Abstract
Quantitative trait loci (QTLs) affecting sugar composition of the cell walls of maize (Zea mays) pericarp were mapped as an approach to the identification of genes involved in cereal wall biosynthesis. Mapping was performed using the IBM (B73 × Mo17) recombinant inbred line population. There were statistically significant differences between B73 and Mo17 in content of xylose (Xyl), arabinose (Ara), galactose (Gal), and glucose. Thirteen QTLs were found, affecting the content of Xyl (two QTLs), Ara (two QTLs), Gal (five QTLs), Glc (two QTLs), Ara + Gal (one QTL), and Xyl + Glc (one QTL). The chromosomal regions corresponding to two of these, affecting Ara + Gal and Ara on maize chromosome 3, could be aligned with a syntenic region on rice (Oryza sativa) chromosome 1, which has been completely sequenced and annotated. The contiguous P1-derived artificial chromosome rice clones covering the QTLs were predicted to encode 117 and 125 proteins, respectively. Two of these genes encode putative glycosyltransferases, displaying similarity to carbohydrate-active enzyme database family GT4 (galactosyltransferases) or to family GT64 (C-terminal domain of animal heparan synthases). The results illustrate the potential of using natural variation, emerging genomic resources, and homeology within the Poaceae to identify candidate genes involved in the essential process of cell wall biosynthesis.
As a defining feature of plants, the cell wall is important to all aspects of their biology. Furthermore, plant cell walls provide fuel, fiber, and food to all human societies. Cell walls are a complex composite of polysaccharides, proteins, and lignin. The polysaccharide components can be classified into three broad categories: pectins, hemicelluloses, and cellulose. In both monocotyledons and dicotyledons, the most abundant polysaccharide in the majority of tissues is the simple polymer cellulose. In contrast, the hemicelluloses are chemically and physically more complex and their monomer composition varies between species and between tissues and cell types within an individual plant (e.g. Fincher, 1992; Doblin et al., 2001). The cell walls of the commelinoid monocotyledons, which includes the grasses and cereals (family Poaceae), differ significantly in composition from other plants in having a larger amount of arabinoxylan and a unique hemicellulose, mixed-linked glucan (β1,3-β1,4-glucan, also known as cereal β-glucan; Carpita and Gibeaut, 1993; Carpita, 1996). Thus, the several stages of hemicellulose biosynthesis (precursor synthesis, polymerization, secretion, and incorporation into the wall) must differ significantly among plant species and be dynamically regulated processes within a particular plant.
A full understanding of the biosynthesis of the cell wall remains a major unsolved problem in plant biology. Biochemical approaches to identification of the enzymes and genes involved have been hindered by the lability of the enzymes and our ignorance of their biosynthetic mechanisms. Although substantial progress has recently been made on the identification and function of cellulose synthases and the corresponding genes (called CESA) and on several non-processive glycosyl transferases such as xylosyl, galactosyl, and fucosyl transferases, little is known about the genes and enzymes involved in synthesis of the backbones of the hemicellulosic polymers (Arioli et al., 1998; Edwards et al., 1999; Perrin et al., 1999; Fagard et al., 2000; Taylor et al., 2000; Faik et al., 2002; Peng et al., 2002; Vanzin et al., 2002).
A genetic approach to cell wall biosynthesis has been successful in identifying CESA genes in different species and also genes involved in hemicellulose precursor biosynthesis (Reiter et al., 1993; Bonin et al., 1997; Arioli et al., 1998). Wall compositional differences in T-DNA-tagged plants of Arabidopsis have also been examined (Gardner et al., 2002). The exploitation of natural variation is an alternative genetic approach to these mutagenesis-based approaches. Traits such as fruit size, flowering time, morphology, and light responsiveness differ among lines, ecotypes, or accessions of a particular plant species, and a number of these have been successfully analyzed genetically. Because they are typically inherited in a quantitative manner, they are more challenging to analyze, and isolation of the responsible genes is more difficult. Nonetheless, in recent years a number of quantitative trait loci (QTLs) have been identified and isolated (Frary et al., 2000; Yano et al., 2000; El-Assal et al., 2001; Maloof et al., 2001). Contrary to some expectations, the QTLs underlying natural variation have turned out to be variant alleles of genes that play a central role in the trait under study, and not minor, secondary genes with an indirect role (Millar, 2001). Thus, the identification of genes controlling natural quantitative differences in cell wall properties might lead to the identification of critical, primary genes involved in hemicellulose biosynthesis.
A number of important agronomic properties of plants are influenced by the properties of their cell walls; for example, nutrient absorption and digestibility by humans and animals, wheat (Triticum aestivum) bread making quality, barley (Hordeum vulgare) brewing quality, and insect resistance (e.g. Martin and Bamforth, 1980; Brice and Morrison, 1982; Hedin et al., 1993; Lundvall et al., 1994; Courtin and Delcour, 1998). The inheritance of the cell wall properties underlying some of these traits has been studied in several species including soybean (Glycine max), wheat, barley, and maize (Zea mays; e.g. Powell et al., 1985; Jung and Buxton, 1994; Lundvall et al., 1994; Saulnier et al., 1995; Lempereur et al., 1997; Stombaugh et al., 2000). These traits typically exhibit a genotype by environment interaction effect and have complex patterns of inheritance indicative of control by many genes. QTLs affecting mixed-linked glucan content in oat (Avena sativa) and barley, fiber content in maize, and ratio of Ara to Xyl in wheat flour have been detected (Han et al., 1995; Lübberstedt et al., 1997, 1998; Martinant et al., 1998; Kianian et al., 2000; Méchin et al., 2000). However, the specific genes underlying the QTLs identified in any of these studies, due to the complexity of the relevant plant genome, the lack of adequate genomic resources, or both.
