Abstract
BeetleBase (http://www.bioinformatics.ksu.edu/BeetleBase/) is an integrated resource for the Tribolium research community. The red flour beetle (Tribolium castaneum) is an important model organism for genetics, developmental biology, toxicology and comparative genomics, the genome of which has recently been sequenced. BeetleBase is constructed to integrate the genomic sequence data with information about genes, mutants, genetic markers, expressed sequence tags and publications. BeetleBase uses the Chado data model and software components developed by the Generic Model Organism Database (GMOD) project. This strategy not only reduces the time required to develop the database query tools but also makes the data structure of BeetleBase compatible with that of other model organism databases. BeetleBase will be useful to the Tribolium research community for genome annotation as well as comparative genomics.
INTRODUCTION
The red flour beetle (Tribolium castaneum) provides an excellent genetic model system for Coleoptera, the largest and most diverse order of eukaryotic organisms. Coleoptera includes many economically important species of crop pests causing major agricultural losses. Similar to Drosophila in the order Diptera, Tribolium has characteristics desired in a genetic model organism including ease of culture, short generation time, large brood sizes and efficacy of genetic manipulation. The potential of Tribolium for genetic analysis has been demonstrated through classical mutational studies (1,2), whole-genome molecular mapping (3) and RNA interference (4–6). Molecular genetic and genomic studies in Tribolium have been greatly facilitated by the recent completion of the genome sequence at the Human Genome Sequencing Center, Baylor College of Medicine. The genome sequence is currently being annotated by the Tribolium research community. In addition, large sets of expressed sequence tags (ESTs) have been generated from stage- and tissue-specific cDNA libraries by members of the Tribolium genome consortium. The sequence data provide useful information for identifying and characterizing the organization and function of beetle genes as well as their orthologues in other insect species. In particular, Tribolium is probably the most efficient model system for performing functional analysis of genes lost in the Drosophila lineage but conserved in other insects. Beetles (Coleoptera) and flies (Diptera) diverged close to 300 million years ago (7). Although Coleoptera is considered to occupy a basal phylogenetic position, Diptera is one of the most advanced insect orders and there is evidence that gene sequences in Drosophila may have evolved rapidly (7,8). As genome sequence data become available for Tribolium and other insect species, comparative genomics may reveal the genetic innovations that accompanied the evolution of higher insects.
The rapid expansion of genomic research in Tribolium calls for a centralized database resource for data curation and integration. BeetleBase is developed to fill this role by providing searchable interfaces to access a variety of Tribolium data, including sequences, genes, genetic markers, mutants, publications and links to other databases. Various datasets have been collected from different sources and integrated in BeetleBase after curation. Importantly, BeetleBase implements the Chado data model developed by the Generic Model Organism Database (GMOD) project (http://www.gmod.org/). This should facilitate the future expansion of BeetleBase to include additional data types (e.g. microarray gene expression data) and enhance its interoperability with other genome databases to support comparative genomics.
DATA ACQUISITION AND ANALYSIS
The current data entries in BeetleBase are summarized in Table 1. The datasets have been collected from public databases and the Tribolium research community. Assembled genomic sequence contigs were obtained from the Human Genome Sequencing Center (HGSC), Baylor College of Medicine. The genome has been sequenced to 7-fold coverage using a whole-genome shotgun approach. Currently, experts of the Tribolium research community are working with HGSC scientists to manually confirm and curate a subset of the predicted genes. We have been participating in the genome annotation, and provided 9162 protein-coding genes that were predicted using the FGENESH program (9). This set of predicted genes are currently stored in BeetleBase, but will be replaced by information transferred from the HGSC at Baylor upon completion of the sequencing project.
Table 1.
Data content in BeetleBase (August 2006)
| • 2341 genomic sequence contigs (226 contigs with >100 kb) |
| • 9162 predicted genes with CDS and protein sequences |
| • 439 GenBank records |
| • 28785 BAC-end sequences |
| • 11254 ESTs aligned to genomic sequence contigs |
| • 424 genetic markers and their mapping results |
| • 81 mutants with phenotype descriptions |
| • 423 genetic stocks |
| • 615 PubMed references |
| • 18327 Gene Ontology terms |
| • 956 Sequence Ontology terms |
Tribolium ESTs and complete cDNA sequences were retrieved from NCBI (http://www.ncbi.nlm.nih.gov/). These expressed sequences were aligned to the genomic sequence contigs using the BLAST program (10). A Perl program was developed to parse the BLAST search results for meaningful alignments. Both the expressed sequence data and the analytical results are stored in BeetleBase. The same approach has also been used to align bacterial artificial chromosome (BAC)-end sequences to the genomic sequence contigs. Although the BLAST-based approach has worked well for EST and BAC-end sequence alignment to genomic sequences, other available tools (11–13) will also be tested and compared with our approach in the future.
