min. After 1 min of chilling on ice, 10 μL of the RNA/primer mixture was added to an equal volume of the master mix. The RT reaction was carried out at 50°C for 30 min followed by a 5-min enzyme inactivation step at 85°C. cDNA products were used directly for PCR amplification without dilution.
PCR
Real-time amplification of cDNA templates was carried out on a RotorGene 2000 Thermal Cycler (Corbett Research, NSW, Australia). Amplifications were carried out in a 20-μL volume using the HotstarTaq PCR kit (Qiagen, Hilden, Germany) and contained 1× PCR buffer; 200 μm each of dATP, dCTP, dGTP, and dTTP; 1 unit of Taq polymerase; 1 μL of cDNA template; and 10 to 20 pmol each of forward and reverse primer. The MgCl2 concentration was supplemented to 2.5 mm. Forty cycles of amplification were used under the following conditions: DNA polymerase activation at 95°C for 15 min, denaturation at 94°C for 20 s, annealing at 57°C for 30 s, product extension at 72°C for 40 s, and signal acquisition at 84°C for 15 s. The accumulation of specific PCR product was monitored by the inclusion of 0.5 μL per reaction of a 10× working solution of SybrGreen1 (a 1,000× dilution of a 10,000× stock solution, Fisher Biotech, Springfield, NJ). PCR reactions were carried out in quadruplicate. Minor genomic DNA contamination resulted in occasional background amplification in minus-RT controls. However, threshold cycle (Ct) values for minus-RT controls were, on average, eight to 14 cycles higher than Ct values from amplified cDNA, depending on the expression level of the transcript (data not shown). PCR amplification for semiquantitative analysis was carried out on a conventional thermal cycler (Mastercycler, Eppendorf, Germany). The conditions were essentially as described above with the exceptions that [Mg2+] was 1.5 mm and no SybrGreen 1 was added to the reactions. All reactions were subjected to 28 cycles of amplification unless stated otherwise.
Analysis of Semiquantitative RT-PCR Products
After PCR amplification, products were electrophoresed on 1.3% (w/v) agarose gels in Tris-borate/EDTA buffer (45 mm Tris, 40 mm boric acid, and 0.5 mm EDTA [pH 9.5]) and stained with ethidium bromide. Gels were scanned at 488-nm excitation with a Fluorimager SI (Vistra Fluorescence, Sunnyvale, CA), and the band intensities were quantified using linked ImageQuant software, with background correction.
Validation of the Method for Real-Time PCR Amplifications
In real-time RT-PCR analysis, the cycle where fluorescence departs from background and product-formation enters the exponential amplification phase has been termed the Ct value. The Ct value is directly proportional to the starting template concentration. The amplification efficiencies for the primer pairs used in this study were determined by measuring the Ct values over a 10-fold dilution series, spanning 7 orders of magnitude, made from gel-purified, gene-specific PCR product. The Ct values were calculated using RotorGene software (version 4.6, Corbett Research) in “Auto-find Threshold” mode and according to the manufacturer's recommendations. The Ct values were plotted against the relative template concentrations, and the slope of the linear regression was used to determine the amplification efficiency according to the equation: E = 10−(1/m), where m = slope of the linear regression. For the primer pairs used in this study, a deviation of less than 10% from the optimal E value of 2 was found. A default value for E of 2 was used in all subsequent quantitation calculations. The specificity of each PCR primer pair was confirmed by electrophoresis on agarose gels (data not shown); however, specificity for real-time RT-PCR analysis was also reconfirmed by melt curve analysis. After the last cycle of amplification, the rotor temperature was ramped from 55°C to 99°C at 0.2°C s−1, and the fluorescence from double-stranded DNA was monitored.
Calculations Used in Real-Time PCR Analysis
Changes in transcript abundance were estimated as fold change (FC) relative to a control condition (e.g. high-Ci conditions) and expressed as percentage increase or decrease. The FC in transcript abundance was calculated according to the equation: FC = 2(−ΔΔCt ± se), where the value 2 is the amplification efficiency, and where ΔΔCt ± se = ΔCttar ± se − ΔCtref ± se (Livak and Schmittgen, 2001). The terms ΔCttar and Ctref represent the differences in mean Ct values between treated and control samples for target genes and the reference gene (i.e. ndhD1), respectively. The ses of the means on these terms were added together to give the se on ΔΔCt. The FCs in transcript abundance for target genes in treated samples, relative to control, were then expressed as a percentage change.
Mass Spectrometric Measurements
Cells were harvested at various periods of induction under CO2-free air by centrifugation (4,800g for 10 min), washed twice with low-Ci assay buffer, and concentrated as described by Sültemeyer et al. (1995). The relative affinity for Ci was determined as the rate at which cellular photosynthesis removed approximately 80 μm Ci from the mass spectrometer cuvette. This was done by monitoring the CO2 (m/e 44) and O2 (m/e 32) in drawdown experiments in the presence of 25 μg mL−1 bovine CA and 400 μmol photons m−2 s−1 irradiance at pH 8. Cells were pre-adapted with 2 min of irradiance. The maximum rate of net O2 evolution (Vmax) was measured in cell suspensions in the presence of 1 to 3 mm NaHCO3 and 25 μg mL−1 bovine CA. Chlorophyll a concentrations were adjusted to between 2.5 and 3 μg mL−1 and were determined by extraction in methanol according to Porra et al. (1989).
