Abstract
The LIPID MAPS Structure Database (LMSD) is a relational database encompassing structures and annotations of biologically relevant lipids. Structures of lipids in the database come from four sources: (i) LIPID MAPS Consortium's core laboratories and partners; (ii) lipids identified by LIPID MAPS experiments; (iii) computationally generated structures for appropriate lipid classes; (iv) biologically relevant lipids manually curated from LIPID BANK, LIPIDAT and other public sources. All the lipid structures in LMSD are drawn in a consistent fashion. In addition to a classification-based retrieval of lipids, users can search LMSD using either text-based or structure-based search options. The text-based search implementation supports data retrieval by any combination of these data fields: LIPID MAPS ID, systematic or common name, mass, formula, category, main class, and subclass data fields. The structure-based search, in conjunction with optional data fields, provides the capability to perform a substructure search or exact match for the structure drawn by the user. Search results, in addition to structure and annotations, also include relevant links to external databases. The LMSD is publicly available at www.lipidmaps.org/data/structure/
INTRODUCTION
Lipids and their metabolites play an important role in the regulation and control of cellular function and disease. To achieve a complete understanding of the involvement of lipids in physiological processes and to develop compounds of therapeutic interest, it is vital not only to identify and characterize existing and novel lipids but also to quantify changes in their metabolites, and develop biochemical pathways and interaction network maps involving lipids. The LIPID metabolites and pathways strategy (LIPID MAPS) consortium, a multi-institutional multi-year effort, is involved in this endeavor.
In addition to developing a variety of computational tools and the underlying infrastructure to integrate, analyze, track and disseminate large volumes of heterogeneous chemical, biological and analytical data generated by multi-disciplinary research groups, we have also developed these two databases: LIPID MAPS Structure Database (LMSD) and LIPID MAPS Proteome Database (LMPD). We present our work on LMSD in this report; LMPD, encompassing information about proteins and genes involved in lipid metabolic pathways, has been described previously (1).
Existing lipid websites include LIPID BANK (2) for Web (www.lipidbank.jp), LIPIDAT (3) (www.lipidat.chemistry.ohio-state.edu), Lipid Library (www.lipidlibrary.co.uk) and Cyberlipids (www.cyberlipid.org). In addition to lipid structures, these websites also provide other relevant chemical and biological information. LMSD, similar to the extant online resources, also contains structures of lipids and other related information. However, unlike the existing LIPID BANK and LIPIDAT databases, all lipids in LMSD are classified systematically using a comprehensive classification scheme proposed by LIPID MAPS (4). Additionally, all the lipid structures are drawn in a consistent fashion based on a defined drawing scheme. We describe these and other distinctions between LMSD and existing lipid databases in detail under the database content and description section.
DATABASE CONTENT AND DESCRIPTION
A classification and nomenclature system for lipids
In order to address the lack of a consistent classification and nomenclature methodology for lipids, LIPID MAPS consortium members have developed a comprehensive classification system for lipids (4). Based on this classification system, lipids have been divided into eight categories: (i) fatty acyls, (ii) glycerolipids, (ii) glycerophospholipids, (iv) sphingolipids, (v) sterol lipids, (vi) prenol lipids, (vii) saccharolipids and (viii) polyketides (Figure 1). Each category is further divided into classes and subclasses. Additionally, following the existing rules and recommendations proposed by the International Union of Biochemistry and Applied Chemists and the International Union of Biochemistry and Molecular Biology (IUPAC-IUBMB) commission on Biochemical Nomenclature (5–19), a consistent nomenclature scheme has also been developed to provide systematic names for various classes and subclasses of lipids.
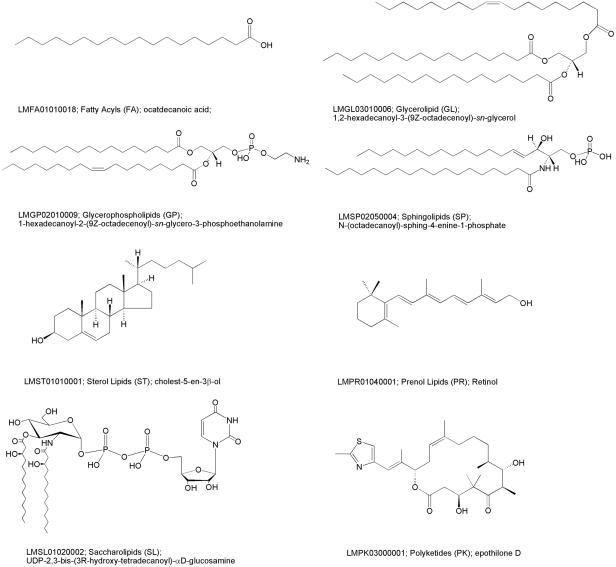
Figure 1.