Besides the sequence data, genetic markers and mapping results were extracted from a recent publication (3). Information about genetic stocks and descriptions of mutant phenotypes were obtained from the Tribolium Mutant Database (http://bru.gmprc.ksu.edu/proj/tribolium/). BeetleBase also stores PubMed references related to Tribolium research. In addition, two sets of controlled vocabulary terms are used, Gene Ontology (GO) terms (http://www.geneontology.org/) for gene functional annotations and Sequence Ontology (SO) terms (http://song.sourceforge.net/) for specifying database object types. Use of standard terms for information indexing is an important component of the Chado data model.
SYSTEM DESIGN AND IMPLEMENTATION
The BeetleBase system consists of web interfaces, Perl CGI programs and a relational database. BeetleBase uses the Chado data model (http://www.gmod.org/schema/) and the MySQL database management system (http://www.mysql.com/). CGI programs have been developed to access the database in response to user queries and then generate web pages to present the query results. In addition, GMOD generic software components are used for data visualization (see below). Use of the GMOD software components allowed us to focus on data processing and management.
Perl programs have been developed to load the various datasets into BeetleBase. Chado is designed as a modular schema so that new modules can be added for new data types. In Beetlebase, data have been populated in five modules, including the Sequence module for all the sequence-related data, the Genetic module for mutant phenotypes, the Organism module for taxonomic data, the Publication module for PubMed references, and the Controlled Vocabulary module for GO and SO terms. For future expansion of BeetleBase, the Expression module will be used to store microarray gene expression data, and the Companalysis module as well as the Organism module may be needed to support comparative genomics. The database was implemented within six months by a bioinformatics specialist assisted by three part time graduate research assistants.
DATABASE QUERY TOOLS
BeetleBase provides search forms for sequences, genes, genetic markers, mutants, stocks and publications. The search results are summarized in a tabular format. Table entries may be clicked to retrieve more information from BeetleBase. The data entries are also linked to external databases if available. For example, published cDNA sequences are referred to GenBank while Tribolium references are linked to PubMed.
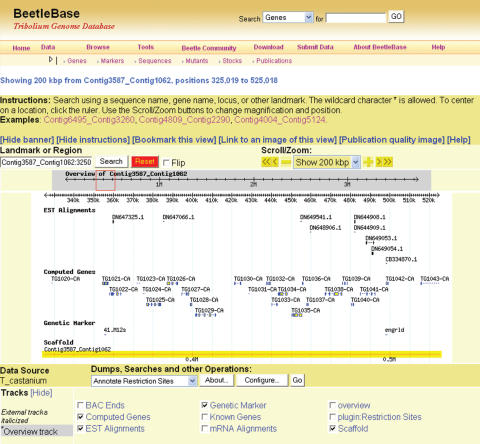
The GMOD software tools, GBrowse (14) and CMap (http://www.gmod.org/cmap/), are used to visualize sequence and mapping data in BeetleBase. As shown in Figure 1, GBrowse is used to provide an integrated view of sequences and genetic markers. The predicted genes, ESTs and genetic markers are aligned to the genomic sequence contig (Contig3587_Contig1062) according to their relative positions, and are clickable for additional information. The GBrowse tool can be queried using sequence or marker identifiers and has been integrated with the BLAST search engine (see below). The graphical representation is useful for genome annotation. CMap is used for browsing the genetic map with information about the linkage groups and map locations of genetic markers.
Figure 1.
An integrated view of BeetleBase data using GBrowse. A part of the contig, Contig3587_Contig1062, is shown together with the aligned ESTs, computed genes and genetic markers.