Substantial progress has been made in recent years in the development of genetic and genomic resources for many plants, including cereals such as rice (Oryza sativa) and maize. This opens the possibility of using natural variation to identify genes involved in a complex process such as wall biosynthesis, with the ultimate goal of elucidating the underlying biochemical mechanisms of hemicellulose backbone synthesis. Here, we have exploited high-throughput cell wall analysis and advanced maize genetic resources to identify QTLs affecting the sugar monomer composition of cell walls. The results illustrate the feasibility of identifying candidate genes involved in hemicellulose biosynthesis when QTL analysis is combined with synteny between rice and maize and with the available partially annotated rice genome.
RESULTS
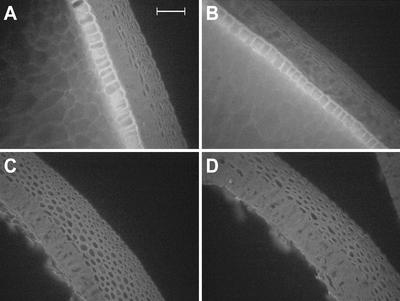
Several criteria were considered for defining a model plant and tissue for the analysis of natural genetic variation of cell wall composition. A major consideration is the existence of rich genetic and genomic resources, which is satisfied for maize in light of recent advances in the development of intermated recombinant inbred lines (RILs) such as the IBM population derived from B73 and Mo17, a large number of genetic markers, and synteny with the completely sequenced genome of rice (Davis et al., 1999, 2001; Goff et al., 2002; Lee et al., 2002; Song et al., 2002). The tissue to be sampled should be easy to collect, have a low level of starch, and its composition should be insensitive to small differences in rates of growth and development among accessions or ecotypes. The maize pericarp satisfies these criteria. It is predicted to have low starch content and, if sampled from a fully mature cob, variation in growth and development should be minimal. Sufficient amounts of tissue can be sampled from a single ear. The pericarp was removed with forceps from seeds that had been soaked in water. When cross-sections of intact seeds were observed by fluorescence microscopy, three types of cells were visible at the edge (Fig. 1, A and B; Kiesselbach, 1949). The pericarp is the outermost layer and is composed of approximately 10 thick-walled cells that fluoresce blue. The aleurone layer is characterized by relatively large round cells that fluoresce bright yellow-green under alkaline conditions. Inside this layer of maternal tissue is the endosperm, composed of large thin-walled cells. Microscopy indicated that the pericarp preparations used in this study contained minimal contamination by aleurone or endosperm walls (Fig. 1, C and D). There were no noticeable differences among B73, Mo17, and two randomly selected RILs from the IBM population in appearance of the pericarp (Fig. 1).
Figure 1.
Fluorescent microscopy of cross sections of intact seeds (caryopses; A and B) and excised pericarp (C and D). A, Mo17; B, B73; C, RIL 356; D, RIL 338. The three layers of cells visible in the whole caryopsis cross section, from inside to outside, are endosperm, aleurone, and pericarp. Only pericarp is present in C and D. Bar = 100 μm.
We could not find any previous reports of the sugar composition of maize pericarp. Our analyses indicate that mature maize pericarp contains an average of 52% Xyl, 32% Ara, 10% Gal, and 5.0% Glc. Rha, Fuc, and Man were present in trace quantities. Arabinoxylan is the major hemicellulose in most cereal cell walls, although there are large differences in the degree of Ara substitution among tissues. Therefore, the high values for Xyl and Ara probably represent a high content of Ara-substituted xylan (arabinoxylan) in the pericarp. The polymeric nature of the pericarp Gal is not known, although in coleoptiles and young seedlings of maize it is mostly nonreducing terminal (Kato and Nevins, 1984; Carpita, 1996). The trace quantities of Rha and Man eliminate galactomannan and rhamnogalacturanan as significant contributors of Gal. The low Glc content indicates that the pericarp contains only small amounts of starch or other acid-digestible Glc polymers such as mixed-linked glucan, which is relatively abundant in barley aleurone and endosperm walls (Bacic and Stone, 1981a, 1981a).
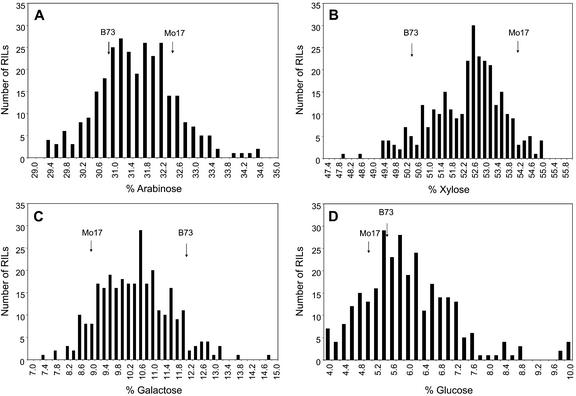
Summary statistics for the cell wall composition of pericarps from the IBM RIL population (Davis et al., 2001; Lee et al., 2002) are given in Table I. There was a continuous range of values for all measured traits among the RILs (Fig. 2). Some RILs had extreme values for Glc (Fig. 2), possibly due to contamination of the pericarp sample with endosperm starch. Normal distributions were observed for all monosaccharides as illustrated by the histograms (Fig. 2). Significant differences among the RILs for monosaccharide content were detected at the 1% level using analysis of variance. The midparent values (i.e. the values halfway between the two parents) and the median of the RILs for Ara, Xyl, and Gal were very similar, but the median for Glc was greater than the midparent value, and the population was slightly skewed to the right (Fig. 2).
Table I.