Representative structures for each lipid category along with LM ID, category name, category abbreviation and systematic name.
All lipids in LMSD are classified and annotated using this comprehensive classification and nomenclature system developed by the LIPID MAPS consortium.
Representation of lipid structures
Currently, different members of the lipids community draw lipid structures in distinct ways. The same lipid structure in one lipid database can appear quite different in another database (2,3). Moreover, large and complex lipids are rather difficult to draw manually which leads to proliferation of shorthand and other abbreviations to represent lipid structures. In order to address these issues, the LIPID MAPS consortium proposed a consistent framework for representing lipid structures (4).
For fatty acids and derivatives, the acid group or its equivalent is drawn on the right side and hydrophobic chain is on the left, except for the eicosanoid class in which the hydrocarbon chain wraps around in a counter clockwise fashion to produce a more compact structure. For glycerolipids and glycerophospholipids, the radyl hydrocarbon chains are drawn to the left; the glycerol group is drawn horizontally with stereochemistry defined at sn carbons; the headgoups for glycerophospholipids are depicted on the right. For sphingolipids, the C1 hydroxyl group of the long-chain base is placed on the right and the alkyl portion on the left; the headgroup of sphingolipids ends up on the right. Sterol structures are drawn using recommendations by IUPAC–IUMBMB (19). The linear prenols and isoprenoids are drawn like fatty acids with the terminal functional groups on the right. A number of structurally complex lipids—acylaminosugar glycans, polycyclic isoprenoids and polyketides—cannot be drawn using these simple rules; these structures are drawn using commonly accepted representations.
Structures of all lipids in LMSD adhere to the structure drawing rules proposed by the LIPID MAPS consortium. Figure 1 shows representative structures for each lipid category.
Populating structure database
Lipid structures in LMSD are obtained from these four sources: (i) LIPID MAPS Consortium's core laboratories and partners; (ii) lipids identified by LIPID MAPS experiments; (iii) computationally generated structures for appropriate lipid classes; (iv) biologically relevant lipids manually curated from LIPID BANK, LIPIDAT and other public sources such as appropriate scientific literature.
After lipids have been selected for inclusion into LMSD, they are classified following the LIPID MAPS classification scheme as explained earlier under the classification and nomenclature system for lipids section. Structures of the lipids are drawn either manually or generated automatically by computational structure drawing tools developed by the LIPID MAPS consortium; the structure representation is consistent and adhere to the rules proposed by LIPID MAPS consortium. Based on its classification, each lipid structure in LMSD is assigned a unique 12 character LIPID MAPS identifier (LM ID). The format of the LM ID (Table 1) not only maintains uniqueness of ID but also provides the capability to add new categories, classes and subclasses as the need arises.
Table 1.
LIPID ID (LM ID) format
| Characters | Position | Description |
|---|---|---|
| LMFA01030001 | 1–2 | Database designation |
| LMFA01030001 | 3–4 | Two-letter category code |
| LMFA01030001 | 5–6 | Two-digit class code |
| LMFA01030001 | 7–8 | Two-digit subclass code |
| LMFA01030001 | 9–12 | Unique four character identified with in a subclass |
In addition to manual curation of biologically relevant lipids from LIPIDAT and LIPID BANK according to LIPID MAPS classification and structure representation schemes, LMSD also maintains their original IDs to enable cross-referencing. LMSD lipid structures are deposited into PubChem database (http://pubchem.ncbi.nlm.nih.gov/) periodically and a link to PubChem substance ID (SID) is also maintained within LMSD. Access to complete set of LMSD lipid structures in PubChem is available at www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pcsubstance&term=LipidMAPS[sourcename]).
Structural representation of positional isomers
LIPID MAPS core laboratories that are engaged in identification, characterization and quantification of known and new lipids using LC and MS experimental techniques; information about various lipid standards developed for these experiments, along with the protocols used, is available on LIPID MAPS website (www.lipidmaps.org/data/standards/). However, for some lipid categories, such as glycerolipids and glycerophospholipids, it is not always straightforward to identify the positions of radyl hydrocarbon chains at the sn carbons on the glycerol group. For example, MS/MS experiments might be able to identify the presence of three radyl hydrocarbons chains in a triacylglycerol but their positions on the glycerol backbone would be unknown. Combinatorial enumeration of the three radyl chains at sn carbons leads to six possible isomeric structures. These positional isomers are stored in LMSD as one structure and it is marked as a computationally generated structure. Structures for all other positional isomers are created on demand. To indicate the positional isomeric nature of the structure, a suffix ‘iso' followed by the number of isomers is also added to the abbreviation used as common name. For example, entry LMGL03010043 in LMSD, with common name TG(16:0/16:1(9Z)/18:1(9Z))[iso6] and systematic name 1-hexadecanoyl-2-(9Z-hexadecenoyl)-3-(9Z-octadecenoyl)-sn-glycerol, represents a lipid structure with six possible positional isomers.
