Abstract
Purified rice (Oryza sativa) mitochondrial proteins have been arrayed by isoelectric focusing/polyacrylamide gel electrophoresis (PAGE), by blue-native (BN) PAGE, and by reverse-phase high-performance liquid chromatography (LC) separation (LC-mass spectrometry [MS]). From these protein arrays, we have identified a range of rice mitochondrial proteins, including hydrophilic/hydrophobic proteins (grand average of hydropathicity = −1.27 to +0.84), highly basic and acid proteins (isoelectric point = 4.0–12.5), and proteins over a large molecular mass range (6.7–252 kD), using proteomic approaches. BN PAGE provided a detailed picture of electron transport chain protein complexes. A total of 232 protein spots from isoelectric focusing/PAGE and BN PAGE separations were excised, trypsin digested, and analyzed by tandem MS (MS/MS). Using this dataset, 149 of the protein spots (the products of 91 nonredundant genes) were identified by searching translated rice open reading frames from genomic sequence and six-frame translated rice expressed sequence tags. Sequence comparison allowed us to assign functions to a subset of 85 proteins, including many of the major function categories expected for this organelle. A further six spots were matched to rice sequences for which no specific function has yet been determined. Complete digestion of mitochondrial proteins with trypsin yielded a peptide mixture that was analyzed directly by reverse-phase LC via organic solvent elution from a C-18 column (LC-MS). These data yielded 170 MS/MS spectra that matched 72 sequence entries from open reading frame and expressed sequence tag databases. Forty-five of these were obtained using LC-MS alone, whereas 28 proteins were identified by both LC-MS and gel-based separations. In total, 136 nonredundant rice proteins were identified, including a new set of 23 proteins of unknown function located in plant mitochondria. We also report the first direct identification, to our knowledge, of PPR (pentatricopeptide repeat) proteins in the plant mitochondrial proteome. This dataset provides the first extensive picture, to our knowledge, of mitochondrial functions in a model monocot plant.
The ATP-synthesizing organelles of eukaryotic cells, mitochondria, can trace their origins back to an event in which one prokaryotic cell was engulfed by another cell to form a new cellular lineage containing two genomes. These genomes became dependent over time, and mitochondria lost the ability to be viable outside the host cell. A significant net transfer of genetic information occurred from the mitochondrial to the nuclear genome during this time. It is predicted that mitochondria now synthesize only a few percent of the proteins required for their function, with the majority of their proteins being encoded in the nucleus, translated in the cytosol, and transferred back to the mitochondria as protein precursors by means of targeting information in the protein sequence (Gray et al., 1999). Biochemical investigations have revealed that isolated plant mitochondria share many features with those from animals and fungi. However, plant mitochondria also contain additional components such as non-phosphorylating bypasses of the electron transport chain, specialized metabolite carriers, and enzymes involved in the synthesis of folate, lipoic acid, and vitamin C (Bartoli et al., 2000; Gueguen et al., 2000; Rebeille et al., 1997). Only a few of the nuclear genes encoding proteins that maintain these unique functions have been identified. The recent increased sequencing of plant genomes (Arabidopsis Genome Initiative, 2000; Goff et al., 2002; Yu et al., 2002) has encouraged the development of bioinformatic tools to predict the subcellular localization of putative plant protein sequences. Programs such as MitoProtII (Claros and Vincens, 1996), TargetP (Emanuelsson et al., 2000), and Predotar (http://www.inra.fr/Internet/ Produits/Predotar/) predict that thousands of nuclear-encoded plant proteins are destined for mitochondria, but the veracity of such predictions still awaits confirmation.
Several reports have recently attempted to identify the protein contents of plant mitochondria from Arabidopsis (Kruft et al., 2001; Millar et al., 2001) and pea (Pisum sativum; Bardel et al., 2002) using isoelectric focusing (IEF)/SDS-PAGE-based separation of proteins. These three reports each identified 40 to 90 proteins using this arraying technique coupled with identification using antibodies, N-terminal sequencing, matrix-assisted laser-desorption/ionization-time-of-flight (TOF) peptide mass fingerprinting, and collision-induced tandem mass spectrometry (MS/MS) spectra of single peptides. Both these species are dicots. Increasing evidence of important differences between monocot and dicot plant lineages at a molecular level have been reported (Qiu et al., 1999; Soltis et al., 1999; Daly et al., 2001). Differences in mitochondrial molecular function between monocots and dicots also have been highlighted recently (Fey and Marechal-Drouard, 1999; Thelen et al., 1999; Considine et al., 2002). Thus, we consider it important to consider the mitochondrial proteome in a model monocot species.
The gel array technique used in the dicot mitochondrial proteome reports has several limitations. In particular, very small (<10 kD) and very large (>100 kD) proteins, basic proteins (pI > 10), and hydrophobic proteins (grand average of hydropathicity [GRAVY] > 0.2) are poorly resolved by this method (Chevallet et al., 1998; Herbert et al., 1998; Santoni et al., 1999, 2000). Incremental improvements in chaotropic agents and nonionic detergents have improved the resolving power of classical denaturing two-dimensional electrophoresis (Chevallet et al., 1998; Herbert et al., 1998). However, a substantial protein set will probably always be excluded due to the physical limitations of this approach. Native gel electrophoresis coupled to second dimension SDS-PAGE provides a flexible solution to the hydrophobic protein exclusion problem. Many such proteins are not found exposed in biological membranes but are core, internalized components of multisubunit complexes. Maintenance of this complex structure avoids precipitation of hydrophobic components. Blue-native (BN) PAGE has been used previously to separate native protein complexes from mitochondria (Schagger et al., 1994). A subsequent denaturing SDS-PAGE dimension allows separation of individual components for analysis from plant mitochondria (Jansch et al., 1996; Werhahn and Braun, 2002). A different solution to the exclusion problem, with an even wider applicability, is non-gel-based separation of complex mixtures. These approaches often use liquid chromatography (LC)-based separations of peptide derived from whole-sample digests that are directly coupled to MS. One approach using a coupled strong cation affinity and reverse-phase C18 separation has been termed multidimensional protein identification technology and is being increasingly applied to a broad range of biological samples (Washburn et al., 2001).
To determine which putative proteins from the rice (Oryza sativa) genome sequence represent constitutively expressed proteins involved in mitochondrial function, we have used two-dimensional gel electrophoresis and LC analysis of purified rice mitochondria coupled with MS identification. Our aim was to identify proteins localized in rice mitochondria, compare and contrast protein display techniques, and identify novel mitochondrial proteins, thereby providing a substantial set of experimentally identified mitochondrial proteins from this model monocot plant.
Results
Integrity and Function of Isolated Rice Mitochondria
The integrity of a subcellular proteome, such as that of mitochondria, is largely dependent on the purification of the isolated compartment away from other cellular contaminants. We have adapted a two-Percoll gradient density separation technique to the isolation of these organelles from dark-grown rice shoots that yields mitochondria largely free of contamination by cytosol, peroxisomes, plastids, and other membranes (Millar et al., 2001). We have assayed the integrity and function of these purified rice mitochondria to ensure that proteins were not being lost by rupture during isolation and that key respiratory functions were maintained. The outer membrane of mitochondrial samples was found to be 96% intact based on the latency of cytochrome c oxidase activity initiated by exogenously added cytochrome c (Table I). Whole-electron transport chain activities showed that O2 consumption by the mitochondria could be supported by succinate, external NADH, or a combination of malate and pyruvate. O2 consumption supported by each of these substrates was significantly and transiently stimulated by the addition of ADP. These data suggested the presence and function of the electron transport chain, the F1F0-ATP synthase, the tricarboxylic acid cycle in the mitochondrial matrix, the integrity of the inner membrane, and the function of external rotenone-insensitive NADH dehydrogenase bypasses of complex I (Table I). Little or no respiration was maintained in the presence of the complex III inhibitor myxothiazol, showing that little or no alternative oxidase (AOX) was present in these mitochondria (Table I). This lack of AOX was also confirmed by addition of pyruvate and dithiothreitol to O2 electrode traces without effect after myxothiazol addition. It was further confirmed by the lack of immunodetectable bands using monoclonal antibodies raised against AOX (data not shown).
Table I.
