Abstract
As knowledge of human genetic polymorphisms grows, so does the opportunity and challenge of identifying those polymorphisms that may impact the health or disease risk of an individual person. A critical need is to organize large-scale polymorphism analyses and to prioritize candidate non-synonymous coding SNPs (nsSNPs) that should be tested in experimental and epidemiological studies to establish their context-specific impacts on protein function. In addition, with emerging high-resolution clinical genetics testing, new polymorphisms must be analyzed in the context of all available protein feature knowledge including other known mutations and polymorphisms. To approach this, we developed PolyDoms (http://polydoms.cchmc.org/) as a database to integrate the results of multiple algorithmic procedures and functional criteria applied to the entire Entrez dbSNP dataset. In addition to predicting structural and functional impacts of all nsSNPs, filtering functions enable group-based identification of potentially harmful nsSNPs among multiple genes associated with specific diseases, anatomies, mammalian phenotypes, gene ontologies, pathways or protein domains. PolyDoms, thus, provides a means to derive a list of candidate SNPs to be evaluated in experimental or epidemiological studies for impact on protein functions and disease risk associations. PolyDoms will continue to be curated to improve its usefulness.
INTRODUCTION
Single nucleotide polymorphisms in coding regions (cSNPs) and regulatory regions have the potential to affect gene function (1–3). Non-synonymous cSNPs (nsSNPs), which change the amino acid sequence of proteins and are likely to affect the structure and function of the proteins, are good candidates for disease-modifying alleles. However, not infrequently molecular epidemiological studies have reported little or no association between cSNPs and disease susceptibility (4–6). Thus, as much as possible, it is essential to identify nsSNPs most likely to have functional effects before undertaking large-scale association studies. Established efforts to predict whether an nsSNP can affect the protein function and structure range from tools to visualize SNPs in their three-dimensional context (7,8), and predict molecular effects and potential impact of nsSNPs (4,9–13), to the recent SNPs3D (14) which integrates a variety of relevant information sources of nsSNPs [for additional details see the recent review by Mooney (15)]. Most of these approaches and analytical methods, however, are divided across various databases and interfaces, and users typically have to go through several web sites to analyze a single nsSNP. To overcome this, we have developed the PolyDoms resource to integrate most of these resources and results for each nsSNP, collating these data along with Gene Ontology, disease and other protein functional annotations in a web-accessible query interface.
DATA SOURCES
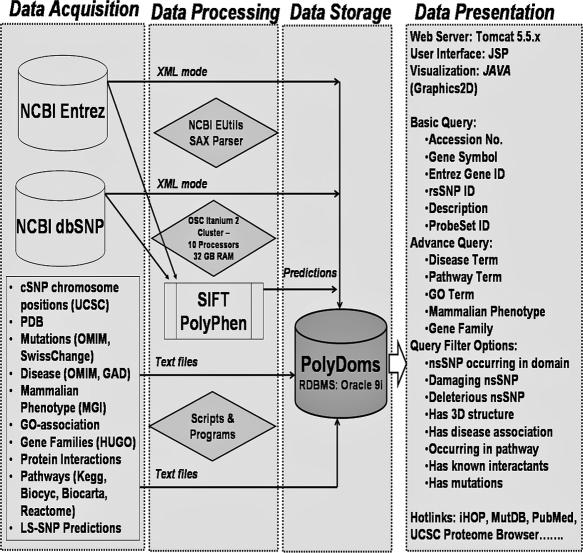
Table 1 and Figure 1 list the various types of data and their sources used for building the PolyDoms database. PolyDoms currently houses a total of 39 325 human RefSeq proteins, representing 26 378 unique RefSeq genes of which 6567 have alternate spliced products. The public repository of SNPs, NCBI's dbSNP database Build 125 (16) is our cSNP resource. We retrieved a total of 47 267 nsSNPs from dbSNP Build 125. To maximize our coverage of potential functional cSNPs, we included all the cSNPs from dbSNP without limiting to validated cSNPs alone. Another reason for this inclusion is that there are many reports of non-validated nsSNPs in the clinical literature [e.g. G1120E in the APC protein in patients with gastric cancer (17)]. The protein sequence data and all associated annotations wereextracted from NCBI's Entrez databases. Other sequence annotations and nsSNP-related information from various sources (see Figure 1) were downloaded as text files from original sources. Supplementary Data 1 summarizes the current status of PolyDoms database.
Table 1.
Data type and sources used in PolyDoms
Figure 1.
Schematic representation of PolyDoms data resources, work-flow and features.