Structural representation of glycans in glycosphingolipids
For structural representation of lipids in neutral and acidic glycosphingolipids—main classes under sphingolipids category—LMSD uses the symbol and text nomenclature as proposed by the Consortium for Functional Glycomics nomenclature committee on symbol and text representation of glycan structures (www.functionalglycomics.org/). In addition to using symbol and text representation for glycans, the last four digits of LM ID are further subdivided into two groups: The first two positions are used to differentiate glycan series within a subclass; the last two positions represent a unique ID. For the first two positions, only letters are used; the last two positions use combinations of numbers and letters.
DATABASE IMPLEMENTATION AND USER INTERFACES
LMSD is implemented as a relational database, using Oracle9i Enterprise Edition Release 9.2.0.2.4 running on a Sun Fire 880 with the Solaris 9 operating system. Perl scripts and Oracle SQL*Loader are used to parse and load data from flat-files and Structure Data Format (SDF) files (described under section MDL CTfile Formats at www.mdli.com) into Oracle database tables. Structure data from SDF files are loaded into Oracle via SQL scripts developed using the Accord Chemical Cartridge available from Accelrys (www.accelrys.com/products/accord/).
The LMSD query and graphical user interface (GUI) is implemented in Perl and PHP using the Apache 1.3.26 Web Server running on a Sun Ultra-80 with the Solaris 9 operating system.
An entity-relationship diagram is available as Supplementary Data at www.lipidmaps.org/data/structure/supplementarymaterial/NAR/2007/ER_diagram.gif.
Generation and storage of lipid structures
LMSD implements storage of lipid structure representations using the following three formats: binary large object (BLOB), ChemDraw Exchange (CDX) format (www.cambridgesoft.com/services/documentation/sdk/chemdraw/cdx/) and Graphics Interchange Format (GIF). LMSD uses BLOB format to store MDL MOL file structural representation via the Accord Chemical Cartridge. CDX format, a richer and flexible format with support for not only structure data but also for visual characteristics and annotations, is stored in LMSD as Character Large Object (CLOB) data; CDX format objects are used to support viewing of structures via ChemDraw viewer. GIF images representing structures are stored in the database table as BLOB objects.
The structures are either drawn manually using Chemdraw Ultra 8.0 (www.cambridgesoft.com/software/ChemDraw/) or generated automatically by structure drawing tools developed by LIPID MAPS consortium. We have developed the structure drawing tools for various subclasses in fatty acyls, glycerolipids, glycerophospholipids, sphingolipids and sterols; these tools are publicly available at www.lipidmaps.org/tools/structuredrawing/. The structure drawing tools are Perl scripts, which can generate a large number of structures relatively quickly via command-line or web-based interface. In addition to consistent structure representations from lipid abbreviations, these scripts also generate ontological information such as number of double bonds, chain lengths at different positions on the glycerol backbone, number of various functional groups and other structural characteristics. The ontological information is also loaded into LMSD.
The GIF files for lipid structures are generated by a custom Visual Basic (VB) executable running under Microsoft Windows XP operating system; The VB executable uses ChemDraw Ultra 8.0 objects (www.cambridgesoft.com/software/ChemDraw/). These structure GIF files are moved to the Sun Fire 880 and loaded into Oracle database tables as binary objects using SQL scripts.
Lipid classification-based browsing
The LMSD browsing page provides the capability to retrieve lipids based on the LIPID MAPS classification scheme. After the user selects one of the main categories of lipids, a listing of all lipids present in the selected category, along with a link to the set of lipids in each main class and subclass, is provided. The user may then select all lipids, which belong to either a main class or a subclass and display the results as a result summary page.
In case of lipids containing multiple functional groups, assignment of a structure to a particular subclass may be somewhat subjective. For example, a fatty acid containing both epoxy and hydroxy groups could be assigned to either epoxy or hydroxy fatty acids subclass. To address this situation, ontology-based search is also provided. The user may choose to search for lipids containing similar functionality and all the lipids with the specific functionality, irrespective of their subclass designation, would be retrieved.
