Abstract
To bridge the gap between the rising information needs of biological and medical researchers and the rapidly growing number of online bioinformatics resources, we have created the Online Bioinformatics Resources Collection (OBRC) at the Health Sciences Library System (HSLS) at the University of Pittsburgh. The OBRC, containing 1542 major online bioinformatics databases and software tools, was constructed using the HSLS content management system built on the Zope® Web application server. To enhance the output of search results, we further implemented the Vivísimo Clustering Engine®, which automatically organizes the search results into categories created dynamically based on the textual information of the retrieved records. As the largest online collection of its kind and the only one with advanced search results clustering, OBRC is aimed at becoming a one-stop guided information gateway to the major bioinformatics databases and software tools on the Web. OBRC is available at the University of Pittsburgh's HSLS Web site (http://www.hsls.pitt.edu/guides/genetics/obrc).
INTRODUCTION
In the past decade, the emergence and rapid advance of genomic and proteomic technologies have generated never-before-seen amounts of genomic and proteomic data. As the genomes of 294 model organisms have been sequenced with 1206 more on the way (1), the amount of nucleotide sequence data alone nearly doubles every year. Such explosive growth of data has spawned hundreds of Web-based, publicly available bioinformatics resources, including databases and software tools, in various fields of biological sciences. The number of the online databases listed in the Nucleic Acids Research (NAR) Molecular Biology Database Collection alone has increased more than 14-fold from 58 in 1996 to 858 in 2006 (2). The majority of these newly emerged online resources are specialized databases and Web servers that provide not only sequence information, but also data on gene expression, macromolecular structures, genotype and phenotype of model organisms, as well as computational tools for analyzing macromolecular sequences/structures and global gene expression. Representing the best state of knowledge in the corresponding fields, these expert curated databases and specialized software tools may greatly assist researchers in designing their own experiments, as well as interpreting and validating their results.
Although the proliferation of bioinformatics databases is a manifestation of collective efforts by the life science community to help individual researchers coping with the phenomenal growth of biological data and information, many researchers find themselves struggling to keep up-to-date with the research in their fields (3,4). The situation is further exacerbated by the fact that locating such large numbers of online resources is anything but an easy task (5). The problem stems from the fact that the information about these online resources is scattered in various life science journals and around the Web, and that few web sites currently provide a guided access point with searchable links to a majority of these resources. Studies suggested that locating bioinformatics resources through literature searches is often very difficult (6–8). One study reported that >50% of the participating researchers use the Web to search for bioinformatics resources (9). However, searches using popular Web search engines, such as Google, are often ineffective. This is because Web search engines rank web sites by popularity rather than their relevance, and that Web search engines do not discriminate between reliable and unreliable web sites. The lack of standard search terms and the fact that Web search engines lump all hits together regardless of the nature of each hit, as long as they all contain the searched terms, further reduces the usefulness of the Web search engines as a mean to locate bioinformatics resources (5).
The urgent need of organizing the bioinformatics resources has recently been raised (5,10). Among the existing efforts to solve the problem are the Molecular Biology Database Collection compiled by the NAR (2), the Bioinformatics Links Directory (11,12), the Expasy Life Sciences Directory (http://www.expasy.org/links.html), the DBcat (13), the Database of Databases (14) and the Pathguide (15). Although these projects are highly valuable, their sole reliance on categorical content structure, limitations in annotation and coverage, and the lack of sophisticated search features may affect their usability and appeal to a wide audiences. For example, the output of search results from the Bioinformatics Links Directory is pages of a scrollable list, which may require users to examine the entire list in order to find the results relevant to their queries. There are also no ranking of the results or indications of any relationships that may exist among the results. Such limitations may pose even bigger problems as the number of the bioinformatics resources is expected to continuously grow at a rapid pace. Different approaches, such as using document clustering techniques (16) to organize search results, may enable users to quickly navigate through a large number of search results (17,18).