Mean and range values for relative and absolute quantity of monosaccharides extracted from pericarp cell walls of the IBM mapping population and parents
| Monosaccharide | Parents
|
RILs
|
|||
|---|---|---|---|---|---|
| B73 (relative chromatograph peak area [RCPA]) | Mo17 (RCPA) | Average (RCPA) | Minimum (RCPA) | Maximum (RCPA) | |
| Ara | 30.98 | 32.48 | 31.49 | 29.26 | 34.47 |
| Xyl | 51.33 | 54.19 | 52.26 | 47.83 | 54.98 |
| Gal | 12.14 | 8.31 | 10.32 | 7.28 | 14.80 |
| Glc | 5.55 | 5.02 | 5.92 | 3.54 | 11.52 |
| B73 | Mo17 | Average | Minimum | Maximum | |
|---|---|---|---|---|---|
| mg g−1 | |||||
| Ara | 156.94 | 161.37 | 153.48 | 113.86 | 188.50 |
| Xyl | 260.20 | 269.70 | 254.97 | 183.85 | 305.43 |
| Gal | 61.51 | 41.25 | 50.31 | 32.59 | 71.26 |
| Glc | 28.13 | 24.81 | 28.91 | 15.91 | 55.04 |
Figure 2.
Distribution of means of monosaccharide composition in the pericarps of the IBM RIL population. A, Ara; B, Xyl; C, Gal; D, Glc. Parental values are indicated by arrows. RIL effects were significant (P < 0.01) for all four sugars.
The proportion of the total variance of each source of variation, RIL, replication, and error (RIL by replication interaction) were calculated from the variance components and are summarized in Table II. Although RIL had a significant effect on monosaccharide content, the proportion of the total variation accounted for by RIL for each trait varied. Error had the largest variance component for Ara, Xyl, and Glc. Glc was the only trait where replicate variance was greater than RIL variance. The magnitude of the variance among replicates for Xyl and Gal was negligible. RIL effects accounted for the greatest proportion of the variance for Gal and the least portion for Glc.
Table II.
Percentage of the total variance for each source of variation for RCPA of cell wall monosaccharide content of the IBM mapping population
| Source of Variation | Monosaccharide
|
|||
|---|---|---|---|---|
| Ara | Xyl | Gal | Glc | |
| RIL | 35.92 | 41.49 | 70.84 | 14.63 |
| Replication | 27.38 | 1.94 | 0.25 | 17.61 |
| Error | 36.69 | 56.57 | 28.90 | 53.16 |
The coefficients of variance for Ara, Xyl, Gal, and Glc were 3.32, 2.33, 6.62, and 28.3, respectively.
Correlation analysis showed a significant and strongly positive relationship between Ara and Xyl content (Table III). Gal content was moderately correlated with Ara and Xyl. No significant correlation at the 5% level was observed between Glc and the other monosaccharides (Table III).
Table III.
Pair-wise Pearson's correlation coefficients of means of absolute monosaccharide content of the IBM mapping population
Significant at P < 0.01.
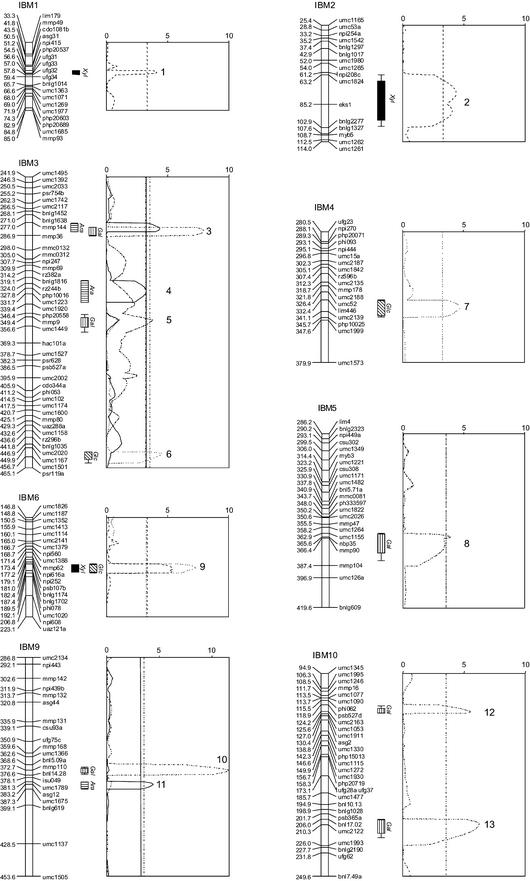
Thirteen QTLs for maize pericarp monosaccharide relative chromotograph peak area (RCPA) were detected. The map location, log of the odds ratio (LOD) score, flanking markers, marker bin, additive effect, and percent of the total variance accounted for by each locus are given in Table IV. Eleven of the loci were associated with single monosaccharides. A locus on chromosome 3 was associated with both Ara and Gal content, and a locus on the short arm of chromosome 6 was associated with both Xyl and Glc content. All of the Mo17 alleles had a positive additive effect on Ara and Xyl content. Nine of the 11 B73 alleles had a positive effect on Gal and Glc content. Four of the QTL accounted for 10.0% or more of the total variance. The size of the genetic map interval for the QTLs ranged from 2.53 cM for QTL 1 to 31.4 cM for QTL 2 (Table IV).
Table IV.