O2 consumption by isolated rice mitochondria following addition of different substrates, effectors, and inhibitors
| Substrate | −ADP | +ADP | +Myxothiazol |
|---|---|---|---|
| nmol O2 min−1 mg−1 protein | |||
| NADH | 87 ± 6 | 199 ± 8 | 3 ± 1 |
| Succinate | 99 ± 7 | 198 ± 17 | 2 ± 1 |
| Malate and pyruvate | 61 ± 9 | 139 ± 11 | 1 ± 1 |
| Cytochrome c | 438 ± 29 | – | – |
| Membrane integrity | 96.1 ± 1% | – | – |
Respiratory assays were performed according to “Materials and Methods.” O2 consumption rates are presented as means ± se (n > 3). Outer membrane integrity is presented as a percentage of intact mitochondria.
IEF and BN Arraying of Rice Mitochondrial Proteins
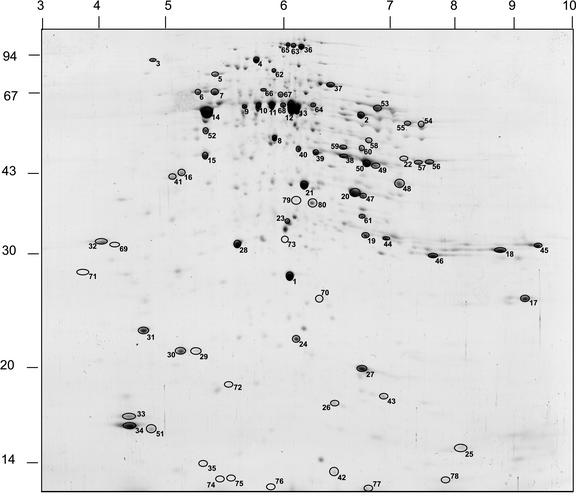
To analyze the complex mixture of proteins present in rice mitochondria, these proteins were separated in gel arrays using the apparent pI and molecular mass of individual polypeptides and the apparent native molecular mass of protein complexes. The IEF/SDS-PAGE separation used the first two of these properties and revealed a set of approximately 250 protein spots (Fig. 1). A set of 145 of the more abundant spots from this set was excised and subjected to in-gel digestion followed by MS/MS-based analysis of the resultant peptides. This analysis lead to the identification of 80 of these protein spots as the products of 63 predicted gene sequences from rice (Table II).
Figure 1.
IEF/SDS-PAGE separation of rice mitochondrial proteins (pI = 3–10). A total of 145 protein spots from this map were excised, digested, and analyzed by MS/MS. Eighty were identified, and these are numbered for comparison with text and Table II. Numbers on the horizontal axis are pI, and numbers on the vertical axis are apparent molecular mass (in kilodaltons).
Table II.
Identification of proteins from purified rice mitochondria using MS/MS peptide spectra matched to rice ORFs translated by the Institute for Genomic Research (TIGR; XXXX.mXXXXX), TIGR-translated expressed sequence tags (ESTs, TCXXXXX), and rice NCBI entries
| Functional Class | 2D-PAGE | BN-PAGE | LC-MS | Mr | pl | GRAVY | TP | PT | MP | Accession No. | Protein | At | Ps |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATP synthase | 11, 12, 64 | 19, 32, 50, 51, 52 | 2 | 55,281 | 5.9 | −0.11 | – | – | – | 2832.m00083, CAA35787 | ATP synthase alpha-subunit | * | * |
| 14 | 18, 31 | 10 | 59,059 | 6.8 | −0.10 | M | M | M | Q01859 | ATP synthase beta-subunit | * | ||
| 17 | 37 | 25,065 | 10.1 | −0.16 | M | M | M | 1903.m00144 | ATP synthase OSCP subunit | * | |||
| 29, 30 | 22 | 3 | 24,359 | 5.0 | −0.71 | – | – | – | TC63644, 3067.m00121 | ATP synthase delta-subunit | * | * | |
| 31 | 38 | 21,213 | 6.0 | 0.15 | M | M | M | 2119.m00096, BAC21366 | ATP synthase delta-subunit | * | * | ||
| 51 | 20, 35 | 4 | 40,955 | 8.5 | −0.10 | – | – | – | 3695.m00193, TC72968 | ATP synthase gamma-subunit | b | ||
| 42, 23 | 6937 | 10.8 | −0.30 | M | M | – | BAB21526 | ATP synthase 6-kD subunit | |||||
| 23 | 7877 | 10.9 | 0.84 | C | C | – | P14863 | ATP synthase subunit 9 | |||||
| 28 | 21 | 2 | 38,217 | 5.0 | −0.44 | M | M | M | 2485.m00122 | ATP synthase FAd subunit | * | * | |
| Complex I | 26 | 2 | 12,492 | 6.7 | −0.99 | – | – | – | AAL67582 | NADH dehydrogenase PDSW subunit | |||
| 76 | 12,033 | 7.6 | −0.22 | M | M | – | 3640.m00126 | NADH dehydrogenase B22 subunit | * | ||||
| 71 | 12 | 23,498 | 4.9 | −0.57 | M | M | – | 3550.m00121 | NADH dehydrogenase (23 kD) | * | |||
| 43 | 57 | 18,518 | 7.1 | −0.90 | M | – | – | 387.m00162 | NADH dehydrogenase B17.2 | ||||
| 4 | 3 | 4 | 81,117 | 6.1 | −0.17 | M | M | M | AAL58200 | NADH dehydrogenase 75-kD subunit | * | ||
| 4, 8, 9 | 1 | 44,309 | 9.6 | −0.08 | M | – | M | 2451.m00084 | NADH dehydrogenase (39 kD) | ||||
| 4, 5 | 56,301 | 8.2 | −0.31 | M | M | M | 2102.m00066 | NADH dehydrogenase (51 kD) | |||||
| 6, 7 | 44,252 | 7.1 | −0.49 | C | – | – | BAC19866 | NADH dehydrogenase subunit 7 | |||||
| 13 | 22,594 | 8.2 | −0.78 | – | – | – | BAA08794 | NADH dehydrogenase subunit 9 | |||||
| 14, 15 | 1 | 21,829 | 4.6 | −0.24 | M | M | M | TC76675, 4381.m00118 | NADH dehydrogenase 13-kD subunit | * | |||
| 16 | 2 | 28,762 | 9.6 | −0.30 | M | M | M | TC79833, 4342.m00113 | NADH dehydrogenase E 16.6-kD subunit | ||||
| Complex III | 24 | – | – | – | – | – | – | TC79968 | Mitochondrial-processing peptidase beta-subunit | * | |||
| 2 | 25 | 53,946 | 7.2 | −0.10 | M | M | – | 2599.m00145, 2709.m00160 | Mitochondrial-processing peptidase alpha-subunit | * | |||
| 27 | 1 | 29,820 | 8.6 | −0.16 | M | M | M | 2341.m00134 | Reiske Fe-S protein | c | |||
| 28 | 16,616 | 9.8 | −0.34 | M | M | M | 2070.m00100, 3403.m00173 | Ubiquinol cytochrome c reductase (14 kD) | * | ||||
| 29, 23 | 13,086 | 10.8 | 0.41 | – | M | M | 3017.m00162, 3017.m00172 | Ubiquinol cytochrome c reductase (8.2 kD) | |||||
| 2 | 33,293 | 6.2 | −0.19 | M | M | M | 2974.m00152 | Cytochrome c1 precursor | c | ||||
| Complex IV | 32 | 18,912 | 4.0 | −1.01 | – | – | – | BAA76393 | Cytochrome c oxidase subunit VIb-1 | ||||
| 33 | 41 | 18,047 | 4.8 | −0.42 | M | M | M | T03033 | Cytochrome c oxidase subunit Vb | * | |||
| 77 | 13,930 | 9.7 | 0.06 | M | M | M | 3550.m00112 | Cytochrome c oxidase subunit Via | |||||
| 44 | 57,767 | 8.6 | 0.69 | – | – | – | P14578 | Cytochrome c oxidase subunit I | |||||
| Pyruvate and | 53 | 47, 48 | 3 | 52,643 | 7.6 | 0.13 | M | M | M | BAB39219, 2726.m00149 | Dihydrolipoamide dehydrogenase (E3) | * | * |
| 2-OG DH | 2 | 55,896 | 7.4 | 0.08 | M | M | M | 3313.m00119 | Dihydrolipoamide dehydrogenase (E3) | * | * | ||
| complexes | 16, 41 | 39,950 | 4.8 | −0.03 | M | – | M | TC57558, 3493.m00141 | Pyruvate dehydrogenase E1 beta-subunit | * | * | ||
| 66 | 59,591 | 8.7 | −0.17 | M | M | M | 3634.m00180 | Dihydrolipoamide acetyltransferase (E2) | a | * | |||
| 52 | 3 | 48,894 | 9.5 | −0.33 | M | M | M | 2307.m00126 | Dihydrolipoamide succinyltransferase (E2) | a | |||
| 2 | 48,283 | 9.1 | −0.30 | – | – | – | CAD40552 | Dihydrolipoamide succinyltransferase (E2) | a | ||||
| 2 | 123,768 | 6.5 | −0.44 | M | M | M | 4591.m00151 | 2-Oxoglurate dehydrogenase E1 | |||||