In order to help biomedical researchers to quickly find the most relevant bioinformatics resources for their specific information needs, we sought to develop a concrete and innovative search strategy as a part of a fledging library-based molecular biology information service at the Universtiy of Pittsburgh (19). For this purpose, we constructed the Online Bioinformatics Resources Collection (OBRC) at the Health Sciences Library System (HSLS), University of Pittsburgh. This collection currently includes 1542 online bioinformatics databases and software tools, most of which have been published by NAR or listed in its Molecular Biology Database Collection (2). In addition, we implemented the Vivísimo Clustering Engine® to OBRC to help users navigate through their search results.
METHODOLOGY
The new search strategy consists of two major components: a centralized collection of the curated information on major online bioinformatics databases and software tools, and the implementation of the Vivísimo Clustering Engine® to enhance the output of search results.
Source materials
The primary sources of OBRC are the databases and software tools published by the NAR (http://nar.oxfordjournals.org/). Specifically, the source materials were mainly the databases published in the NAR Annual Database Issues from 2001 to 2006, and the software tools published in the NAR Annual Web Server Issues from 2004 to 2006. Other databases listed in the NAR Molecular Biology Database Collection, including those published by NAR before 2001 and those not published by the NAR, were also selected. Selected databases and software tools described in other peer-reviewed journals, such as Bioinformatics and BMC Bioinformatics, were included in the collections. In addition, a number of unpublished but popular online software tools were also entered.
Collection construction, organization and maintenance
Information on each resource was entered using the HSLS content management system built on the Zope® Web application server. For each entry, the information for the following fields was entered: URL to the resource; name of the resource; a one-sentence description of the major functions; URL to the relevant PubMed abstract(s); last modification date of the entry; highlights of the resource; and keywords. The title, description and highlights for each entry were generated based on the PubMed abstract(s), as well as the content and scope of the resource. Together with the keywords, the textual information in these fields are automatically indexed by the Zope® Zcatalog and subsequently processed by the Zope®-based search engine.
As a major part of curation efforts, keywords were generated based on the information in the PubMed abstract(s), the MESH terms of the abstract(s), the information posted on corresponding web site, as well as the domain knowledge in molecular biology. Standard terminologies, commonly used by researchers in their publications, were used. The main types of keywords include biological concepts, entities, organism names, widely studied gene and protein names, and common molecular biology tasks. Whenever possible, common synonyms of the most important keywords were included as a conscious effort to improve the recall.
We implemented a categorical structure and basic classification theme that were derived from those used in the NAR Molecular Biology Database Collection (2). To facilitate users to browse OBRC, we consolidated the category structure and limited it to three levels. We also expanded the category names to make them more self-evident.
To ensure the up-to-dateness and running status of each entry, we perform link analysis and content verification at least every 6 months. The results are used to update the URLs and remove the entries that are no longer available.
Vivísimo Clustering Engine® implementation
The Vivísimo Clustering Engine® is based on a novel, intricate three-pass algorithm that is augmented with hundreds of special processing heuristics and endowed with thousands of specific facts and general patterns of English and other languages (http://Vivisimo.com/). It automatically organizes large number of search results into different groups and enables users to quickly survey and identify relevant groups. The Vivísimo Clustering Engine® has been successfully applied on the Web by search engines such as the Clusty (http://clusty.com) and ClusterMed™ (http://www.clustermed.info).
Queries can be formed with basic Boolean operators. Queries are first processed by the Zope®-based search engine that leverages on Zope® search tools. The results are then processed by the Vivísimo Clustering Engine® on-the-fly using the textual information from a set of fields selected from the following fields: title, descriptions, highlights and keywords. The search results organized by the Vivísimo Clustering Engine® are finally presented to the users.