BeetleBase also provides a BLAST server for searching Tribolium sequences. On the results page of a BLAST search, each hit is linked to the GBrowse view of the sequence. This feature allows non-Tribolium sequences to be mapped on to the Tribolium genome for comparative analysis. Thus, the BLAST server is useful not only for the Tribolium search community but also for other scientists who are interested in identifying the Tribolium homologues for their sequences. In addition, we have set up a FTP site (ftp://bioinformatics.ksu.edu/pub/beetlebase/) for downloading various datasets and software programs from BeetleBase.
FUTURE DIRECTIONS
BeetleBase will be continually updated and expanded in the future. We plan monthly updates of the database, depending on the availability of new data. The immediate effort will be to populate the database with a comprehensive set of genes that have been predicted using different software tools and are currently being curated by the Tribolium research community. Additional ESTs that are being generated by the Tribolium genome consortium will soon be available in BeetleBase. The large amount of ESTs will be assembled into non-redundant contigs, and then the EST contigs will be aligned to the genome sequence to assist gene functional annotation. BeetleBase will also be expanded to include microarray gene expression data from the Tribolium search community and genome sequences from other model insects to support comparative genomics. We will utilize the additional modules of the GMOD schema, and where necessary, develop new interfaces and tools to make the information accessible in an effective way.
Acknowledgments
We thank Drs Richard W. Beeman, Marce D. Lorenzen and Yoonseong Park for helpful discussions and for providing data to BeetleBase; and Youping Deng for his help to initiate this project. This work is supported by the K-INBRE Bioinformatics Core (NIH grant number P20 RR016475). Funding to pay the Open Access publication charges for this article was provided by NIH grant number P20 RR016475.
Conflict of interest Statement. None declared.
REFERENCES
- 1.Brown S.J., DeCamillis M., Gonzalez-Charneco K., Denell M., Beeman R., Nie W., Denell R. Implications of the Tribolium Deformed mutant phenotype for the evolution of Hox gene function. Proc. Natl Acad. Sci. USA. 2000;97:4510–4514. doi: 10.1073/pnas.97.9.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorenzen M.D., Berghammer A.J., Brown S.J., Denell R.E., Klingler M., Beeman R.W. piggyBac-mediated germline transformation in the beetle Tribolium castaneum. Insect Mol. Biol. 2003;12:433–440. doi: 10.1046/j.1365-2583.2003.00427.x. [DOI] [PubMed] [Google Scholar]
- 3.Lorenzen M.D., Doyungan Z., Savard J., Snow K., Crumly L.R., Shippy T.D., Stuart J.J., Brown S.J., Beeman R.W. Genetic linkage maps of the red flour beetle, Tribolium castaneum, based on bacterial artificial chromosomes and expressed sequence tags. Genetics. 2005;170:741–747. doi: 10.1534/genetics.104.032227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown S.J., Mahaffey J.P., Lorenzen M.L., Denell R.E., Mahaffey J.W. Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evol. Dev. 1999;1:11–15. doi: 10.1046/j.1525-142x.1999.99013.x. [DOI] [PubMed] [Google Scholar]
- 5.Bucher G., Scholten J., Klingler M. Parental RNAi in Tribolium (Coleoptera) Curr. Biol. 2002;12:R85–R86. doi: 10.1016/s0960-9822(02)00666-8. [DOI] [PubMed] [Google Scholar]
- 6.Tomoyasu Y., Denell R.E. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev. Genes Evol. 2004;214:575–578. doi: 10.1007/s00427-004-0434-0. [DOI] [PubMed] [Google Scholar]
- 7.Savard J., Tautz D., Lercher M.J. Genome-wide acceleration of protein evolution in flies (Diptera) BMC Evol. Biol. 2006;6:7. doi: 10.1186/1471-2148-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Gregorio E., Lemaitre B. The mosquito genome: the post-genomic era opens. Nature. 2002;419:496–497. doi: 10.1038/419496a. [DOI] [PubMed] [Google Scholar]
- 9.Salamov A.A., Solovyev V.V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mott R. EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA. Comput. Appl. Biosci. 1997;13:477–478. doi: 10.1093/bioinformatics/13.4.477. [DOI] [PubMed] [Google Scholar]
- 12.Florea L., Hartzell G., Zhang Z., Rubin G.M., Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheelan S.J., Church D.M., Ostell J.M. Spidey: a tool for mRNA-to-genomic alignments. Genome Res. 2001;11:1952–1957. doi: 10.1101/gr.195301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]