| Succinyl-CoA | 48, 22 | 34,246 | 8.4 | 0.08 | M | M | M | 2046.m00126 | Succinyl-CoA ligase alpha-subunit | * | |||
| ligase | 15 | 49 | 3 | 45,091 | 6.2 | −0.03 | M | M | M | 2466.m00139 | Succinyl-CoA ligase beta-subunit | * | |
| Isocitrate DH | 40, 49 | 40,629 | 8.1 | −0.06 | M | M | M | 2447.m00175 | NAD-dependent isocitrate dehydrogenase | * | |||
| Malate DH | 20, 21, 47 | 2 | 35,461 | 8.9 | 0.08 | M | M | M | BAB55686, 2720.m00059 | Malate dehydrogenase | * | * | |
| 53 | 4 | 35,436 | 8.3 | 0.14 | M | – | M | 3321.m00193, 2990.m00151 | Putative malate dehydrogenase | * | * | ||
| Citrate synthase | 58 | 2 | 52,121 | 7.2 | −0.07 | M | M | M | AAG28777, 2942.m00114 | Citrate synthase | * | * | |
| Malic enzyme | 30 | 70,721 | 5.9 | −0.19 | M | M | M | 2119.m00105 | NAD-dependent malic enzyme 62-kD subunit | ||||
| 44 | 57,143 | 8.3 | −0.16 | C | – | – | AAG13628 | NAD-dependent malic enzyme 59-kD subunit | |||||
| Complex II | 37 | 59,103 | 6.2 | −0.40 | – | – | – | 2338.m00147 | Succinate dehydrogenase FP alpha-subunit | * | |||
| Aconitase | 36, 63 | 107,452 | 7.3 | −0.18 | C | – | M | TC61943, 4161.m00311 | Aconitate hydratase | * | |||
| 65 | 98,397 | 5.9−0.15 | – | – | – | TC68934, 5178.m00213 | Aconitate hydratase | * | |||||
| GDC | 34 | 4 | 17,367 | 4.7 | −0.23 | M | M | M | AAK39594 | Gly decarboxylase H protein | * | ||
| 73, 80, 56, 57 | 3 | 29,224 | 6.5 | −0.16 | – | – | – | 2329.m00191 | Aminomethyltransferase precursor (T protein) | * | |||
| 54, 55 | 54,979 | 8.4 | −0.29 | M | M | M | TC52724, 4124.m00171 | Ser hydroxymethyltransferase (SHMT) | * | ||||
| General | 59 | 47,875 | 6.6 | −0.15 | M | M | – | 1207.m00066 | Glutamate dehydrogenase 2 | * | * | ||
| metabolism | 60 | 33 | 40,631 | 6.7 | −0.19 | M | M | M | 2275.m00146 | Glutamate dehydrogenase | * | * | |
| 70 | 2 | 43,762 | 6.8 | −0.18 | M | M | M | 1904.m00155 | NAD-dependent formate dehydrogenase | * | |||
| 50 | 41,243 | 7.4 | −0.21 | M | M | M | BAA77337 | NAD-dependent formate dehydrogenase | * | ||||
| 68 | 1 | 45,255 | 9.6 | −0.21 | – | – | – | 1902.m00219 | Aldehyde dehydrogenase | * | * | ||
| 13 | 59,307 | 6.8 | −0.04 | M | M | M | BAB19052 | Aldehyde dehydrogenase | * | * | |||
| 67 | 2 | 87,073 | 8.4 | −0.10 | M | M | M | 4976.m00233 | Aldehyde dehydrogenase | * | * | ||
| 1 | 56,106 | 5.8 | −0.02 | C | C | – | 4694.m00128 | Aldehyde dehydrogenase | * | * | |||
| 9, 10 | 56,980 | 6.3 | −0.03 | M | M | M | AAC03055 | Methylmalionate semi-aldehyde dehydrogenase | * | * | |||
| 79 | – | – | – | – | – | – | TC59081, TC66764 | Thiosulfate sulfurtransferase | * | ||||
| 38 | 47,495 | 7.8 | −0.25 | M | M | M | JC5125 | Asp transaminase | * | ||||
| 39 | – | – | – | – | – | – | TC54297 | Isovaleryl-CoA dehydrogenase | * | ||||
| 27 | 25,425 | 9.4 | −0.09 | C | C | – | TC62983, 4134.m00168 | Inosine-5′-monophosphate dehydrogenase | * | ||||
| 2 | 252,808 | 6.4 | −0.28 | – | – | – | 4000.m00165 | Acetyl-CoA carboxylase | |||||
| 2 | 36,812 | 8.9 | −0.14 | M | M | M | 2884.m00186 | Formyl transferase | |||||
| 2 | 80,029 | 6.6 | −0.21 | C | – | M | 3385.m00173, 3006.m00143 | Transketolase | |||||
| 2 | 29,775 | 10.6 | −0.32 | – | – | – | 2541.m00142 | Triacylglycerol lipase | |||||
| 46 | 35,327 | 8.2 | −0.09 | – | – | – | 2329.m00193 | (S)-2-hydroxy-acid oxidase | |||||
| Import | 75 | 10,843 | 5.9 | −0.59 | – | – | – | Q9XGX7 | Translocase of inner membrane 9 (TIM9) | ||||
| 57 | 15,677 | 9.0 | 0.12 | – | – | – | 2461.m00155 2506.m00124 | Translocase of inner membrane 17 (TIM17) | |||||
| 34 | 37,956 | 6.8 | −0.13 | – | – | – | 2733.m00140 | Translocase of outer membrane 40 (TOM40) | * | ||||
| 42 | 9665 | 10.2 | −0.01 | M | – | – | 2494.m00117 3845.m00238 | Translocase of outer membrane 9 (TOM9) | |||||
| Transport | 46 | 29,220 | 7.9 | −0.16 | – | M | – | TC48593, CAB82853 | Voltage-dependent anion channel | * | |||
| 45 | – | – | – | – | – | – | TC65294 | Voltage-dependent anion channel | * | ||||
| 18 | 39, 36, 26, 54 | 27,815 | 8.7 | −0.05 | – | C | – | CAC80850, 2981.m00134 | Voltage-dependent anion channel | * | |||
| 45 | 2 | 24,534 | 7.7 | 0.01 | C | C | M | 5186.m00209 | Voltage-dependent anion channel | * | |||
| 2 | 33,663 | 9.8 | −0.12 | – | – | – | 3497.m00176 | Voltage-dependent anion channel | * | ||||
| 36 | 3 | 41,510 | 10.3 | −0.10 | C | – | – | 2506.m00136 2986.m00110 | Adenine nucleotide translocator ANT | b | |||
| 2 | 33,021 | 9.4 | 0.14 | C | – | – | 3698.m00164, 3631.m00202 | Phosphate transporter | b | ||||
| 3 | 32,802 | 10.1 | 0.10 | – | – | – | 3317.m00075 | 2-Oxoglutarate/malate translocator | b | ||||
| Ox stress | 1 | 55 | 3 | 24,947 | 7.0 | −0.10 | S | M | M | AAA57130, Q43008 | Manganese superoxide dismutase | * | * |
| 24 | 20,874 | 8.2 | 0.01 | M | M | M | BAA90363 | Peroxiredoxin | * | ||||
| 57 | 17,146 | 7.5 | −0.03 | M | M | M | 3606.m00327, 3602.m00254 | Adrenodoxin precursor | * | ||||
| Chaperones | 5 | 2 | 83,380 | 8.1 | −0.42 | M | M | M | TC51015, 4256.m00159 | Heat shock protein 70 | * | * | |
| 6, 7 | 5 | 60,850 | 5.6 | −0.08 | M | M | M | 1379.m00089, 5187.m00183 | Chaperonin 60 | * | * | ||
| 3 | 65,276 | 4.8 | −0.10 | – | – | – | 4781.m00152 | Chaperonin 60 | * | ||||
| 78, 25 | 3 | 10,643 | 9.0 | −0.09 | M | C | M | 3664.m00136, T03585 | Chaperonin 10 | * | |||
| 2 | 13,996 | 10.2 | −0.39 | M | M | M | 5140.m00090 | Chaperonin 10 | * | ||||
| 19 | 11 | 30,643 | 7.1 | 0.03 | S | – | – | 2952.m00055 | Prohibitin | * | |||
| 1, 2 | 31,413 | 10.1 | −0.24 | S | – | – | 4127.m00167, BAC07170 | Prohibitin | * | ||||
| Transcription/ | 8 | 48,423 | 6.4 | −0.06 | M | M | M | AAL5626t | Translational elongation factor T0 | * | * | ||
| translation | 35 | 15,547 | 8.9 | −0.59 | C | – | M | CAA05729 | Gly-rich RNA-binding protein | * | |||
| 2 | 97,919 | 10.8 | −0.89 | – | – | – | 3373.m00119 | DEAD box RNA helicase | * | ||||
| 3 | 127,634 | 6.3 | −0.53 | C | – | M | 3952.m00081 | DEAD box RNA helicase | * | ||||
| 2 | 172,773 | 9.1 | −0.58 | – | M | – | AAK54292 | Putative helicase | * | ||||
| 2 | 31,230 | 9.2 | −0.46 | C | – | – | 5028.m00163 | C2H2-type zinc finger protein | |||||
