Abstract
We have developed the database, TMBETA-GENOME, for annotated β-barrel membrane proteins in genomic sequences using statistical methods and machine learning algorithms. The statistical methods are based on amino acid composition, reside pair preference and motifs. In machine learning techniques, the combination of amino acid and dipeptide compositions has been used as main attributes. In addition, annotations have been made using the criterion based on the identification of β-barrel membrane proteins and exclusion of globular and transmembrane helical proteins. A web interface has been developed for identifying the annotated β-barrel membrane proteins in all known genomes. The users have the feasibility of selecting the genome from the three kingdoms of life, archaea, bacteria and eukaryote, and five different methods. Further, the statistics for all genomes have been provided along with the links to different algorithms and related databases. It is freely available at http://tmbeta-genome.cbrc.jp/annotation/.
INTRODUCTION
The β-barrel membrane proteins perform a variety of functions, such as pore formation, membrane anchoring, enzyme activity, bacterial virulence, mediating non-specific, passive transport of ions and small molecules, selectively passing the molecules such as maltose and sucrose and are involved in voltage-dependent anion channels (1). The annotation of β-barrel membrane proteins in genomic sequences will be helpful for understanding their functions. In our earlier works, we have developed statistical methods and machine learning techniques for discriminating transmembrane β-barrel proteins (TMBs) from globular and transmembrane helical (TMH) proteins (2–5) and we obtained the accuracy in the range of 89–95% in discriminating TMBs. These methods are mainly based on amino acid composition, residue pair preference/dipeptide composition, motifs and the combinations of them. On the other hand, different methods, such as hidden Markov models, neural networks and nearest neighbor algorithms, have been proposed for discriminating TMBs and screening them in genome sequences (6–12). However, there is no electronically available database for the annotated β-barrel membrane proteins in genomes.
In this work, we have developed a database, TMBETA-GENOME, which has the annotated β-barrel membrane proteins for all the completed genomes. The annotation has been carried out with several statistical methods and machine learning techniques along with a new approach based on detecting β-barrel membrane proteins and eliminating other folding types of globular and membrane proteins. The users have the feasibility of selecting the method and the genome. The database is freely available at http://tmbeta-genome.cbrc.jp/annotation/.
CONTENTS OF THE DATABASE
TMBETA-GENOME contains the annotated β-barrel membrane proteins for 275 completed genomes, including 23 genomes from archaea, 237 from bacteria and 15 from eukaryote. It may be noted that very few TMBs are annotated experimentally in the eukaryote proteomes. Further, for a genome that have different chromosomes (e.g. human genome has 24 chromosomes) the data for each chormosome is given individually. This increased the total number of entries into 24, 254 and 149, respectively, for archaea, bacteria and eukaryote. The total number of proteins in these three kingdoms of life is 52 241, 686 562 and 165 186, respectively, with the total of 903 989 sequences. The amino acid sequences of all the genomes have been taken from the NCBI database (http://www.ncbi.nih.gov/). In addition, we have provided the statistics for the annotated β-barrel membrane proteins in all the genomes by different discrimination methods (see below).
DISCRIMINATION METHODS
The annotation results are accumulated for different statistical methods and machine learning techniques. The statistical methods include the composition of amino acid residues (2), residue pair preference (3) and motifs (4). In these methods the compositions of amino acid residues/residue pairs/residue pairs with a gap (motif) have been computed for a training set of 674 globular and 377 TMB proteins obtained from Protein Data Bank (PDB) and PSORT database, respectively (2–4,13,14). For a new protein X, we have calculated the deviations of the amino acid composition between protein X and globular/TMBs. The protein is said to be a TMB if the deviation is the lowest for TMB and vice versa (2). For residue pair preference and motif, the compositional difference between globular and TMBs have been calculated (σTMB-glob). The weighted average of σTMB-glob with the dipeptide/motif composition of protein X discriminates the TMB and globular protein (3,4). We have also applied support vector machines (5) for discriminating TMBs, which uses the combinations of amino acid composition and residue pair preference. Further, the program, SOSUI (15) has been used for finding the TMH in genomic sequences. In the new approach we have used the following criteria: (i) identification of TMBs using the preference of residue pairs in globular, TMH and TMBs, (ii) elimination of globular/TMH proteins that show the sequence identity of >70% for the coverage of 80% residues with known structures in PDB, (iii) elimination of globular/TMH proteins that have the sequence identity of >60% with known sequences in SWISS-PROT, and (iv) exclusion of TMH proteins using SOSUI, a prediction system for TMH proteins. This method also showed good agreement with experimental observations.