Text-based search
The text-based query page allows the user to search LMSD by any combination of these data fields: LM ID, common or systematic name, mass along with a tolerance value, formula, category, main class and subclass. Selecting a category from the category drop-down menu causes the corresponding set of main classes to appear in the main class drop-down menu. Selecting a main class then shows the corresponding set of subclasses in the subclass menu.
Before performing the database search, an SQL query statement is constructed by joining all the specified data fields by an AND operation. Initiating the search using the default page, without specifying any search parameters, retrieves all the records in LMSD.
Structure-based search
The structure-based query page provides the capability to search LMSD by performing a substructure or exact match using the structure drawn by the user. Two supported structure drawing tools are MarvinSketch (www.chemaxon.com/marvin/) and JME (www.molinspiration.com/jme/index.html). Both of these structure drawing tools are Java applets and require only applet support in the browser. In addition to structure, the user can also specify LM ID and common or systematic name for the search. The default search page, without specification of any structural and textual parameters, retrieves all the records in LMSD; the results summary page displays up to 50 records per page.
Results summary
The results summary page displays a table containing all the lipids matching specified search criteria. The results table contains the following columns for text-based and structure-based queries: LM ID, common name, systematic name, main class, subclass and exact mass. For lipid classification-based browsing, instead of displaying main class and subclass columns, the results table displays a formula column. From the results page, the user can select the LM ID of any lipid and display the record details page for the selected lipid.
Record details
The record details page, in addition to displaying the structure for the selected lipid, also contains the following data fields: LM ID, common name, systematic name, formula, category, main class, subclass, synonyms and status. Appropriate links to LIPID BANK ID, PubChem SID and SphinGOMAP ID (20) are also provided.
The default page uses a GIF image for representing structure of the lipid. These additional structure viewing tools are also supported: MarvinView (www.chemaxon.com/products.html), JMol (http://jmol.sourceforge.net/) and Chem Draw.ActiveX/Plugin Viewer (www.cambridgesoft.com/software/ChemDraw/). Both MarvinView and JMol are Java applets and require applet support in the browser; ChemDraw structure view uses ChemDraw ActiveX Control/Plugin Viewer, which must be downloaded and installed by the user for the appropriate browser environment supported by CambridgeSoft.
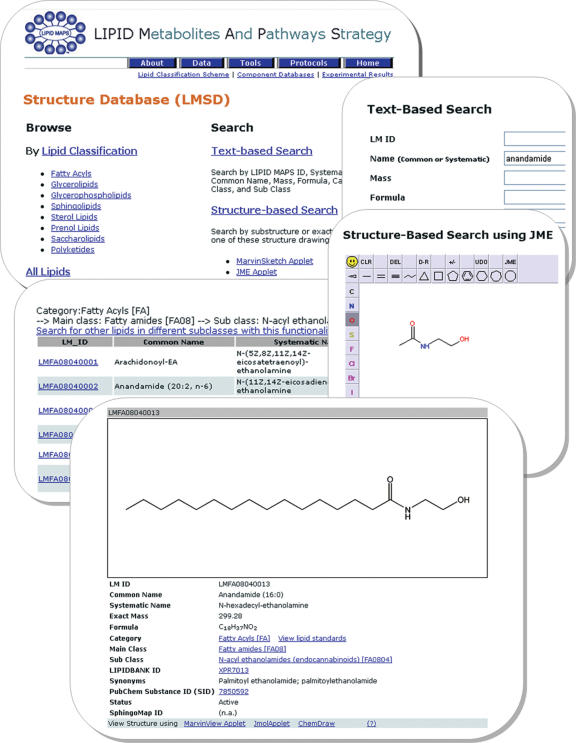
Figure 2 shows screen shots of LMSD user interface for lipid classification-based browsing, text-based and structure-based search; a screen of record details view is also provided.
Figure 2.
A montage of LMSD screen shots showing different ways to retrieve information for anandamide.
SUMMARY AND FUTURE WORK
LMSD is a database encompassing structures and annotations of biologically relevant lipids. Unlike other existing databases of lipids such as LIPID BANK and LIPIDAT, LMSD has these distinct features:
Usage of hierarchical classification and consistent nomenclature based on a comprehensive classification scheme proposed by LIPID MAPS.
A unique LM ID assignment to all structures.
No duplicate structures.
Ability to search database using structure.
LMSD is a growing database of lipids. Data about biologically relevant lipids from existing databases and appropriate scientific literature, after manual and automatic curation to generate consistent structural and ontological information, are added to LMSD database regularly. As the LIPID MAPS core laboratories identify and characterize novel lipids, these are also added to LMSD. As of November 2006, LMSD contains 9143 lipids.