| 2 | 62,012 | 8.8 | 0.04 | M | M | M | AAL59047 | PPR protein | |||||
| 2 | 79,219 | 8.5 | −0.06 | M | M | – | BAB64813 | PPR protein | |||||
| 2 | 184,237 | 8.1 | −0.70 | – | – | – | 2474.m00123, 5076.m00090 | CREB-binding protein | |||||
| Miscellaneous | 69 | 29,437 | 4.2 | −0.53 | M | – | M | 2772.m00163 | Mitochondrial glycoprotein | ||||
| function | 23 | 27,499 | 7.9 | 0.12 | M | M | M | TC65865, 4358.m00206 | Ferripyochelin-binding protein | * | |||
| 44 | 11 | 26,455 | 6.4 | −0.02 | M | M | M | 2030.m00175 | Ferripyochelin-binding protein | * | |||
| 61 | 10 | 23,214 | 6.4 | −0.19 | M | – | – | TC48467, 2868.m00167 | Ferripyochelin-binding protein | * | |||
| 3 | 43 | 67,681 | 5.0 | −0.60 | M | M | M | 2249.m00144 | Mitofilin motor protein | ||||
| 47 | 60,729 | 4.4 | −0.74 | S | – | – | 2296.m00078 | Calnexin-like protein | |||||
| 1 | 72,778 | 4.8 | −0.46 | C | – | – | 2673.m00127 | ADP-ribosylation factor binding protein | |||||
| 2 | 53,280 | 6.5 | −0.30 | – | – | – | 2500.m00126 | Actin | |||||
| 2 | 39,196 | 6.1 | −0.27 | S | – | – | AAM01174 | Wall-associated kinase 2-like protein | |||||
| 2 | 28,078 | 8.3 | −0.37 | – | – | – | BAB92895 | sec14-Like protein | |||||
| 3 | 98,221 | 6.6 | −0.28 | – | – | – | 2251.m00196 | Disease resistance protein | |||||
| 2 | 94,632 | 7.6 | −0.27 | S | – | – | 4193.m00176 | Disease resistance protein | |||||
| Unknown | 42, 23 | 10,945 | 9.4 | −0.40 | – | M | – | 2419.m00150 | Unknown protein | ||||
| function | 56 | 12,914 | 7.3 | −0.21 | C | C | – | 4127.m00174 | Unknown protein | * | |||
| protein | 17 | 7466 | 10.7 | −0.55 | M | M | – | 2468.m00122, 2520.m00121 | Unknown protein | ||||
| 72 | 27,479 | 9.2 | −0.03 | M | M | M | 2284.m00104 | Unknown protein | * | ||||
| 74 | 13,273 | 8.7 | −0.13 | M | C | M | 32.m00149 | Unknown protein | |||||
| 42 | – | – | – | – | – | – | TC48363 | Unknown protein | |||||
| Unknown function | 1 | 134,187 | 5.6 | −0.20 | – | – | – | 3273.m00129 | Unknown protein | ||||
| protein | 1 | 8786 | 9.2 | −0.17 | S | – | – | 3018.m00158 | Unknown protein | ||||
| (continued) | 1 | 8101 | 9.8 | −0.50 | C | – | – | 1987.m00120 | Unknown protein | ||||
| 1 | 19,438 | 5.9 | −0.33 | – | – | – | 4958.m00111 | Unknown protein | |||||
| 3 | 126,829 | 6.7 | −0.21 | – | – | – | 4983.m00113 | Unknown protein | |||||
| 2 | 100,723 | 9.2 | −0.41 | M | M | – | 4638.m00135 | Unknown protein | |||||
| 2 | 78,515 | 10.7 | −0.72 | M | M | M | BAA84788 | Unknown protein | |||||
| 2 | 12,966 | 12.1 | −0.30 | – | C | – | AAL58157 | Unknown protein | |||||
| 3 | 75,587 | 5.8 | 0.01 | – | – | – | AAN06831 | Unknown protein | |||||
| 2 | 57,805 | 5.7 | −0.46 | M | C | – | 2366.m00159, 2275.m00133 | Unknown protein | |||||
| 2 | 48,104 | 8.7 | −0.07 | – | – | – | 2284.m00111 | Unknown protein | |||||
| 2 | 171,963 | 7.4 | −0.63 | – | M | – | 4183.m00149, 3872.m00127 | Unknown protein | |||||
| 2 | 69,967 | 8.7 | −0.37 | C | – | – | 2475.m00089 | Unknown protein | |||||
| 2 | 48,779 | 8.7 | −0.21 | M | M | M | 2631.m00097 | Unknown protein | |||||
| 2 | 129,607 | 8.7 | −0.87 | M | – | – | BAC20757 | Unknown protein | |||||
| 3 | 84,126 | 5.0 | −0.65 | M | – | – | 5113.m00174 | Unknown protein | |||||
| 3 | 21,831 | 12.5 | −1.27 | M | – | – | AAM22746 | Unknown protein |
Proteins were identified either from gel spots derived from IEF/SDS-PAGE (2-D PAGE), BN SDS-PAGE (BN-PAGE), or HPLC separation of total digested mitochondrial samples (LC-MS). Numerals in 2D-PAGE and BN-PAGE columns are spot nos. for reference to Figures 1 and 2. Numerals in the LC-MS column are the no. of peptides matches to each predicted protein sequence. The Mr, pI, and hydropathicity (GRAVY) for each predicted protein sequence are shown along with prediction of targeting by TargetP (TP), Predotar (PT), and MitoProtII (MP). Asterisks in final columns indicate the presence of orthologs in the IEF/SDS-PAGE proteome reports from Arabidopsis (At; Kruft et al., 2001; Millar et al., 2001) and pea (Ps; Bardel et al., 2002). Several other Arabidopsis proteins identified elsewhere by a variety of different electrophoresis techniques have also been included for comparison: a, Sweetlove et al. (2002); b, Millar and Heazlewood (2003); and c, Werhahn and Braun (2002). Spot 62 from IEF/SDS-PAGE was identified as bovine serum albumin, used in the preparation of mitochondria, and has not been included in the table.
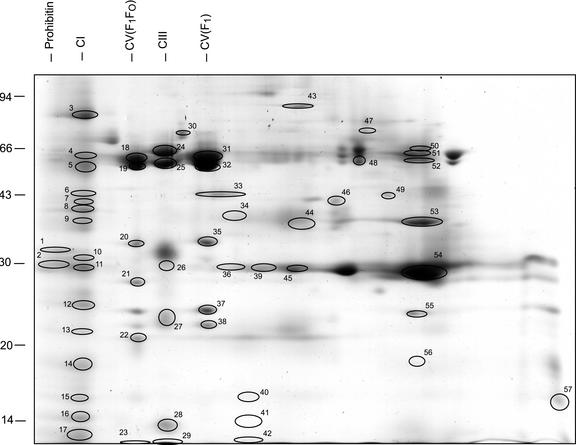
The BN SDS-PAGE uses a combination of native masses of complexes in the first dimension and the denatured masses of polypeptides in the second dimension (Fig. 2). This analysis was only performed on the membrane fraction from rice mitochondria that contains a large number of hydrophobic proteins that were unlikely to be soluble under IEF conditions. Protein staining revealed a series of large complexes of comigrating subunits that constitute the major electron transport chain complexes: complexes I, III, and V (Fig. 2). A set of approximately 100 spots was visible, a set of 89 spots was excised, and a subset of 57 spots from this set was identified as the products of 49 rice genes. There are several examples where a single protein sample contained peptides derived from two different and unrelated gene products; note spot 44 (cytochrome oxidase subunit I and malic enzyme), spot 57 (adrenodoxin and TIM17), and spot 42 (TOM9 and ATP synthase, 6 kD). In all these cases, the MS evidence for the presence of both proteins in very convincing. Because streaking of some proteins occurs during native gel electrophoresis, it is possible that the same proteins do exist more discretely elsewhere on the gel and that our identification point (i.e. the spot excision point) is just coincidently the point of overlap. Comparison of these identified sets from IEF/SDS-PAGE and BN SDS-PAGE revealed that there were 21 nonredundant gene matches in common, whereas together, they represent a combined nonredundant set of 91 gene products. A set of 18 well-known components of mitochondrial inner and outer membrane complexes was identified in the BN SDS-PAGE analysis that was not identified from the IEF/SDS-PAGE analysis (Table II).