Culture [Ci] Measurements
Cells were harvested and subjected to aeration with CO2-free air for up to 40 min according to the protocol outlined above. At 10-min intervals, approximately 0.8 mL of culture medium was withdrawn through a 0.45-μm filter into a sterile, 1-mL syringe. This was immediately placed into a 0.5-mL tube with no remaining headspace. Three 100-μL aliquots were injected into 4 mL of 0.02 n HCl (pH 2.4) with a gas-tight syringe in the mass spectrometer cuvette, and the rise in the 12CO2 signal was recorded. The mass spectrometer was calibrated by injecting known amounts of NaHCO3 into 0.02 n HCl.
ACKNOWLEDGMENT
The authors gratefully acknowledge Dr. Fiona Woodger (Research School of Biological Sciences, Australian National University) for critical reading of portions of the manuscript.
Footnotes
This work was supported by the Research School of Biological Sciences and by the Australian National University (to P.J.M.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.019349.
LITERATURE CITED
- Badger MR, Gallagher A. Adaptation of photosynthetic CO2 and HCO3− accumulation by the cyanobacterium Synechococcus PCC6301 to growth at different inorganic carbon concentrations. Aust J Plant Physiol. 1987;14:189–201. [Google Scholar]
- Benschop J, Badger MR, Price GD (2003) Characterisation of CO2 and HCO3− uptake in the cyanobacterium, Synechocystis sp. PCC6803. Photosynth Res (in press) [DOI] [PubMed]
- Bonfil DJ, Ronentarazi M, Sültemeyer D, Liemanhurwitz J, Schatz D, Kaplan A. A putative HCO3− transporter in the cyanobacterium Synechococcus sp. strain PCC7942. FEBS Lett. 1998;430:236–240. doi: 10.1016/s0014-5793(98)00662-0. [DOI] [PubMed] [Google Scholar]
- Figge RM, Cassier-Chauvat C, Chauvat F, Cerff R. Characterisation and analysis of an NAD(P) H dehydrogenase transcriptional regulator critical for survival of cyanobacteria facing inorganic carbon starvation and osmotic stress. Mol Microbiol. 2001;39:455–468. doi: 10.1046/j.1365-2958.2001.02239.x. [DOI] [PubMed] [Google Scholar]
- Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell. 2001;13:793–806. doi: 10.1105/tpc.13.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan A, Badger MR, Berry J. Photosynthesis and intracellular inorganic carbon pool in the blue-green algae Anabaena variabilis: response to external CO2 concentration. Planta. 1980;149:219–226. doi: 10.1007/BF00384557. [DOI] [PubMed] [Google Scholar]
- Kaplan A, Reinhold L. CO2-concentrating mechanisms in photosynthetic microorganisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:539–570. doi: 10.1146/annurev.arplant.50.1.539. [DOI] [PubMed] [Google Scholar]
- Klughammer B, Sültemeyer D, Badger MR, Price GD. The involvement of NAD(P) H dehydrogenase subunits NdhD3 and NdhF3, in high affinity CO2 uptake in Synechococcus sp. PCC7002 gives evidence for multiple NDH-1 complexes with specific roles in cyanobacteria. Mol Microbiol. 1999;32:1305–1315. doi: 10.1046/j.1365-2958.1999.01457.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maeda S, Badger MR, Price GD. Novel gene products associated with NdhD3/D4-containing NDH-1 complexes are involved in photosynthetic CO2 hydration in the cyanobacterium, Synechococcus PCC7942. Mol Microbiol. 2002;43:425–436. doi: 10.1046/j.1365-2958.2002.02753.x. [DOI] [PubMed] [Google Scholar]
- Marcus Y, Zenvirth D, Harel E, Kaplan A. Induction of HCO3− transporting capability and high photosynthetic affinity to inorganic carbon by low concentration of CO2 in Anabaena variabilis. Plant Physiol. 1982;69:1008–1012. doi: 10.1104/pp.69.5.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo WP, Elrifi IR, Turpin DH. The relationship between ribulose bisphosphate concentration, dissolved inorganic carbon (DIC) transport and DIC-limited photosynthesis in the cyanobacterium Synechococcus leopoliensis grown at different concentrations of inorganic carbon. Plant Physiol. 1989;90:720–727. doi: 10.1104/pp.90.2.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo WP, Williams TG, Birch DG, Turpin DH. Photosynthetic adaptation by Synechococcus leopoliensis in response to exogenous dissolved inorganic carbon. Plant Physiol. 1986;80:1038–1040. doi: 10.1104/pp.80.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay RML, Gibbs SP, Espie GS. Effect of dissolved inorganic carbon on the expression of carboxysomes, localization of Rubisco and the mode of inorganic carbon transport in cells of the cyanobacterium Synechococcus UTEX 625. Arch Microbiol. 1993;159:21–29. [Google Scholar]