In addition to maintenance and addition of new data to LMSD, future work also involves tighter integration and cross-referencing of lipids in LMSD with lipid-associated proteins and genes in LIPID MAPS proteome database (LMPD). Association of lipids in LMSD with lipid metabolic pathways is also under active consideration.
Acknowledgments
This work was supported by National Institutes of Health (NIH) and National Institute of General Medical Sciences (NIGMS) Glue Grant NIH/NIGMS Grant 1 U54 GM69338. Funding to pay the Open Access publication charged was also provided by Glue Grant NIH/NIGMS Grant 1 U54 GM69338.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cotter D., Maer A., Guda C., Saunders B., Subramaniam S. LMPD: LIPID MAPS proteome database. Nucleic Acids Res. 2006;34:D507–D510. doi: 10.1093/nar/gkj122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watanabe K., Yasugi E., Ohshima M. How to search the glycolipid data in ‘Lipidbank for web’, the newly-developed lipid database in Japan. Trends Glycosci. Glycotechnol. 2000;12:175–184. [Google Scholar]
- 3.Caffrey M., Hogan J. LIPIDAT: a database of lipid phase transition temperatures and enthalpy changes. DMPC data subset analysis. Chem. Phys. Lipids. 1992;61:1–109. doi: 10.1016/0009-3084(92)90002-7. [DOI] [PubMed] [Google Scholar]
- 4.Fahy E., Subramaniam S., Brown H.A., Glass C.K., Merrill A.H., Jr, Murphy R.C., Raetz C.R.H., Russell D.W., Seyama Y., Shaw W., et al. A comprehensive classification system for lipids. J. Lipid Res. 2005;46:839–862. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.IUPAC-IUB Commission on Biochemical Nomenclature (CBN). The nomenclature of lipids (Recommendations 1976) Eur. J. Biochem. 1977;79:11–21. [Google Scholar]
- 6.IUPAC-IUB Commission on Biochemical Nomenclature (CBN). The nomenclature of lipids (Recommendations 1976) Hoppe-Seylers Z. Physiol. Chem. 1977;358:617–631. [PubMed] [Google Scholar]
- 7.IUPAC-IUB Commission on Biochemical Nomenclature (CBN). The nomenclature of lipids (Recommendations 1976) Lipids. 1977;12:455–468. [PubMed] [Google Scholar]
- 8.IUPAC-IUB Commission on Biochemical Nomenclature (CBN). The nomenclature of lipids (Recommendations 1976) Mol. Cell. Biochem. 1977;17:157–171. [Google Scholar]
- 9.IUPAC-IUB Commission on Biochemical Nomenclature (CBN). The nomenclature of lipids (Recommendations 1976) Chem. Phys. Lipids. 1978;21:159–173. [Google Scholar]
- 10.IUPAC-IUB Commission on Biochemical Nomenclature (CBN). The nomenclature of lipids (Recommendations 1976) J. Lipid. Res. 1978;19:114–128. [PubMed] [Google Scholar]
- 11.IUPAC-IUB Commission on Biochemical Nomenclature (CBN). The nomenclature of lipids (Recommendations 1976) Biochem. J. 1978;171:21–35. doi: 10.1042/bj1710021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of glycolipids (Recommendations 1997). 2000. Adv. Carbohydr. Chem. Biochem. 2000;55:311–326. [Google Scholar]
- 13.IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of glycolipids. Carbohydr. Res. 1988;312:167–175. [Google Scholar]
- 14.IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of glycolipids. Eur. J. Biochem. 1998;257:293–298. doi: 10.1046/j.1432-1327.1998.2570293.x. [DOI] [PubMed] [Google Scholar]
- 15.IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of glycolipids. Glycoconj. J. 1999;16:1–6. [Google Scholar]
- 16.IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of glycolipids. J. Mol. Biol. 1999;286:963–970. doi: 10.1006/jmbi.1998.2485. [DOI] [PubMed] [Google Scholar]
- 17.IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of glycolipids. Pure Appl. Chem. 1997;69:2475–2487. [Google Scholar]
- 18.IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of prenols (Recommendations 1987) Eur. J. Biochem. 1987;167:181–184. [PubMed] [Google Scholar]
- 19.IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of steroids (Recommendations 1989) Eur. J. Biochem. 1989;186:429–458. [PubMed] [Google Scholar]
- 20.Merrill A.H., Jr SphinGOMAP—a web-based biosynthetic pathway map of sphingolipids and glycosphingolipids. Glycobiology. 2005;15:15G. doi: 10.1093/glycob/cwi070. [DOI] [PubMed] [Google Scholar]