Summary of QTLs affecting sugar composition of maize pericarp
| QTL | Chromosome Position | QTL Map Position | LOD | Trait | Flanking Markers | Bin | Additive Effect | Variance |
|---|---|---|---|---|---|---|---|---|
| % | ||||||||
| 1 | 1 | 54.52–57.05 | 4.10 | Xyl | php20537-ufg33 | 1.01 | −0.2972 | 5.56 |
| 2 | 2 | 67.19–98.23 | 4.22 | Xyl | umc1824-bnl2g277 | 2.02 | −0.3773 | 16.2 |
| 3 | 3 | 277.70–284.00 | 4.37 | Ara | mmp144-mmp36 | 3.04 | −0.2263 | 5.50 |
| 3 | 3 | 281.00–287.04 | 7.98 | Gal | mmp144-mmc0132 | 3.04 | 0.3362 | 8.54 |
| 4 | 3 | 328.70–331.70 | 3.27 | Ara | bnlg1816-umc1920 | 3.04 | −0.2215 | 5.15 |
| 5 | 3 | 351.55–353.84 | 3.79 | Gal | mmp9-umc1449 | 3.04 | 0.2229 | 3.67 |
| 6 | 3 | 453.05–459.15 | 4.53 | Glc | umc1167-psr119a | 3.05 | 0.2907 | 5.45 |
| 7 | 4 | 332.44–345.10 | 4.73 | Glc | lim446-php10025 | 4.09 | 0.3167 | 6.60 |
| 8 | 5 | 364.89–365.64 | 3.44 | Gal | umc1155-nbp35 | 5.05 | 0.2502 | 4.51 |
| 9 | 6 | 182.42–189.44 | 7.33 | Glc | bnlg1174-phi078 | 6.05 | 0.3931 | 10.0 |
| 9 | 6 | 182.42–189.44 | 5.37 | Xyl | bnlg1174-phi078 | 6.05 | −0.3483 | 14.5 |
| 10 | 9 | 368.57–375.66 | 11.63 | Gal | bnl5.09a-bnl14.28 | 9.06 | 0.4110 | 12.4 |
| 11 | 9 | 381.32–387.22 | 4.41 | Ara | umc1789-umc1675 | 9.06 | −0.2262 | 5.43 |
| 12 | 10 | 119.86–125.10 | 5.63 | Gal | psb527d-umc1053 | 10.04 | −0.2864 | 5.91 |
| 13 | 10 | 205.96–221.34 | 6.25 | Gal | psb365a-umc1993 | 10.06 | 0.4110 | 6.95 |
A positive additive effect indicates that the B73 allele was associated with a higher value, whereas a negative effect indicates that the Mo17 allele was associated with a higher value.
Several approaches were taken to identify known nucleic acid sequences that might plausibly account for the observed variation associated with the QTLs. Some genes of known or putative function mapped near some of the QTLs. Other candidates were identified by BLAST search of molecular marker partial nucleic acid sequences. For example, the microsatellite locus umc1111 maps to bin 1.11, and its flanking sequences have sequence similarity to a gene encoding a sterol-methyltransferase. Sterols have recently been shown to play a role as intermediates in cellulose biosynthesis (Peng et al., 2002). Marker umc1366, which maps to bin 9.06 along with Gal and Ara QTLs 10 and 11, is predicted to encode a β1,3-glucanase, an enzyme with a potential role in degradative cell wall metabolism. A QTL for Glc content maps to bin 3.05 as does Suc phosphate synthase (UDP-Glc-Fru-phosphate glucosyltransferase, encoded by sps2; Causse et al., 1995). The nearest flanking marker to the 5.63-cM Gal QTL interval on chromosome 10, umc1053, has high sequence homology to a gene encoding a wall-localized invertase (Shanker et al., 1995).
Searching for gene candidates in maize is currently limited by the lack of genomic sequence. Therefore, syntenic regions for the QTLs were identified within the rice genome using MapSearch at the Gramene Web site (http://www.gramene.org; Ware et al., 2002). Rice chromosome 1 is syntenic to much of the apical portion of maize chromosome 3 (Gale and Devos, 1998). Because rice chromosome 1 is completely sequenced and annotated, two contiguous segments of DNA could be identified that correspond to QTL 3 (Ara plus Gal), QTL 4 (Ara), and QTL 5 (Gal) on maize chromosome 3 (Table III). A flanking marker, mmp36, of QTL 3 has significant homology to sequences on overlapping rice P1-derived artificial chromosome (PACs) AP002872 and AP002540 (http://rgp.dna.affrc.go.jp/). These two PACs map to the short arm of rice chromosome 1. The nearest maize/rice marker on the other flank of the QTL, umc1392, is 45 cM proximal to mmp36 and has homology to rice PAC AP003214 (Fig. 3). Assuming that map distances and physical distances are roughly proportional, but erring on the side of inclusion, AP002540 and five PACs in the direction of umc1392 (i.e. AP002872, AP003311, AP002483, AP002872, and AP002913) were selected as most likely completely spanning QTL3. Together, these six PACs form a contig of 638 kb and were predicted to encode 117 proteins. A second contig chosen to include the proximal Ara and Gal QTLs 4 and 5 (flanking markers mmp9 and umc1449) includes PACs AP001073, AP001081, AP000837, AP000836, and AP001072, is 658 kb, and is predicted to encode 125 proteins.
Figure 3.
Chromosome line graphs and QTL likelihood plots of relative pericarp cell wall monosaccharide content (RCPA) in the IBM population based on composite interval mapping. The horizontal axes in each graph indicate LOD scores, and the vertical lines indicate the empirically derived LOD threshold (P < 0.05) for each monosaccharide. The length of the bars indicates an LOD score of 3.0 for each QTL, and the line extensions of each bar, when present, indicate an LOD score of 2.0.