- Ohkawa H, Pakrasi HB, Ogawa T. Two types of functionally distinct NAD(P) H dehydrogenases in Synechocystis sp. strain PCC6803. J Biol Chem. 2000a;275:31630–31634. doi: 10.1074/jbc.M003706200. [DOI] [PubMed] [Google Scholar]
- Ohkawa H, Price GD, Badger MR, Ogawa T. Mutation of ndh genes leads to inhibition of CO2 uptake rather than HCO3− uptake in Synechocystis sp. strain PCC6803. J Bacteriol. 2000b;182:2591–2596. doi: 10.1128/jb.182.9.2591-2596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa H, Sonoda M, Katoh H, Ogawa T. The use of mutants in the analysis of the CO2-concentrating mechanisms in cyanobacteria. Can J Bot. 1998;76:1035–1042. [Google Scholar]
- Omata T, Gohta S, Takahashi Y, Harano Y, Maeda S. Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J Bacteriol. 2001;183:1891–1898. doi: 10.1128/JB.183.6.1891-1898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata T, Price GD, Badger MR, Okamura M, Gohta S, Ogawa T. Identification of an ATP-binding cassette transporter involved in HCO3− uptake in the cyanobacterium Synechococcus sp. strain PCC7942. Proc Natl Acad Sci USA. 1999;96:13571–13576. doi: 10.1073/pnas.96.23.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata T, Takahashi Y, Yamaguchi O, Nishimura T. Structure, function and regulation of the cyanobacterial high-affinity bicarbonate transporter, BCT-1. Funct Plant Biol. 2002;29:151–159. doi: 10.1071/PP01215. [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. Determinations of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- Price GD, Maeda S, Omata T, Badger MR. Modes of active inorganic carbon uptake in the cyanobacterium Synechococcus sp. PCC7942. Funct Plant Biol. 2002;29:131–149. doi: 10.1071/PP01229. [DOI] [PubMed] [Google Scholar]
- Price GD, Sültemeyer D, Klughammer B, Ludwig M, Badger MR. The functioning of the CO2 concentrating mechanism in several cyanobacterial strains: a review of general physiological characteristics, genes, proteins, and recent advances. Can J Bot. 1998;76:973–1002. [Google Scholar]
- Reddy KJ, Masamoto K, Sherman DM, Sherman LA. DNA sequence and regulation of the gene (cbpA) encoding the 42-kilodalton cytoplasmic membrane carotenoprotein of the cyanobacterium Synechococcus sp. strain PCC7942. J Bacteriol. 1989;171:3486–3493. doi: 10.1128/jb.171.6.3486-3493.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Katoh H, Sonoda M, Ohkawa H, Shimoyama M, Fukuzawa H, Kaplan A, Ogawa T. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria: function and phylogenetic analysis. J Biol Chem. 2002a;277:18658–18664. doi: 10.1074/jbc.M112468200. [DOI] [PubMed] [Google Scholar]
- Shibata M, Ohkawa H, Kaneko T, Fukuzawa H, Tabata S, Kaplan A, Ogawa T. Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc Natl Acad Sci USA. 2001;98:11789–11794. doi: 10.1073/pnas.191258298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Ohkawa H, Katoh H, Shimoyama M, Ogawa T. Two CO2 uptake systems in cyanobacteria: four systems for inorganic carbon acquisition in Synechocystis sp. strain PCC6803. Funct Plant Biol. 2002b;29:123–129. doi: 10.1071/PP01188. [DOI] [PubMed] [Google Scholar]
- So AKC, John-McKay M, Espie GS. Characterization of a mutant lacking carboxysomal carbonic anhydrase from the cyanobacterium Synechocystis PCC6803. Planta. 2002;214:456–467. doi: 10.1007/s004250100638. [DOI] [PubMed] [Google Scholar]
- Sültemeyer D, Klughammer B, Badger MR, Price GD. Fast induction of high-affinity HCO3− transport in cyanobacteria. Plant Physiol. 1998;116:183–192. [Google Scholar]
- Sültemeyer D, Price GD, Badger MR. Characterization of carbon dioxide and bicarbonate uptake during steady-state photosynthesis in the marine cyanobacterium Synechococcus PCC7002. Planta. 1995;197:597–607. [Google Scholar]
- Sültemeyer D, Price GD, Bryant DA, Badger MR. PsaE and NdhF-mediated electron transport affect bicarbonate transport rather than carbon dioxide uptake in the cyanobacterium Synechococcus sp. PCC7002. Planta. 1997;201:36–42. [Google Scholar]
- Turpin DH, Miller AG, Canvin DT. Carboxysome content of Synechococcus leopoliensis (Cyanophyta) in response to inorganic carbon. J Phycol. 1984;20:249–253. [Google Scholar]
- Yu J-W, Price GD, Badger MR. Characterisation of CO2 and HCO3− uptake during steady-state photosynthesis in the cyanobacterium Synechococcus PCC7942. Aust J Plant Physiol. 1994;21:185–195. [Google Scholar]