RESULTS
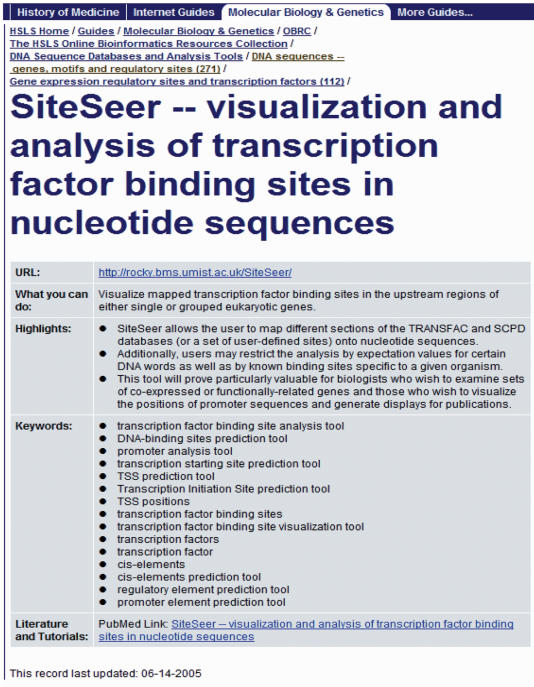
Figure 1 shows a sample record display of OBRC.
Figure 1.
The screenshot of a sample record display of OBRC.
There are a total of 1542 unique online bioinformatics resources in the current version of OBRC. The databases (475) and software tools (397) published in NAR Annual Database Issues (2001–2006) and Web Server Issues (2004–2006) contribute to ∼30.8 and 25.7% of the total entries in OBRC, respectively. The resources published in other journals (488) contribute to ∼31.6%. In addition, all the valid databases listed in the latest NAR Molecular Biology Database Collection (2) are included.
Organized with a three-level hierarchical category classification, OBRC was divided into 13 major categories, 40 secondary-categories and 12 tertiary-categories to assist users browsing the entire collection (Supplementary Table 1). The top five main categories are ‘DNA Sequence Databases and Analysis Tools’ (325), ‘Protein Sequence Databases and Analysis Tools’ (306), ‘Genomic Databases and Analysis Tools’ (270), ‘Structure Databases and Analysis Tools’ (244) and ‘RNA Databases and Tools’ (130). The top five specific topics are ‘Protein structures’ (214), ‘Regulatory sites and transcription factors’ (112), ‘Protein sequence motifs, active or functional sites, and functional annotations’ (77), ‘Human mutations and diseases’ (76) and ‘General protein sequence databases, sequence similarity search, analysis, and alignment tools’ (68). Some resources were listed in multiple categories.
DISCUSSION
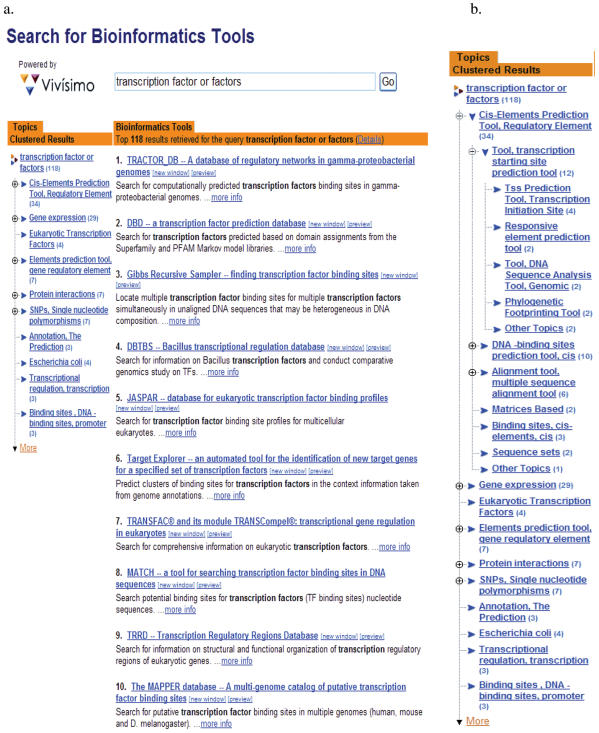
Studies have shown that the clustered results display is more efficient and user friendly than the traditional sequential search results display (20,21). Applying the Vivísimo Clustering Engine® to the search results offers the users not only a quick overview of all the search results requiring little scrolling, but also shows how the search results are related to each other, as represented by the themes (Figure 2). This advantage becomes compelling in cases where a large number of search results are returned, as the clustered results display drastically reduce the effort needed to navigate through the results set in order to locate the most relevant ones. The sequential display, as employed by popular Web search engines, requires users to scroll down page by page in order to find the results specific to their needs. Another benefit brought by the Vivísimo Clustering Engine® is that users can use relatively broad query terms and may still able to find specific results quickly. This could be particularly helpful to users during their searches as it may reduce the efforts on query reformulation. Furthermore, with Vivísimo's document clustering, there is little need for the expensive and laborious tasks of creating a controlled vocabulary and/or to extensively indexing or pre-labeling the documents.