The predicted protein sequences were analyzed by standard BLAST against the nonredundant database of the National Center for Biotechnology Information and also against the carbohydrate-active enzyme database (CAZy) of glycosyltransferases (Campbell et al., 1997; Coutinho and Henrissat, 1999). A single candidate glycosyltransferase was found in each contig. One candidate, BAB40110, encodes a predicted member of glycosyltranserase family GT4, which contains a variety of transferases that form alpha-linkages, including Suc synthase, GDP-Man α-mannosyltransferase, α-1,3-rhamnosyltransferase, trehalose phosphorylase, and digalactosyldiacylglycerol synthase. A second predicted protein, BAA90366, is a member of family GT64. This family includes plant proteins that are related to the C-terminal domain of animal heparan synthases, which are themselves bifunctional glycosyltransferases that use UDP-GlcNAc and UDP-GlcA to synthesize polysaccharides with alternating α1,4-GlcNAc and β1,4-GlcA residues. Plant genes related to the N-terminal domain or the C-terminal domain of animal heparan synthetases (families GT47 and GT64, respectively) are abundant, Arabidopsis having at least 44 members and rice more than 30. The function of at least one member of plant family GT47 has been elucidated (pectin β-glucuronyltransferase; Iwai et al., 2002). The precise function of family GT64 in plants is not known, but similarity to the C-terminal domain (α-GlcNAc transferase) of animal heparan synthetases suggests it could be an α-glycosyltransferase.
DISCUSSION
Analysis of the IBM population of RILs led to the identification of 13 QTLs affecting the sugar monomer composition of maize pericarp walls. Using markers that bridge maize and rice, it was possible to identify syntenic regions in rice corresponding to some of the maize QTLs and, thus, identify corresponding candidate rice genes. Due to the nature of QTLs, the regions for searching were rather large. Nonetheless, within this collection, the number of proteins with a plausible role in wall biosynthesis was small. Therefore, it should be practical to study the candidate genes further for a role in wall biosynthesis; for example, by detailed studies on their pattern of expression (e.g. are they expressed in the pericarp?) or biochemical function by heterologous expression (e.g. Faik et al., 2002).
It was fortuitous to find genetically influenced differences in cell wall composition between B73 and Mo17, thereby making the genetic reference map a tool for cell wall discovery. The IBM mapping population was instrumental in mapping these QTL with high resolution because it has nearly a 4-fold increase in recombination events relative to a conventional RIL population that is not intermated, in addition to a high level of saturation of over 1,800 molecular markers (Lee et al., 2002). The large population size of over 300 RILs and the heritability of the traits also contributed to the fine level of mapping. Several small effect QTLs were identified for each trait. Although all but four of the QTL accounted for less than 10.0% of the total variation, many were mapped to small intervals. The maize genome is estimated to be 2,500 Mb (Laurie and Bennett, 1985), and the IBM genetic linkage map reported at MaizeDB has a total of 6,246 cM (Coe et al., 2002). Therefore, the approximate physical distance of 1 cM is 400 kb. If the maize genome contains 50,000 genes, the gene density is predicted to be 8 genes cM−1. Therefore, the individual QTLs identified in this study are predicted to contain 20 to 250 genes.
Continuous and natural genetic variation for cell wall properties has been documented in various cereals and legumes (e.g. Powell et al., 1985; Lundvall et al., 1994; Saulnier et al., 1995; Lempereur et al., 1997; Stombaugh et al., 2000). Environmental effects and genotype by environment interaction effects also influence cell wall composition. The range of significant phenotypic variation within the IBM population and the differences between the parents imply that the quantity of the individual monosaccharides is under the control of several genes, each having a small effect. Our results indicate that a large portion of the total variance of Gal and Xyl content is controlled by at least nine loci. A smaller portion of the Ara and Glc variability can be attributed to genetic effects. However, several QTLs were identified for each trait.
No differences in pericarp cell morphology or pericarp cell counts were observed between samples, and the content of total monosaccharides was also not significantly different within the recombinant population. Therefore, the genetic variation that we observed is likely due to differences in cell wall composition, in either quantity of backbone sugars and/or the degree of their substitution, rather than cell size or shape. The differences in composition could, in turn, be due to any of a number of allelic variations in the genes controlling the multiple steps along the pathway of cell wall biosynthesis, from precursor synthesis to final incorporation into the wall.
Two QTLs were each found to be associated with differences in the levels of two sugars, Ara and Gal on chromosome 3 (QTL 3) and Glc and Xyl on chromosome 6 (QTL 9). The gene underlying QTL3 might be one that affects synthesis of a polysaccharide containing both Ara and Gal, such as the arabinogalactan attached to membrane-associated proteins (arabinogalactan proteins). QTL9 might plausibly be associated with synthesis of xyloglucan, which is present in cereals (Kato et al., 1982; Carpita, 1996). On the other hand, although there was a strong positive correlation between Ara and Xyl content, no QTLs were identified that influence the quantity of those two monosaccharides together. One possible explanation for this is that B73 and Mo17 do not have any genetic differences influencing overall arabinoxylan biosynthesis.
The ultimate goal of the study of natural variation in maize cell wall composition is to identify the exact nucleotide polymorphism that is responsible for each mapped phenotypic variation. Based on current resources, we were able unequivocally to identify only two candidate genes in the genomic regions corresponding to QTLs 3 to 5. Further analyses were limited by the lack of sufficient markers that bridge maize and rice and the incomplete annotation of the rice genome. It is expected that these resources will become available within the next year. The combined use of maize mapping populations and the completed rice genome is also potentially limited by a lack of synteny in critical regions. It is too early to know if this will create insurmountable obstacles.
Additional genetic resources can be used to make further progress. For example, it should be possible to map some of the QTLs to smaller regions using other near-isogenic RILs, such as the BC3-derived near-isogenic lines representing introgression of Tx303 chromosomal regions into the B73 genetic background and BC3-derived near-isogenic lines representing introgression of Oh43 alleles into the Mo17 genetic background (Stuber, 1998).