Figure 2.
BN SDS-PAGE separation of rice membrane mitochondrial proteins. A total of 89 protein spots from this map were excised, digested, and analyzed by MS/MS. Fifty-seven were identified, and these are numbered for comparison with text and Table II. Numbers on vertical axis are apparent molecular mass of protein subunits (kilodaltons), and annotations on horizontal axis are known mitochondrial protein complexes identified in this study.
Matching of MS/MS spectra from peptides from IEF/SDS-PAGE and BN SDS-PAGE spots to predicted gene products was performed using ProID software (Applied Biosystems, Sydney) with custom-made databases using TIGR rice open reading frame and EST data. Many of the rice proteins identified were not represented at National Center for Biotechnology Information; thus, Mascot on-line (http://www.matrixscience.com) could not be used routinely for analysis. ProID was used with mass accuracy cutoffs of peptide mass ± 0.15 and MS/MS ions ± 0.05, which are significantly more stringent than Mascot default settings. In most cases, more than three matching MS/MS spectra were identified by ProID, but in the case of a number of proteins with low masses (<15 kD), single or pairs of matching MS/MS spectra with very high probabilities of matching (>95%) derived by ProID were evaluated and included in Table II.
LC-MS/MS Analysis of Whole Mitochondrial Protein Extracts
Gel-based arraying of protein samples can exclude proteins from analysis due to hydrophobicity, size, and pI that physically prevents separation in these media. Gel arraying also prevents analysis of peptides that are not easily eluted from polyacrylamide gel samples. The use of reverse-phase chromatography using C18 media provides a gel-free separation of peptides from complex samples. Coupled with on-line MS/MS analysis, the gradual elution of peptides from reverse-phase C18 with organic solvent can be used as a powerful proteomic identification tool. We have taken this approach with a whole-rice mitochondrial protein extract. The identified peptides are matched back to predicted gene products, and the total number of peptides matched provides increasing confidence that the intact gene product was present in the original sample before digestion. Matching of peptides to predicted gene products was again performed using ProID (Applied Biosystems) using mass accuracy cutoffs of peptide mass ± 0.15 and MS/MS ± 0.05. This analysis matched a set of 170 MS/MS spectra back to a set of 72 predicted rice gene products (Table II). For multipeptide matches to a single gene product, a probability cutoff of 85% in ProID was used to filter matches; for single peptide matches, a probability cutoff of 99% in ProID was used for filtering.
A Wide Range of Mitochondrial Functions Identified in Rice
Extensive discussions of many of the components identified here and their biological roles can be found in the earlier reports on the proteomes of Arabidopsis and pea (Kruft et al., 2001; Millar et al., 2001; Bardel et al., 2002). Here, we have largely restricted any extensive comment to components that were not identified in these earlier studies (Table II). The protein sequences for each entry were subjected to BLAST-based similarity analysis to confirm annotation or propose appropriate putative functional annotation for each entry.
Electron Transport Chain
Over 30 components of the five electron transport chain complexes from rice were identified. Many of these samples were from BN SDS-PAGE (Fig. 2). This has now allowed the clear identification of the native complexes separating by this technique and noted on the horizontal axis of Figure 2. A significant number of these identifications, although clear orthologs to components of these complexes in other eukaryotic species, have not been reported previously in plant proteome mapping studies. These newly identified components include two subunits of ATP synthase, seven subunits of complex I, and three subunits of complex IV (Table II).
Carbon Metabolism
At least one component of each of the seven tricarboxylic acid cycle enzymes were identified in Table II along with components of the associated pyruvate dehydrogenase complex and the NAD-malic enzyme. The two distinct NAD-malic enzyme subunits have been identified by antibody reaction on IEF/SDS-PAGE from potato (Solanum tuberosum; Jenner et al., 2001) but were not revealed by proteome mapping studies in Arabidopsis or pea. Here, the two subunits were both found on BN SDS-PAGE and appear to be present in separate high-molecular mass complexes. The larger subunit is in a complex with a mass over 300 kD based on its presence between complex III and separated FO of the ATP synthase (Fig. 2). The E1 subunit of 2-oxoglutarate dehydrogenase complex was not identified in the Arabidopsis or pea reports, perhaps due to its large mass (Table II). Three components associated with Gly metabolism in mitochondria, the H and T subunits of Gly decarboxylase and the Ser hydroxymethyltransferases, were identified, even though these rice mitochondria were isolated from dark-grown shoots and the photorespiratory requirement for Gly metabolism was absent. A series of 18 enzymes involved in amino acid metabolism (Glu dehydrogenase, Asp transaminase, aldehyde dehydrogenases, and methylmalonate semi-aldehyde dehydrogenase), C1 metabolism (formate dehydrogenase), nucleotide metabolism (inosine-5-monophosphate dehydrogenase and formyl transferase), and miscellaneous metabolic functions were identified. Based on sequence similarity, the formyl transferase is mostly likely a phosphoribosylglycinamide formyl transferase (GART) involved in purine biosynthesis. The potential for plant mitochondria to facilitate nucleotide metabolism has been reported previously (Atkins et al., 1997; Smith et al., 1998). The acetyl-CoA carboxylase is normally involved in long-chain fatty acid synthesis, and although this is commonly considered to be a plastidic function in plants, both mammalian and yeast (Saccharomyces cerevisiae) mitochondria contain this enzyme (Schon, 2001).
Protein and Metabolite Transport
Four components of the import apparatus were identified, two TIM components (subunits 9 and 17) and two from the outer membrane TOM complex (subunits 9 and 40). The latter comigrated during BN electrophoresis (Fig. 2). In Arabidopsis, the TOM complex has to be extensively purified to see its subunits on BN-PAGE (Werhahn and Braun, 2002). Interestingly, the TOM9 and TOM40 subunits represent the core of the TOM complex in yeast, to which other subunits are peripherally attached. A series of specific gene products for the outer membrane voltage-dependent anion channels (porins) were identified, along with three members of the carrier family on the inner mitochondrial membrane responsible for the exchange of inorganic phosphate, ATP/ADP, and dicarboxylates. The Arabidopsis ortholog of the dicarboxylate carrier has been shown recently to be a broad dicarboxylate/tricarboxylate carrier capable of transporting many of the tricarboxylic acid cycle intermediates that enter and exit plant mitochondria (Picault et al., 2002). These carriers were identified mainly through LC-MS/MS, which is in agreement with the difficulty of identifying these proteins in two-dimensional gel separations from Arabidopsis (Millar and Heazlewood, 2003). None of these carrier proteins were identified in the IEF/SDS-PAGE approaches in Arabidopsis or pea (Millar et al., 2001; Bardel et al., 2002).
Miscellaneous Functions
The mitochondrial-specific superoxide dismutase and the redox carriers, peroxiredoxin and adrenodoxin, identified in Arabidopsis mitochondrial preparations, were also identified here in rice. Members of the major mitochondrial molecular chaperone classes (70, 60, and 10) and the prohibitins were also found. A series of transcription/translation-related proteins were identified, including a translation elongation factor, a Gly-rich RNA-binding protein, and a set of RNA helicases, like those observed in Arabidopsis (Kruft et al., 2001; Millar et al., 2001). In addition, two members of the PPR (pentatricopeptide repeat) family have been found, along with members of the CH2H2 zinc finger and CREB-binding families putatively involved in protein-RNA interactions. Proteins from the PPR class have been implicated in nuclear restoration of cytoplasmic male sterility in plants (Bentolila et al., 2002) and belong to a large family of proteins that may be responsible for the control of RNA stability and editing in plant mitochondria (Small and Peeters, 2000). To our knowledge, this is the first time these proteins have been definitively identified in plant mitochondrial isolates. A series of miscellaneous function proteins include homologs of the ferripyochelin-binding proteins that were also identified in Arabidopsis (Millar et al., 2001). Homologs of the known mitochondrial acid glycoprotein in yeast, Mam33p (Seytter et al., 1998), the coiled-coil mitofilin motor protein for cytoskeleton attachment to mitochondria (Odgren et al., 1996), along with actin itself, were also identified. A set of 23 unknown function proteins have been found for which no clear similarity based on sequence can be proposed, but a very significant number contain predicted mitochondrial-targeting presequences (Table II).