DATA PROCESSING AND STORAGE
Data processing
The NCBI's Entrez Programming Utilities (EUtils) (http://eutils.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html) were used to download the protein (including protein domain information) and the cSNP-related data. The results fetched using EUtils XML mode were parsed using SAX parser (available as part of J2SDK 5.0). For nsSNPs in genes with more than one mRNA transcript, individual entries were recorded for each unique transcript to reflect potential differences in amino acid numbering. Individual entries were also recorded where more than one allele frequency submission was available. For example, an nsSNP with three mRNA transcripts and four different submissions resulted in a total of twelve separate entries.
JAVA programs were written to parse and normalize other downloaded text files (GO-gene associations, protein–protein interactions, OMIM/SwissChange mutations, LS-SNP predictions, mammalian phenotype gene associations) and uploaded to PolyDoms database.
Prediction of nsSNP implication
We used two sequence homology-based tools, SIFT (Sort Intolerant from Tolerant; version 2.1) (9) and PolyPhen (Polymorphism Phenotype; version 1.1) (4), to predict the potential impact of nsSNP on protein function. Additionally, when available, we have included the LS-SNP predictions (11). LS-SNP predicts positions where nsSNPs destabilize proteins, interfere with the formation of domain–domain interfaces, have an effect on protein–ligand binding or severely impact human health (11). In cases, due to data-related errors, where an amino acid residue position in the dbSNP record did not match with the amino acid residue at the same position in the corresponding protein record from RefSeq database, SIFT/PolyPhen analysis returned errors. For example, rs11557865 denotes nsSNP Ser551Pro; but the corresponding protein sequence (NP_061872; KIAA1128) has aspartic acid at position 551. Similarly, rs10891338 represents nsSNP Pro208Leu whereas the corresponding protein, BCDO2 (NP_114144), has lysine at position 208.
SIFT uses sequence homology among related genes and domains across species to predict the impact of all 20 possible amino acids at a given position, allowing users to determine which nsSNPs would be of most interest to study. The SIFT algorithm has been shown to predict a phenotype for an nsSNP more accurately than previously used substitution scoring matrices, such as BLOSUM62, as these matrices do not incorporate information specific to the protein of interest (18,19). Another advantage of using SIFT is the potential to analyze a larger number of nsSNPs than methods that are dependent on the availability of protein structure alone (19,20). The PolyPhen algorithm, such as SIFT, takes an evolutionary approach in distinguishing deleterious nsSNPs from functionally neutral ones. However, it also takes into account the data from protein structure databases, such as PDB (Protein Data Bank) and PQS (Protein Quarternary Structure), DSSP (Dictionary of Secondary Structure in Proteins), and three-dimensional structure databases to determine if a variant may have an effect on the secondary structure of the protein, interchain contacts, functional sites and binding sites (4).
SIFT and PolyPhen analyses were performed on Ohio Supercomputer Center's (OSC) Itanium 2 Cluster (http://www.osc.edu/hpc/computing/it2/), configured in shared memory parallel running mode with a maximum of 10 processors and 32 GB RAM. Under this configuration, ∼50 SIFT or ∼600 PolyPhen jobs can be processed in an hour. The LS-SNP predictions were downloaded from the original source, parsed and uploaded to PolyDoms database.
Data storage
The PolyDoms database is implemented in Oracle 9i. The central table is ‘Gene’ that has an up-to-date list of all human RefSeq genes. The Gene table is linked to several other master tables. The cSNP table, apart from annotations, contains the SIFT and PolyPhen predictions. Other tables linked to the Gene table are as follows: the Transcript table (RefSeq mRNAs); the Protein table (RefSeq proteins); the ProbeSets; the Mutation table (OMIM and SwissChange) the Disease tables (OMIM and GAD); Mammalian Phenotype; Pathway (KEGG, Biocarta, BioCyc and Reactome), Protein–protein interactions (BIND, HPRD and Reactome); and Protein function (GO).
ACCESS AND INTERFACE
The main access to PolyDoms is through its web interface at http://polydoms.cchmc.org, by querying with sequence accession numbers, gene symbols, Entrez Gene IDs, rsSNP IDs, description or probeset IDs (Illumina; Affymetrix). Additionally, it is possible to retrieve a list of genes and associated cSNPs using a GO term, disease term (OMIM or GAD), pathway term (KEGG, Biocarta BioCyc or Reactome), mammalian phenotype or gene family (Figure 1). The output of a search presents the user with an option to view synonymous SNPs or nsSNPs. cSNPs are represented graphically in the context of protein sequence and domains (Figure 2). The results of the SIFT and PolyPhen predictions along with the LS-SNP extracted predictions for all nsSNPs of a protein are provided as a table below the image. Where available the mutant allele information from OMIM and SwissChange, and the protein–protein interactions are also provided. Up-to-date literature references implicating polymorphisms in disease are also provided. An expandable list provides links to various GO terms, pathways, diseases and phenotypes associated with the queried protein. Apart from these, the resource page is supplemented with cross-references to PDB, iHOP, MutDB and the UCSC Proteome Browser. All cross-references to data sources are hyperlinked enabling the original data to be viewed.