FEATURES OF TMBETA-GENOME
TMBETA-GENOME includes several features, such as, the service for detecting TMBs in genomic sequences using various methods, related references, statistics for the detected TMBs by different methods for each genome, details about all algorithms used to detect TMBs, relative links to other databases and a help page. The ‘help’ section illustrates the details to perform the search and to obtain the results.
ACCESS TO TMBETA-GENOME
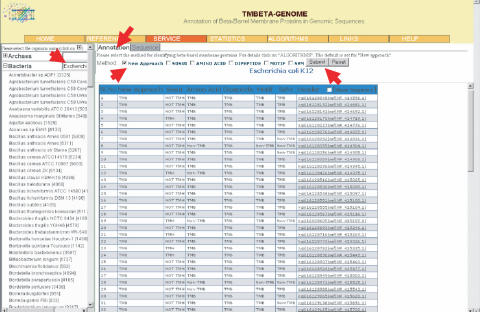
TMBETA-GENOME can be directly accessed through the web at http://tmbeta-genome.cbrc.jp/annotation/. The users have the feasibility to select the method of annotation and name of the genome. This server provides the annotated TMBs using our previous methods, such as, statistical, dipeptide, motif and SVM as well as the ‘New Approach’. The new approach considerably reduced the number of false positives and it has the ability of picking up most of the real TMBs. An example is shown in Figure 1. In this figure the results are shown for Escherichia coli K12 genome. This can be obtained by clicking on the button + Bacteria and selecting the name of the genome. It is also possible to get the data by entering the name of the genome.
Figure 1.
Illustration for searching the database. The arrows indicate the selected items, (i) name of the genome (Escherichia coli K12), (ii) method (New Approach) and (iii) annotation (set as default). Each change can be taken into account by clicking on the ‘Submit’ button. The displayed results are also shown in this figure.
We have selected the method, ‘New Approach’ for obtaining the annotated TMBs. This search picked up 87 entries and the TMBs identified by the new approach are shown with the identification number. In addition, the results obtained with other methods are also given for comparison. The method SVM yielded 337 TMBs and the combination of ‘Amino acid’ and ‘Dipeptide’ showed 501 TMBs. It is noteworthy that several discrimination methods including statistical, dipeptide and motifs have a tendency of providing many false positives. The new approach results are reasonable that it identified just 2.05% of the proteins as TMBs. Further, the comparison between identified TMBs and experimentally known TMBs revealed that the new approach could correctly pick up all the 11 TMBs of known three-dimensional structures obtained from E.coli and representative proteins in all the families of Transport Classification Database (TCDB, 16). Hence, we suggest to use the new approach for obtaining TMBs in genomic sequences.
Few other methods have also been proposed for detecting TMBs and these methods have their own advantages and shortcomings (6–12). Zhai and Saier (10) developed a program, β-barrel finder, based on secondary structure, hydropathy and amphipathicity for identifying β-barrel membrane proteins in prokaryotic genomes. This method detected 118 TMBs in E.coli genome and it could identify representative proteins from nine among 15 families available in TCDB. The method based on k-nearest neighbor missed few TMBs with the E-value > 3 although these proteins have been used in the training set to develop the method (9). The BOMB server based on (i) C-terminal pattern typical of many integral β-barrel proteins and (ii) integral β-barrel score based on the extent to which the sequence contains stretches of amino acids typical of transmembrane β-strands missed eight proteins (11). The profile based hidden Markov model (12) identified TMBs belonging to 12 families in TCDB. This analysis reveals that the results obtained with new approach are better than other methods in the literature.
For more details about the protein the appropriate links to NCBI protein sequences have been provided for each annotated protein. Further, the users have the feasibility of downloading the annotated TMBs in FASTA format, which can be used for further analysis. The complete sequences of the genome have been obtained by selecting the ‘Sequence’ button. It is also possible to download all the protein sequences of the specific genome in FASTA format.
LINKS TO OTHER DATABASES
Each protein in the specified genome as well as the annotated TMBs are directly linked with NCBI protein sequences. Further, TMBETA-GENOME is linked with several related genome, structure and sequence databases, such as, Genome Online Database [GOLD; (17)], NCBI (http://www.ncbi.nlm.nih.gov/Genomes/), KEGG (http://www.genome.jp/), PDB (13), SWISS-PROT (http://www.expasy.org/sprot/), Protein Information Resource (PIR; http://pir.georgetown.edu), Uniprot (18), Protein Data Bank of Transmembrane proteins [PDBTM; (19)], Transport Classification Database [TCDB; (16)] etc.