Discussion
Comparison of the Three Proteome Datasets
Each separation technique identified a series of proteins contributing to a total of 136 nonredundant rice proteins (Table II). Within the total set, each separation technique matched 38% to 53% of the identified proteins. Using the predicted protein sequences, a series of parameters were calculated that included physical characteristics such as molecular mass, pI, and hydrophobicity (GRAVY), and predicted intercellular targeting based on N-terminal sequence as determined by Predotar, TargetP, and MitoProtII. These data were then used for further analysis of the proteome set as outlined below.
More Hydrophobic, Large, and Basic Proteins Were Identified by BN SDS-PAGE and LC-MS Than by IEF/SDS-PAGE Separations
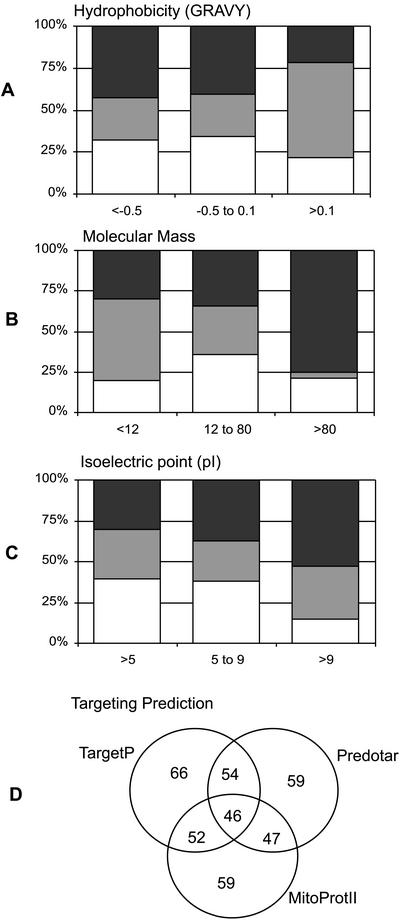
When the protein identifications made by each technique are considered in the context of physical properties of the predicted proteins, such as hydrophobicity, molecular mass, and pI, an interesting pattern emerges (Fig. 3, A–C). A GRAVY score is a single-value indication of the overall hydropathicity of a protein sequence based on the Kyte and Doolittle algorithms; the more positive the score, the more hydrophobic the overall sequence (Kyte and Doolittle, 1982). The addition of thiourea as a chaotropic agent, the advent of new nonionic sulfobetaine detergents, and the use of the non-charged reductant, tributylphosphine, have all been credited with allowing more hydrophobic proteins to enter two-dimensional gels for separation and analysis (Chevallet et al., 1998; Rabilloud, 1998). To date, proteins with GRAVY scores of approximately +0.25 to +0.30 are considered to represent the upper limit of these two-dimensional gel solubility improvements (Herbert, 1999). Our analysis has revealed that the highest GRAVY score for a rice mitochondrial protein identified from IEF/SDS-PAGE was +0.14 for the ATP synthase delta subunit. In BN SDS-PAGE, GRAVY scores reached +0.84 in the case of subunit 9 of the ATP synthase. Overall, there was little difference in the proportion of identifications achieved by the three separation techniques at GRAVY < +0.1, but above this level, more than 60% of the identifications were achieved from BN SDS-PAGE (Fig. 3A).
Figure 3.
Comparison of identified rice protein sets. Proportion of rice proteins identified by the three separation techniques (IEF/SDS-PAGE, clear; BN SDS-PAGE, gray; and LC-MS, black) in different classes of hydrophobicity (A), molecular mass (B), and pI (C). D, Overlapping of targeting prediction of the rice proteins identified by TargetP, Predotar, and MitoProtII. Numbers in each circle indicate numbers predicted either by one, two, or all three predictors.
Proteins with large molecular masses can be separated by single-dimension SDS-PAGE. Proteins as large as 200 to 300 kD can be routinely separated using gradient gels. However, such large proteins are not very soluble during IEF and as a result, large proteins (>100 kD) are rarely seen on IEF/SDS-PAGE gels. The proportion of identifications of proteins greater than 80 kD was very significantly shifted toward the LC-MS/MS separations, where size of the parent protein was not an impediment to separation and analysis (Fig. 3B). The basicity of proteins has often restricted identification from IEF/SDS-PAGE due to the difficulty of preparing stable gradients in excess of pH 10 for separation of such proteins. Clearly, in our analysis, it is both the non-IEF gel-separating technique of BN SDS-PAGE and the non-gel LC MS/MS analysis that dominate the identifications of proteins with pI values greater than pH 9 (Fig. 3C).
These analyses clearly indicate that to fully characterize mitochondrial proteomes, a series of experimental approaches will be required to maximize the representation of all proteins without bias based on physical characteristics of size, charge, and hydropathicity. LC MS/MS does not appear to be the complete answer, although when many biases are eliminated, a great deal of information about the parent protein from which peptides are derived is lost. Furthermore this technique appears to selectively not identify some proteins present in two-dimensional gel separation of the same material (Table II). Using different gel-based approaches, information on possible posttranslational modifications (such as the multiple spots for malate dehydrogenase, T protein of GDC, alpha-subunit of ATP synthase, and E1β-subunit of pyruvate dehydrogenase subunit; Fig. 1; Table II) and protein-protein interactions (such as components in the electron transport chain and import translocase complexes; Fig. 2; Table II) can be uncovered.
Targeting Prediction of the Rice Proteome Set
The predicted protein sequences of the identified rice proteome were analyzed by three targeting prediction programs commonly used to assess mitochondrial localization in plants (Table II). Five entries in Table II were matched only to rice TCs. It is unknown whether these represent full or partial sequences; thus, they have been excluded from the analysis. Each prediction program assessed that approximately 50 to 60 of the protein products contained N-terminal extensions for mitochondrial targeting. In total, 77 of the 130 protein sequences were predicted to be mitochondrial by at least one targeting program. Looking at the overlapping predictions (Fig. 3D), TargetP identified the largest number of proteins, and the combination of TargetP and Predotar provided the best pair for prediction in rice. In the list of 39 proteins not assessed to be mitochondrial targeted by any program, a large number (19) are well-known mitochondrial proteins involved in the electron transport chain, tricarboxylic acid cycle, import apparatus, and mitochondrial transport. This lack of prediction may be explained either by the absence of targeting presequences (as has been documented for carrier and import components; Millar and Heazlewood, 2003), cryptic presequences not recognized by these program, and/or the lack of presequences due to incorrect annotation of the exons making up N-terminal sequences of these predicted proteins (Frishman et al., 1999). A further three proteins are products of mitochondrial-encoded genes (α-subunit ATP synthase, COXI, and NAD9); thus, they do not require targeting presequences. A total of 11 of the unknown function proteins identified were predicted as mitochondrial targeted by at least one prediction program.
Comparison of Rice, Arabidopsis, and Pea Mitochondrial Proteomes
A comparison between the proteins found here in rice mitochondria and those previously discovered in Arabidopsis (Kruft et al., 2001; Millar et al., 2001) and pea mitochondria (Bardel et al., 2002) is presented in the final two columns of Table II. An asterisk in the second-to-last column indicates that an ortholog was identified in Arabidopsis among the 91 nonredundant set defined by the two Arabidopsis proteome reports and several recent publications containing small sets of proteins. Overall, this analysis indicates that 71 proteins have been found in the mitochondrial proteomes of both species, including two of unknown function. A further set of 28 of the identified rice proteins represent orthologs of well-known mitochondrial proteins in other eukaryotic systems but have not be identified in the published Arabidopsis proteome to date. An asterisk in the last column indicates that an ortholog was found in the nonredundant set of 48 proteins found to date in pea mitochondria. A total of 28 proteins were found in both rice and pea mitochondria. Six of these proteins were not observed in Arabidopsis mitochondria. This leaves a set of 35 proteins, of which 21 have no predicted function, that have been exclusively identified in rice mitochondria to date. This demonstrates the necessity to compare mitochondrial proteomes from different plant species, and most certainly between the major lineages of higher plants, to better understand the complex role of these organelles in the plant cell. However, until a much more complete survey of the proteomes of mitochondria in several plants is conducted in similar tissue types using more similar protein arraying and identification technology, it will be difficult to truly discuss the presence and/or absence of specific proteins between plant species with any accuracy. Currently, many absences in the published Arabidopsis and pea sets reflect technical issues rather than biological absence. Here, we highlight the rice mitochondrial protein complement and present the tools that will be required for such comparative plant proteome investigations in the future.