MATERIALS AND METHODS
Genetic Resources
We measured the cell wall monosaccharide profile of the intermated RIL IBM population derived from B73 and Mo17 (Davis et al., 2001; Coe et al., 2002). B73 and Mo17 represent two of the major heterotic U.S. maize (Zea mays) germplasm pools, Iowa Stiff Stalk Synthetic and Lancaster Sure Crop, respectively. The F2 were intermated for five generations and subsequently self-pollinated to make RILs. The RILs (302 total) were used to make a high-density genetic linkage map with >1,850 molecular markers. These data are available at MaizeDB (www.agron.missouri.edu/maps.html). A subset of the population was obtained from the Maize Genetic Consortium Stock Center and the complete population from the University of Missouri Maize Mapping Project. Seed were collected from a single location and year and used directly.
Wall Analysis
For each replicated sample, two seeds were soaked overnight in 2 mL of tap water in 24-well microtiter plates. The pericarp was removed using forceps. All tissue samples were stored in 70% (v/v) ethanol at 4°C. Only the pericarp was removed from the seed. For microscopy, maize seeds or pericarp were sectioned by hand and mounted on glass slides in 0.1 m ammonium hydroxide (pH 9.8). Slides were observed using an Axiophot Fluorescence Microscope (HBO 100 Hg vapor bulb, Zeiss, Jena, Germany) with a 365- ± 12.5-nm exciter and a 450-nm long-pass filter, using the 10× and 20× objectives. Wall-bound ferulic acid fluoresces green at high pH (Rudall and Caddick, 1994).
Cell wall monosaccharides were analyzed as the alditol acetates after acid hydrolysis as described by Reiter et al. (1993) and Blakeney et al. (1983) with modification. The samples were washed for 1 h at 70°C with 70% (v/v) ethanol, with one change of ethanol after 30 min, washed once with acetone, and dried. Aliquots of each sample (7–15 mg) were hydrolyzed with 1 m H2SO4 for 1 h at 121°C. The released sugars were reduced to alditols by addition of 100 μL of 9 m ammonium hydroxide followed by 1 mL of 2% (w/v) NaBH4 in dimethyl sulfoxide, and incubated at 40°C for 90 min. The alditols were acetylated by the addition of 250 μL of acetic acid, 250 μL of 1-methylimidazole, and 4 mL of acetic anhydride. After addition of 8 mL of water, the alditol acetates were extracted with 2 mL of dichloromethane and separated using an Agilent 6890 series GC system equipped with an Agilent DB-225 capillary column (Agilent, Palo Alto, CA). Detection was by flame ionization. Quantitation used the integration software in GC ChemStation (Agilent). The temperature program was: 1 min at 160°C, 20°C min−1 to 200°C, hold for 5 min, 20°C min−1 to 240°C, hold for 11.5 min, 25°C min−1 to 150°C, and hold for 1 min.
Peak areas were adjusted relative to an internal inositol standard. Rha, Fuc, and Man were present in maize pericarp in only trace amounts and were not included in the QTL analysis. Sugars are reported as both RCPA of the four principal monosaccharides (Ara, Xyl, Gal, and Glc) and total quantity (milligram of sugar per gram of tissue).
Statistical Analyses
The seeds were sampled and analyzed within each replication in a completely random manner. Three independent pericarp samples were collected from a pair of seeds for each RIL. Analyses of variance were performed using PROC GLM with inbred as the single random effect (SAS Institute, 2000). Variance components were calculated to estimate the percentage of the total variance originating from RIL, replication, and error using PROC VARCOMP under the assumption that all sources of variation were random.
The map score data and genetic linkage maps were provided by the Maize Mapping Project (University of Missouri, Columbia). Map distances were confirmed using MapMaker version 3.0 (Lander et al., 1987) with an LOD score threshold of 3.0 and a maximum recombination frequency of 0.50. The Kosambi function was used to transform recombination frequencies into centiMorgans. Cosegregation of phenotypic properties and genetic markers was determined using QTL Cartographer version 1.15 (Basten et al., 2001). QTLs were identified using means and a framework genetic linkage map with a marker density of approximately one per 10 cM. Areas where QTLs were identified were saturated with previously mapped markers to a density of one per 0.10 cM and reanalyzed. In both cases, we employed composite interval mapping to increase resolution and reduce background marker effect (Zeng, 1994). LOD thresholds were determined by computing 1,000 permutations for each trait (Churchill and Doerge, 1994). The levels of significance for Ara, Xyl, Gal, and Glc levels at P < 0.05 were LOD 3.22, 3.32, 3.54, and 3.27, respectively, and at P < 0.01, the LOD were 4.17, 4.17, 4.24, and 4.12, respectively. The graphical representation of the linkage maps and QTLs were prepared using MapChart (Voorrips, 2002).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank the Maize Genetic Cooperation Stock Center for germplasm; the Rice Genome Program (Tsukuba, Japan) for making their excellent resources publicly and efficiently available; Judith Kolkman (Department of Crop and Soil Science, Oregon State University, Corvallis) and Danielle Trebbi (Department of Crop and Soil Sciences, Michigan State University) for assistance with the QTL analyses; Shirley Owens and Joanne Whallon (Michigan State University Center for Advanced Microscopy) and Anthony Sanderfoot (Michigan State University-Department of Energy-Plant Research Laboratory) for assistance with the microscopy; and Emeline Deleury (Architecture et Function des Macromolécules Biologiques, Marseille, France) for her help with the CAZy database searches.
Footnotes
This work was supported by the U.S. Department of Energy, Division of Energy Biosciences, by the National Science Foundation Plant Genome Research Program, and by the European Commission (grant no. QLK5–CT–2001–00443).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.020016.