Figure 2.
(a) The screenshot of the first page results for the testing query ‘transcription factor or factors’ from searching the OBRC using the Zope®-based search engine coupled with the Vivísimo Clustering Engine®. (b) The expanded view of the major clusters of the search results.
Our preliminary evaluation study suggests that OBRC search strategy performs much better than Web search engine based strategy, largely attributed to its centralized collection and curated keywords (data not shown). However, the recall and precision are still imperfect. A close examination of the search results indicates that the false negatives, which lower the recall, are primarily due to the synonym problems that have long plagued information retrieval in the biological literatures (22). Another main cause is the singular or plural form of terminologies. Such problems can be largely circumvented by implementing a special online thesaurus or synonym mapping protocol in OBRC. The false positives, which lower the precision, are mainly attributed to the fact that the Zope®-based search engine searches all the text fields of each OBRC entry, and sometimes words in some of the fields match with the queries despite their irrelevance to the major content/function of the corresponding database/software tool. Such false positives could be entirely eliminated if the Zope®-based search engine searched only the keyword field of each OBRC entry. A tradeoff of such strategy is that the keywords are generated to represent only the main concepts, contents and functions of an underlying database/software tool, thus restricting the search to only the keywords field may result in lower recall as the less relevant database/software tools are likely to be left out.
CONCLUSIONS
We have created the OBRC, covering the most widely used and authoritative open source bioinformatics databases and software tools on the Web. The implementation of the Vivísimo Clustering Engine® in OBRC enhances the output of search results and may help users to navigate through large numbers of results with ease. The rich content in OBRC coupled with the advance search features represents a novel search solution for online bioinformatics resources that will benefit biomedical researchers at large. Its aggregated content may also be useful as part of an integrated biological information system.
A future direction will be to continue to expand OBRC to include databases and software tools published in other journals. We will also explore new methods, such as constructing an embedded synonym mapping protocol, implementing the Vivísimo domain-specific controlled vocabularies to further boost the recall and precision, as well as to enhance the results clustering process. Additionally, we will improve the usability of OBRC by studying user experiences and implementing other features, such as adding RSS feed and user/curator preferences/ratings of each resource. We welcome any comments and suggestions on further improvement of OBRC.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors wish to thank Ms Barbara Epstein for her support of this project, and Ms Jill Foust for Web maintenance of OBRC. This research was supported by the National Library of Medicine Biomedical Informatics Training Grant to Y.-B.C. under contract no. 5T15LM007059-18. Funding to pay the Open Access publication charges for this article was provided by the Health Sciences Library System, University of Pittsburgh.