MATERIALS AND METHODS
Growth of Rice Seedlings
Batches of 200 g of rice (Oryza sativa cv Amaroo) seed were washed in 1% (w/v) bleach for 10 min, rinsed in distilled water, and grown in the dark in vermiculite trays (30 × 40cm) at a constant 30°C, watered daily, and shoot tissue harvested at 7 d for mitochondrial isolation.
Rice Mitochondrial Isolation
Rice shoots were harvested using scissors and cut into 5- to 10-mm lengths into a beaker in ice. Cut shoots (100 g) were ground in a precooled mortar and pestle using acid-washed sand in 300 mL of homogenization solution (0.3 m Suc, 25 mm tetrasodiumpyrophosphate, 2 mm EDTA, 10 mm KH2PO4, 1% [w/v] polyvinylpyrrolidone-40, 1% [w/v] bovine serum albumin, and 20 mm ascorbate [pH 7.5]). After filtering homogenate through four layers of Miracloth (Calbiochem, San Diego), it was centrifuged for 5 min at 1,000g, and the supernatant was centrifuged again at 20,000g for 20 min. The resultant organelle pellet was resuspended in wash buffer (0.3 m Suc, 10 mm TES-KOH [pH 7.5], and 0.1% [w/v] bovine serum albumin), and the two centrifugations steps were repeated to produce a washed organelle pellet. This resuspended pellet was layered over 0% to 4.4% (w/v) polyvinylpyrrolidone-40 preformed gradient in a 28% (v/v) Percoll self-forming gradient in wash buffer and centrifuged at 30,000g for 45 min. Mitochondria formed a band toward the bottom of the gradient, the upper plastid material was discarded, and the mitochondrial band was removed in an attempt to minimize contamination of peroxisomal material from the bottom of the gradient. After two wash centrifugation steps at 20,000g for 15 min, the pellet was layered over a second self-forming gradient consisting of 45% (v/v) Percoll in wash buffer and centrifuged for 30 min at 30,000g. Mitochondria remained near the top of the gradient, whereas peroxisomal material migrated to the bottom of the gradient. The mitochondrial layer was aspirated and concentrated by wash centrifugation steps at 20,000g for 15 min.
Respiratory Measurements
O2 consumption was measured in a Clark-type O2 electrode in 1 mL of reaction medium containing: 0.3 m mannitol, 10 mm TES-KOH (pH 7.5), 5 mm KH2PO4, 10 mm NaCl, 2 mm MgSO4 and 0.1% (w/v) bovine serum albumin. Pyruvate (5 mm), malate (0.5 mm), succinate (10 mm), NADH (1 mm), ADP (0.5 mm), and myxothiazol (5 μm) were added as indicated to modulate O2 consumption rates. Cytochrome c oxidase activity was measured as ascorbate (5 mm) and cytochrome c (25 μm)-dependent O2 consumption in the presence of 0.05% (w/v) Triton X-100. Outer membrane integrity was assayed as the latency of cytochrome c oxidase activity (Neuberger, 1985).
Two-Dimensional Gel Electrophoresis
Mitochondria protein samples (500 μg) were acetone extracted by addition of acetone to a final concentration of 80% (v/v) at −80°C. Samples were stored at −20°C for 4 h and then centrifuged at 20,000g for 15 min. The pellets were resuspended in an IEF sample buffer consisting of: 6 m urea, 2 m thiourea, 2% (w/v) CHAPS, 2% (v/v) ampholytes (pH 3–10), 2 mm tributylphosphine, and 0.001% (w/v) bromphenol blue. Aliquots of 360 μL were used to reswell dried 180-mm, nonlinear immobilized pH gradient strips (pH 3–10; Immobiline DryStrips, APBiotech, Sydney) overnight, and then IEF was performed for 19.5 h, reaching a total of 49 kV h−1 at 20°C on a flat-bed electrophoresis unit (Multiphor II, APBiotech). Immobilized pH gradient strips were then transferred to an equilibration buffer consisting of 50 mm Tris-HCl (pH 6.8), 4 m urea, 2% (w/v) SDS, 0.001% (w/v) Bromphenol blue, and 100 mm mercaptoethanol and incubated for 20 min with rocking. The equilibrated strips were then slotted into central single wells of 4% (w/v) acrylamide stacking gels above 0.1- × 18.5- × 20-cm, 12% (w/v) acrylamide, 0.1% (w/v) SDS-polyacrylamide gels. Strips were overlaid with 0.5% (w/v) agarose in SDS-PAGE running buffer. Gel electrophoresis was performed at 25 mA per gel with circulating cooling and completed in 5 h. Proteins were visualized by colloidal Coomassie (G250) staining. Mr and pI standards from APBiotech were used to confirm fixed pH gradient positioning on first dimension separation and to identify apparent molecular masses on second dimension separation.
BN PAGE/SDS-PAGE
BN PAGE was performed largely according to Jansch et al. (1996). Gels consisted of a separating gel (5%–15% [w/v] acrylamide) and a stacking gel (4% [w/v] acrylamide) were formed in a solution of 0.25 m ε-amino-n-caproic acid and 25 mm Bis-Tris-HCl (pH 7.0). The anode buffer consisted of 50 mm Tricine, 15 mm Bis-Tris-HCl, 0.02% (w/v) Blue G250 (pH 7.0), and the cathode buffer of 50 mm Bis-Tris-HCl (pH 7.0). Aliquots of 1 mg of membrane protein were suspended in 75 μL of an amino caproic acid buffer solution (containing 0.75 m ε-amino-n-caproic acid, 0.5 mm Na2EDTA, and 50 mm Bis-Tris-HCl [pH 7.0]) followed by the addition of 15 μL of a freshly prepared solution of 10% (w/v) n-dodecylmaltoside. Afterward, 10-min centrifugation at 20,000g was performed. Fifteen microliters of 5% (w/v) Blue G250 was added to the supernatants (dissolved in amino caproic acid buffer). Gels were run at 4°C in a precooled apparatus with all samples, and buffers were precooled to 4°C. Approximately 30 min before the run, 0.03% (w/v) n-dodecylmaltoside was added to the cathode buffer only. Electrophoresis was commenced at 100-V constant voltage for 45 min and then increased to 15 mA without voltage limitation for 5 h. BN lanes were cut from gels, equilibrated in standard SDS-PAGE sample buffer for 30 min, laid horizontally on 12% (w/v) acrylamide SDS-PAGE separating gels, sealed in with 0.5% (w/v) agarose, and electrophoresis was continued for 5 h under standard conditions.
Trypsin Digestion of Gel Plugs and Whole Mitochondrial Extracts
Protein spots to be analyzed were cut from two-dimensional PAGE gels, destained, dried at 50°C, and digested with trypsin according to Sweetlove et al. (2001). Aliquots of 50 μg of purified rice mitochondrial protein were acetone precipitated, and the protein pellets were air dried. A digestion solution consisting of 100 mm Tris-HCl (pH 8.6) and 50 μg mL−1 trypsin was added to a total volume of 45 μL, and the digestion mixture was incubated for 16 h at 37°C.
Quadrupole TOF MS
Quadrupole TOF MS/MS was performed on an Applied Biosystems Q-STAR Pulsar using an IonSpray source. Samples from gel plugs were directly injected into the electrospray source in 50% (v/v) methanol/0.1% (v/v) formic acid, and selected doubly charged peptides, identified in MS-TOF mode, were fragmented by N2 collision and analyzed by MS/MS using IDA software (Applied Biosystems, Foster City, CA). Samples from digestion of whole extracts were bound onto a microbore HPLC C18 column (Agilent, Palo Alto, CA) and eluted over 6 h with a linear acetonitrile gradient from 2% to 80% (v/v) in water. Mass spectra and collision MS/MS data were analyzed with Analyst QS and BioAnalyst software (Applied Biosystems).