LITERATURE CITED
- Arioli T, Peng LC, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Bacic A, Stone BA. Isolation and ultrastructure of aleurone cell walls from wheat and barley. Aust J Plant Physiol. 1981a;8:453–474. [Google Scholar]
- Bacic A, Stone BA. Chemistry and organization of aleurone cell wall components from wheat and barley. Aust J Plant Physiol. 1981b;8:475–495. [Google Scholar]
- Basten CJ, Weir BS, Zeng Z-B (2001) QTL cartographer, version 1.15. Department of Statistics, North Carolina State University, Raleigh. http://statgen.ncsu.edu/qtlcart/cartographer.html
- Blakeney AB, Harris PJ, Henry RJ, Stone BA. A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr Res. 1983;113:291–299. [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter WD. The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-d-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-l-fucose. Proc Natl Acad Sci USA. 1997;94:2085–2089. doi: 10.1073/pnas.94.5.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice RE, Morrison IM. The degradation of isolated hemicellulose and lignin-hemicellulose complexes by cell free rumen hemicellulases. Carbohydr Res. 1982;101:93–100. [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC. Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:445–474. doi: 10.1146/annurev.arplant.47.1.445. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plant: consistency of molecular structure with physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Causse M, Rocher JP, Pelleschi S, Barriere Y, de Vienne D, Prioul JL. Sucrose phosphate synthase: an enzyme with heterotic activity correlated with maize growth. Crop Sci. 1995;35:995–1001. [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe E, Cone K, McMullen M, Chen SS, Davis G, Gardiner J, Liscum E, Polacco M, Paterson A, Sanchez-Villeda H et al. Access to the maize genome: an integrated physical and genetic map. Plant Physiol. 2002;128:9–12. doi: 10.1104/pp.128.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtin CM, Delcour JA. Physicochemical and bread-making properties of low molecular weight wheat-derived arabinoxylans. J Agric Food Chem. 1998;46:4066–4073. [Google Scholar]
- Coutinho PM, Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert HJ, Davies G, Henrissat B, Svensson B, editors. Recent Advances in Carbohydrate Bioengineering. Cambridge, UK: The Royal Society of Chemistry; 1999. pp. 3–12.http://afmb.cnrs-mrs.fr/CAZY/ [Google Scholar]
- Davis GL, McMullen MD, Baysdorfer C, Musket T, Grant D, Staebell M, Xu G, Polacco M, Koster L, Melia-Hancock S et al. A maize map standard with sequenced core markers, grass genome reference points and 932 expressed sequence tagged sites (ESTs) in a 1736-locus map. Genetics. 1999;152:1137–1172. doi: 10.1093/genetics/152.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis G, Musket T, Melia-Hancock S, Duru N, Sharopova N, Schultz L, McMullen M, Sanchez H, Schroeder S, Garcia AA. The intermated B73 × Mo17 genetic map: a community resource. Maize Genet Conf Abst. 2001;43:W15. , P62. [Google Scholar]
- Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM. Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiol. 2001;125:2040–2052. doi: 10.1104/pp.125.4.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards ME, Dickson CA, Chengappa S, Sidebottom C, Gidley MJ, Reid JSG. Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 1999;19:691–697. doi: 10.1046/j.1365-313x.1999.00566.x. [DOI] [PubMed] [Google Scholar]
- El-Assal SE, Alonso-Blanco C, Peters AJM, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000;12:2409–2423. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K. An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci USA. 2002;99:7797–7802. doi: 10.1073/pnas.102644799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher GB. Cell wall metabolism in barley. In: Shewry PR, editor. Barley: Genetics, Biochemistry, Molecular Biology and Biotechnology. Oxford: Alden Press Ltd.; 1992. pp. 413–440. [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM. Comparative genetics in the grasses. Proc Natl Acad Sci USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SL, Burrell MM, Fry SC. Screening of Arabidopsis thaliana stems for variation in cell wall polysaccharides. Phytochemistry. 2002;60:241–254. doi: 10.1016/s0031-9422(02)00046-8. [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Prestig G, Wang R, Dunn M, Glazebrook J, Sessions A, Deller P, Varma H et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Han F, Ullrich SE, Chirat S, Menteur S, Jestin L, Sarrafi A, Hayes PM, Jones BL, Blake TK, Wesenberg DM et al. Mapping of β-glucan content and β-glucanase activity loci in barley grain and malt. Theor Appl Genet. 1995;91:921–927. doi: 10.1007/BF00223901. [DOI] [PubMed] [Google Scholar]
- Hedin PA, Williams WP, Buckley PM, Da Vis FM. Arrestant responses of southwestern corn borer larvae to free amino acids: structure-activity relationships. J Chem Ecol. 1993;19:301–311. doi: 10.1007/BF00993697. [DOI] [PubMed] [Google Scholar]
- Iwai H, Masaoka N, Ishii T, Satoh S. A pectin glucuronyltransferase gene is essential for intercellular attachment in the plant meristem. Proc Natl Acad Sci USA. 2002;99:16319–16324. doi: 10.1073/pnas.252530499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HJG, Buxton DR. Forage quality variation among maize inbreds: relationships of cell-wall composition and in-vitro degradability for stem internodes. J Sci Food Agric. 1994;66:313–322. [Google Scholar]