REFERENCES
- 1.Liolios A., Tavernarakis N., Kyrpides N.C. The Genomes On Line Database (GOLD) v.2: a monitor of genome projects world-wide. Nucleic Acids Res. 2006;34:D332–D334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galperin M.Y. The Molecular Biology Database Collection: 2006 update. Nucleic Acids Res. 2006;34:D3–D5. doi: 10.1093/nar/gkj162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kostoff R. The extraction of useful information from the biomedical literature. Acad. Med. 2001;76:1265–1270. doi: 10.1097/00001888-200112000-00025. [DOI] [PubMed] [Google Scholar]
- 4.Kostoff R. Role of technical literature in science and technology development and exploitation. J. Inform. Sci. 2003;29:223–228. [Google Scholar]
- 5.Cannata N., Merelli E., Altman R.B. Time to organize the Bioinformatics Resourceome. PLoS Comput. Biol. 2005;1:e76. doi: 10.1371/journal.pcbi.0010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grivell L. Mining the bibliome: searching for a needle in a haystack? EMBO Rep. 2002;3:200–203. doi: 10.1093/embo-reports/kvf059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schilling L.M., Wren J.D., Dellavalle R.P. Letter to the editor: Bioinformatics leads charge by publishing more Internet addresses in abstracts than any other journal. Bioinformatics. 2004;20:2903. doi: 10.1093/bioinformatics/bth385. [DOI] [PubMed] [Google Scholar]
- 8.Wren J.D. 404 not found: the stability and persistence of URLs published in MEDLINE. Bioinformatics. 2004;20:668–672. doi: 10.1093/bioinformatics/btg465. [DOI] [PubMed] [Google Scholar]
- 9.Lu D. Information needs of biologists for online bioinformatics resources: implications for health science information professionals. Proceedings, 105th Annual Meetings Medical Library Association, Inc., 94, E21; May 14–19, 2005; San Antonio, TX. 2006. [Google Scholar]
- 10.Teufel A., Krupp M., Weinmann A., Galle P.R. Current bioinformatics tools in genomic biomedical research. Int. J. Mol. Med. 2006;17:967–973. [PubMed] [Google Scholar]
- 11.Fox J.A., Butland S.L., McMillan S., Campbell G., Ouellette B.F.F. The Bioinformatics Links Directory: a compilation of molecular biology web servers. Nucleic Acids Res. 2005;33:W3–W24. doi: 10.1093/nar/gki594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox J.A., Butland S.L., McMillan S., Ouellette B.F.F. A compilation of molecular biology web servers: 2006 update on the Bioinformatics Links Directory. Nucleic Acids Res. 2006;34:W3–W5. doi: 10.1093/nar/gkl379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Discala C., Benigni X., Barillot E., Vaysseix G. DBcat: a catalog of 500 biological databases. Nucleic Acids Res. 2000;28:8–9. doi: 10.1093/nar/28.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babu P.D., Boddepalli R., Lakshmi V.V., Rao G.N. DoD: Database of Databases—updated molecular biology databases. In Silico Biol. 2005;5:605–610. [PubMed] [Google Scholar]
- 15.Bader G.D., Cary M.P., Sander C. Pathguide: a Pathway Resources List. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salton G. Cluster search strategies and the optimization of retrieval effectiveness. In: Salton G., editor. The SMART Retrieval System. NJ: Prentice Hall, Englewood Cliffs; 1971. pp. 223–242. [Google Scholar]
- 17.Zamir O., Etzioni O. Web document clustering: a feasibility demonstration. Proceedings of the 21st International ACM SIGIR Conference on Research and Development of Information Retrieval (SIGIR'98); August 24–28; Melbourne, Australia. NY: ACM Press; 1998. pp. 46–54. [Google Scholar]
- 18.Zeng H., He Q., Chen Z., Ma W., Ma J. Learning to cluster Web search results. Proceedings of the 27th annual international conference on research and development in information retrieval (SIGIR'04); July 25–29; Sheffield, UK. NY: ACM Press; 2004. pp. 210–217. [Google Scholar]
- 19.Chattopadhyay A., Tannery N.H., Silverman D.A.L., Bergen P., Epstein B.A. Design and implementation of a library-based information service in molecular biology and genetics at the University of Pittsburgh. J. Med. Libr. Assoc. 2006;94:307–313. [PMC free article] [PubMed] [Google Scholar]
- 20.Leuski A. Evaluating document clustering for interactive information retrieval. Proceedings of 10th International Conference on Information and Knowledge Management (CIKM'01); November 5–10; Atlanta, GA. NY: ACM Press; 2001. pp. 33–40. [Google Scholar]
- 21.Wu M., Fuller M., Wilkinson R. Using clustering and classification approaches in interactive retrieval. Inf. Proc. Manage. 2001;37:459–484. [Google Scholar]
- 22.Shatkay H., Feldman R. Mining the biomedical literature in the genomic era: an overview. J. Comput. Biol. 2003;10:821–855. doi: 10.1089/106652703322756104. [DOI] [PubMed] [Google Scholar]