Identification of Database Entry Matches and Sequence Analysis
TIGR draft rice genome and rice EST gene indices and rice National Center for Biotechnology Information entries were used for sequence matches. Predotar (http://www.inra.fr/Internet/Produits/Predotar/), TargetP (http://www.cbs.dtu.dk/services/TargetP/), and MitoProtII (http://www.mips.biochem.mpg.de/cgi-bin/proj/medgen/mitofilter) predictions were determined using full-length predicted protein sequences. Molecular mass, pI, and GRAVY scores were determined using the ProtParam program on an ExPASy Web site (http://au.expasy.org).
Footnotes
This work was supported by the Australian Research Council Discovery Program (to A.H.M. and J.W.). K.A.H. was a recipient of the Eric Cyril Lawrence Medical Research Scholarship.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.018986.
LITERATURE CITED
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Atkins CA, Smith PMC, Storer PJ. Re-examination of the intracellular localisation of de novo purine synthesis in cowpea nodules. Plant Physiol. 1997;113:127–135. doi: 10.1104/pp.113.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardel J, Louwagie M, Jaquinod M, Jourdain A, Luche S, Rabilloud T, Macherel D, Garin J, Bourguignon J. A survey of the plant mitochondrial proteome in relation to development. Proteomics. 2002;2:880–898. doi: 10.1002/1615-9861(200207)2:7<880::AID-PROT880>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Bartoli C, Pastori G, Foyer C. Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol. 2000;123:335–343. doi: 10.1104/pp.123.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Alfonso AA, Hanson MR. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci USA. 2002;99:10887–10892. doi: 10.1073/pnas.102301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallet M, Santoni V, Poinas A, Rouquie D, Fuchs A, Kieffer S, Rossignol M, Lunardi J, Garin J, Rabilloud T. New zwitterionic detergents improve the analysis of membrane proteins by two-dimensional electrophoresis. Electrophoresis. 1998;19:1901–1909. doi: 10.1002/elps.1150191108. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Considine M, Holtzapffel RC, Day DA, Whelan J, Millar AH. Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiol. 2002;129:949–953. doi: 10.1104/pp.004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly DC, Cameron KM, Stevenson DW. Plant systematics in the age of genomics. Plant Physiol. 2001;127:1328–1333. [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Fey J, Marechal-Drouard L. Compilation and analysis of plant mitochondrial promoter sequences: an illustration of a divergent evolution between monocot and dicot mitochondria. Biochem Biophys Res Commun. 1999;256:409–414. doi: 10.1006/bbrc.1999.0349. [DOI] [PubMed] [Google Scholar]
- Frishman D, Mironov A, Gelfand M. Starts of bacterial genes: estimating the reliability of computer predictions. Gene. 1999;234:257–265. doi: 10.1016/s0378-1119(99)00200-0. [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Prestig G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al. A draft sequence of the rice genome (Oryza sativaL. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Gray M, Burger G, Lang F. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Gueguen V, Macherel D, Jaquinod M, Douce R, Bourguignon J. Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J Biol Chem. 2000;275:5016–5025. doi: 10.1074/jbc.275.7.5016. [DOI] [PubMed] [Google Scholar]
- Herbert B. Advances in protein solubilisation for two-dimensional electrophoresis. Electrophoresis. 1999;20:660–663. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<660::AID-ELPS660>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Herbert BR, Molloy MP, Gooley AA, Walsh BJ, Bryson WG, Williams KL. Improved protein solubility in two-dimensional electrophoresis using tributyl phosphine as reducing agent. Electrophoresis. 1998;19:845–851. doi: 10.1002/elps.1150190540. [DOI] [PubMed] [Google Scholar]
- Jansch L, Kruft V, Schmitz UK, Braun HP. New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J. 1996;9:357–368. doi: 10.1046/j.1365-313x.1996.09030357.x. [DOI] [PubMed] [Google Scholar]
- Jenner HL, Winning BM, Millar AH, Tomlinson KL, Leaver CJ, Hill SA. NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol. 2001;126:1139–1149. doi: 10.1104/pp.126.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruft V, Eubel H, Jansch L, Werhahn W, Braun HP. Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 2001;127:1694–1710. [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle R. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Millar AH, Heazlewood JL. Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis thaliana. Plant Physiol. 2003;131:443–453. doi: 10.1104/pp.009985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giege P, Leaver CJ. Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 2001;127:1711–1727. [PMC free article] [PubMed] [Google Scholar]
- Neuberger M. Preparation of plant mitochondria, criteria for assessment of mitochondrial integrity and purity, survival in vitro. In: Douce R, Day D, editors. Higher Plant Cell Respiration. Berlin: Springer-Verlag; 1985. pp. 7–24. [Google Scholar]
- Odgren PR, Toukatly G, Bangs PL, Gilmore R, Fey EG. Molecular characterisation of mitofilin (HMP), a mitochondrial associated protein with predicted coiled coil and intermembrane space targeting domains. J Cell Sci. 1996;109:2253–2264. doi: 10.1242/jcs.109.9.2253. [DOI] [PubMed] [Google Scholar]
- Picault N, Palmieri L, Pisano I, Hodges M, Palmieri F. Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria. J Biol Chem. 2002;277:24204–24211. doi: 10.1074/jbc.M202702200. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Lee J, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, Zimmer EA, Chen Z, Savolainen V, Chase MW. The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes. Nature. 1999;402:404–407. doi: 10.1038/46536. [DOI] [PubMed] [Google Scholar]
- Rabilloud T. Use of thiourea to increase the solubility of membrane proteins in two-dimensional electrophoresis. Electrophoresis. 1998;19:758–760. doi: 10.1002/elps.1150190526. [DOI] [PubMed] [Google Scholar]
- Rebeille F, Macherel D, Mouillon JM, Garin J, Douce R. Folate biosynthesis in higher plants: purification and molecular cloning of a bifunctional 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase/7,8-dihydropteroate synthase localized in mitochondria. EMBO J. 1997;16:947–957. doi: 10.1093/emboj/16.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni V, Kieffer S, Desclaux D, Masson F, Rabilloud T. Membrane proteomics: use of additive main effects with multiplicative interaction model to classify plasma membrane proteins according to their solubility and electrophoretic properties. Electrophoresis. 2000;21:3329–3344. doi: 10.1002/1522-2683(20001001)21:16<3329::AID-ELPS3329>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Santoni V, Rabilloud T, Doumas P, Rouquie D, Mansion M, Kieffer S, Garin J, Rossignol M. Towards the recovery of hydrophobic proteins on two-dimensional electrophoresis gels. Electrophoresis. 1999;20:705–711. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<705::AID-ELPS705>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Schagger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligometric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- Schon EA. Gene products present in mitochondria of yeast and animal cells. In: Pon LA, Schon EA, editors. Mitochondria. 65, Methods in Cell Biology. San Diego: Academic Press; 2001. pp. 463–482. [DOI] [PubMed] [Google Scholar]
- Seytter T, Lottspeich F, Neupert W, Schwarz E. Mam33p, an oligomeric, acidic protein in the mitochondrial matrix of Saccharomyces cerevisiaeis related to the human complement receptor gC1q-R. Yeast. 1998;14:303–310. doi: 10.1002/(SICI)1097-0061(19980315)14:4<303::AID-YEA217>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Small ID, Peeters N. The PPR motif: a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- Smith PMC, Mann AJ, Goggin DE, Atkins CA. Air synthetase in cowpea nodules: a single gene product targeted to two organelles. Plant Mol Biol. 1998;36:811–820. doi: 10.1023/a:1005969830314. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE, Chase MW. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature. 1999;402:402–404. doi: 10.1038/46528. [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH. The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 2002;32:891–904. doi: 10.1046/j.1365-313x.2002.01474.x. [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Mowday B, Hebestreit HF, Leaver CJ, Millar AH. Nucleoside diphosphate kinase III is localized to the inter-membrane space in plant mitochondria. FEBS Lett. 2001;16:272–276. doi: 10.1016/s0014-5793(01)03069-1. [DOI] [PubMed] [Google Scholar]
- Thelen JJ, Muszynski MG, David NR, Luethy MH, Elthon TE, Miernyk JA, Randall DD. The dihydrolipoamide S-acetyltransferase subunit of the mitochondrial pyruvate dehydrogenase complex from maize contains a single lipoyl domain. J Biol Chem. 1999;274:21769–21775. doi: 10.1074/jbc.274.31.21769. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR. Large scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Werhahn W, Braun HP. Biochemical dissection of the mitochondrial proteome from Arabidopsis thalianaby three-dimensional gel electrophoresis. Electrophoresis. 2002;23:640–646. doi: 10.1002/1522-2683(200202)23:4<640::AID-ELPS640>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al. A draft sequence of the rice genome (Oryza sativaL. ssp indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]