AVAILABILITY AND CITATION OF TMBETA-GENOME
The database can be freely accessible at http://tmbeta-genome.cbrc.jp/annotation/. If this database is used as a tool in your published research work, please cite this article including the URL. Suggestions and comments are welcome and should be sent to michael-gromiha@aist.go.jp.
Acknowledgments
We thank Dr Yutaka Akiyama for encouragement. Funding to pay the Open Access publication charges for this article was provided by the National Institute of Advanced Industrial Science and Technology (AIST).
Conflict of interest statement. None declared.
REFERENCES
- 1.Koebnik R., Locher K.P., Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 2.Gromiha M.M., Suwa M. A simple statistical method for discriminating outer membrane proteins with better accuracy. Bioinformatics. 2005;21:961–968. doi: 10.1093/bioinformatics/bti126. [DOI] [PubMed] [Google Scholar]
- 3.Gromiha M.M., Ahmad S., Suwa M. Application of residue distribution along the sequence for discriminating outer membrane proteins. Comput. Biol. Chem. 2005;29:135–142. doi: 10.1016/j.compbiolchem.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Gromiha M.M. Motifs in outer membrane protein sequences: applications for discrimination. Biophys. Chem. 2005;117:65–71. doi: 10.1016/j.bpc.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Park K.J., Gromiha M.M., Horton P., Suwa M. Discrimination of outer membrane proteins using support vector machines. Bioinformatics. 2005;21:4223–4229. doi: 10.1093/bioinformatics/bti697. [DOI] [PubMed] [Google Scholar]
- 6.Martelli P.L., Fariselli P., Krogh A., Casadio R. A sequence-profile-based HMM for predicting and discriminating beta barrel membrane proteins. Bioinformatics. 2002;18:S46–S53. doi: 10.1093/bioinformatics/18.suppl_1.s46. [DOI] [PubMed] [Google Scholar]
- 7.Bagos P.G., Liakopoulos T.D., Spyropoulos I.C., Hamodrakas S.J. A hidden Markov model method, capable of predicting and discriminating beta-barrel outer membrane proteins. BMC Bioinformatics. 2004;5:29. doi: 10.1186/1471-2105-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Natt N.K., Kaur H., Raghava G.P. Prediction of transmembrane regions of beta-barrel proteins using ANN- and SVM-based methods. Proteins. 2004;56:11–18. doi: 10.1002/prot.20092. [DOI] [PubMed] [Google Scholar]
- 9.Garrow A.G., Agnew A., Westhead D.R. TMB-Hunt: a web server to screen sequence sets for transmembrane beta-barrel proteins. Nucleic Acids Res. 2005;33:W188–W192. doi: 10.1093/nar/gki384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhai Y., Saier M.H., Jr The β-barrel finder (BBF) program, allowing identification of outer membrane β-barrel proteins encoded within prokaryotic genomes. Protein Sci. 2002;11:2196–2207. doi: 10.1110/ps.0209002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berven F.S., Flikka K., Jensen H.B., Eidhammer I. BOMP: a program to predict integral β-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res. 2004;32:W394–W399. doi: 10.1093/nar/gkh351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bigelow H.R., Petrey D.S., Liu J., Przybylski D., Rost B. Predicting transmembrane beta-barrels in proteomes. Nucleic Acids Res. 2004;32:2566–2577. doi: 10.1093/nar/gkh580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardy J.L., Spencer C., Wang K., Ester M., Tusnady G.E., Simon I., Hua S., de Fays K., Lambert C., Nakai K., Brinkman F.S. PSORT-B: Improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 2003;31:3613–3617. doi: 10.1093/nar/gkg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirokawa T., Boon-Chieng S., Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 16.Busch W., Saier M.H., Jr The transporter classification (TC) system. Crit Rev Biochem Mol Biol. 2002;37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 17.Liolios K., Tavernarakis N., Hugenholtz P., Kyrpides N.C. The Genomes On Line Database (GOLD) v.2: a monitor of genome projects worldwide. Nucleic Acids Res. 2006;34:D332–D334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The universal protein resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusnady G.E., Dosztanyi Z., Simon I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic Acids Res. 2005;33:D275–D278. doi: 10.1093/nar/gki002. [DOI] [PMC free article] [PubMed] [Google Scholar]