- Kato Y, Ito S, Iki K, Matsuda K. Xyloglucan and β-d-glucan in cell walls of rice seedlings. Plant Cell Physiol. 1982;23:351–364. [Google Scholar]
- Kato Y, Nevins DJ. Enzymic dissociation of Zea shoot cell wall polysaccharides. Plant Physiol. 1984;75:740–744. doi: 10.1104/pp.75.3.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kianian SF, Phillips RL, Rines HW, Fulcher RG, Webster FH, Struthman DD. Quantitative trait loci influencing β-glucan content in oat (Avena sativa, 2n = 6x = 42) Theor Appl Genet. 2000;101:1039–1048. [Google Scholar]
- Kiesselbach TA. The Structure and Reproduction of Corn. Cold Spring Harbor, New York: Cold Spring Harbor Press; 1949. [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Laurie DA, Bennett MD. Nuclear DNA content in the genera Zea and Sorghum: intergeneric, interspecific and intraspecific variation. Heredity. 1985;55:307–313. [Google Scholar]
- Lee M, Sharopova N, Beavis WD, Grant D, Katt M, Blair D, Hallauer A. Expanding the genetic map of maize with the intermated B73 × Mo17 (IBM) population. Plant Mol Biol. 2002;48:453–461. doi: 10.1023/a:1014893521186. [DOI] [PubMed] [Google Scholar]
- Lempereur I, Rouau X, Abecassis J. Genetic and agronomic variation in arabinoxylan and ferulic acid contents of durum wheat (Triticum durum L.) grain and its milling fractions. J Cereal Sci. 1997;25:103–110. [Google Scholar]
- Lübberstedt T, Melchinger AE, Fähr S, Klein D, Dally A, Westhoff P. QTL mapping in testcrosses of flint lines of maize: III. Comparison across populations for forage traits. Crop Sci. 1998;38:1278–1289. [Google Scholar]
- Lübberstedt T, Melchinger AE, Klein D, Degenhardt H, Paul C. QTL mapping in testcrosses of European flint lines of maize: II. Comparison of different testers for forage quality traits. Crop Sci. 1997;37:1913–1922. [Google Scholar]
- Lundvall JP, Buxton DR, Hallauer AR, George JR. Forage quality variation among maize inbreds: in-vitro digestibility and cell-wall components. Crop Sci. 1994;34:1672–1678. [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC et al. Natural variation in light sensitivity of Arabidopsis. Nat Genet. 2001;29:441–446. doi: 10.1038/ng777. [DOI] [PubMed] [Google Scholar]
- Martin HL, Bamforth CW. The relationship between β-glucan solubilase, barley autolysis and malting potential. J Inst Brew. 1980;86:216–221. [Google Scholar]
- Martinant JP, Cadalen T, Billot A, Chartier S, Leroy P, Bernard M, Saulnier L, Branlard G. Genetic analysis of water extractable arabinoxylans in bread wheat endosperm. Theor Appl Genet. 1998;97:1069–1075. [Google Scholar]
- Méchin V, Argillier O, Menanteau V, Barriere Y, Mila I, Pollet B, Lapierre C. Relationship of cell wall composition to in vitro cell wall digestibility of maize inbred line stems. J Sci Food Agric. 2000;80:574–580. [Google Scholar]
- Millar AJ. Light responses of a plastic plant. Nat Genet. 2001;29:357–358. doi: 10.1038/ng1201-357. [DOI] [PubMed] [Google Scholar]
- Peng L, Kawagoe Y, Hogan P, Delmer D. Sitosterol-β-glucoside as primer for cellulose synthesis in plants. Science. 2002;295:147–150. doi: 10.1126/science.1064281. [DOI] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng WQ, Norambuena L, Orellana A, Raikhel NV, Keegstra K. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science. 1999;284:1976–1979. doi: 10.1126/science.284.5422.1976. [DOI] [PubMed] [Google Scholar]
- Powell W, Caligari PDS, Swanston JS, Jink JL. Genetical investigation into β-glucan content in barley. Theor Appl Genet. 1985;71:461–466. doi: 10.1007/BF00251188. [DOI] [PubMed] [Google Scholar]
- Reiter WD, Chapple CCS, Somerville CR. Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science. 1993;261:1032–1035. doi: 10.1126/science.261.5124.1032. [DOI] [PubMed] [Google Scholar]
- Rudall PJ, Caddick LR. Investigation of the presence of phenolic compounds in monocotyledonous cell walls, using UV fluorescence microscopy. Ann Bot. 1994;74:483–491. [Google Scholar]
- SAS Institute. SAS/STAT User's Guide: Statistics. Version 8. Ed 1. Cary, NC: SAS Institute Inc.; 2000. [Google Scholar]
- Saulnier L, Peneau N, Thibault JF. Variability in grain extract viscosity and water soluble arabinoxylan content in wheat. J Cereal Sci. 1995;22:259–264. [Google Scholar]
- Shanker S, Salazar RW, Taliercio EW, Chourney PS. Cloning and characterization of full-length cDNA encoding cell-wall invertase from maize. Plant Physiol. 1995;108:873–874. doi: 10.1104/pp.108.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Llaca V, Messing J. Mosaic organization of orthologous sequences in grass genomes. Genome Res. 2002;12:1549–1555. doi: 10.1101/gr.268302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stombaugh SK, Jung HG, Orf JH, Somers DA. Genotype and environmental variation in soybean seed cell wall polysaccharides. Crop Sci. 2000;40:408–412. [Google Scholar]
- Stuber CW. Case history in crop improvement: yield heterosis in maize. In: Paterson AH, editor. Molecular Dissection of Complex Traits. Boca Raton, FL: CRC Press; 1998. pp. 197–206. [Google Scholar]
- Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell. 2000;12:2529–2539. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD. The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA. 2002;99:3340–3345. doi: 10.1073/pnas.052450699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Ware D, Jaiswal P, Ni J, Pan X, Chang K, Clark K, Teytelman L, Schmidt S, Zhao W, Cartinhour S et al. Gramene: a resource for comparative grass genomics. Nucleic Acids Res. 2002;30:103–105. doi: 10.1093/nar/30.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]