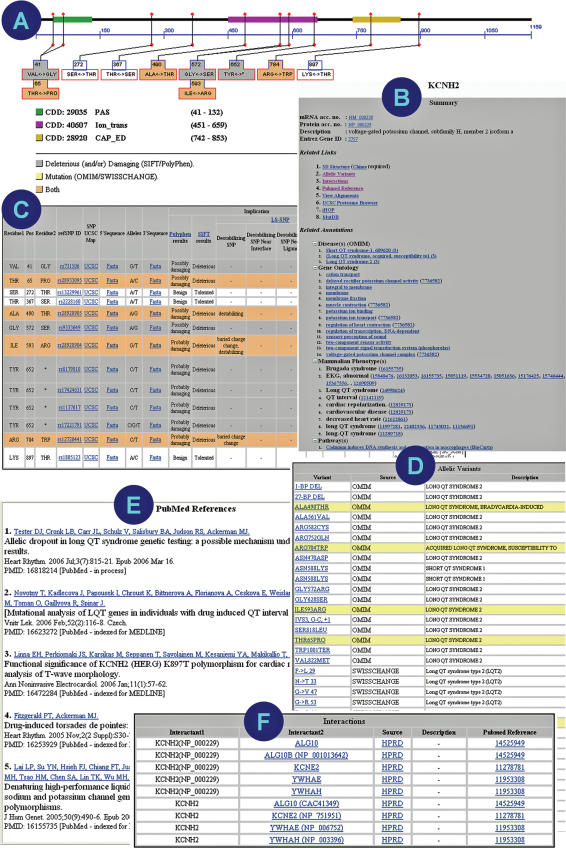
Figure 2.
PolyDoms feature displays. (A) PolyDoms image of a nsSNP model of the protein KCNH2. Numbers in the image indicate the amino acid residue positions from the corresponding RefSeq protein sequence. The pink, yellow and green blocks over the protein sequence represent the three known domains derived from the NCBI's CDD. Vertical lines represent nsSNPs. The color codes indicate the predictions—gray represents an nsSNP predicted as deleterious and/or damaging; yellow indicates mutation (based on OMIM/SwissChange); orange indicates an nsSNP that has been predicted as deleterious and/or damaging and also reported as a mutation. (B) The summary view gives the basic sequence annotations along with an expandable list of diseases, GO terms, mammalian phenotypes and Pathways associated with the queried gene. (C) Tabular description of nsSNP predictions based on PolyPhen and SIFT analysis and LS-SNP annotations (refer to B above for descriptions of color codes). (D) Tabular list of allelic variants derived from OMIM and SwissChange. (E) Top five relevant abstracts, when available, related to queried gene polymorphisms and disease association. The list is generated dynamically and therefore is up-to-date with current literature. (F) List of protein–protein interactions (from NCBI Entrez Gene).
UTILITY
We present the utility and various features of PolyDoms through one case study using mammalian phenotype as an example. Since knowledge of complex diseases is limited, a comprehensive list of candidate genes and a method of ranking those genes by their disease-relevance is important in designing a good association study (14). Using NCBI's OMIM (21) and NIA's GAD database (22) and the mammalian phenotype (23), we provide a query interface through which a user can select any disease/phenotype term and create an nsSNP list based on the candidate genes associated with that particular disease/phenotype term. Although, we can assume the complexity of phenotype based on the number of genes associated with it, it may also partly reflect the current state of knowledge for that particular phenotype. Additional examples illustrating the utility and contents of database can be accessed through various case studies from Supplementary Data 8.
Case study: using the mammalian phenotype to investigate SNP–phenotype relationships
Aim: To obtain a list of human orthologous genes based on mouse genes associated with the phenotype ‘abnormal podocytes’.
From the homepage, click on the ‘Phenotype Selector’ (under section ‘Search by disease, gene ontology, pathway, or gene family’).
A new window (‘Search for Mammalian Phenotype’) opens up. Enter the search term ‘abnormal podocytes’ (or ‘podocyte’) and hit ‘Search’. Select the term ‘abnormal podocytes’ from the search results window and hit ‘use this phenotype for search’ button to populate the ‘Phenotype already selected’ window. Click ‘Done’ to return to the PolyDoms query page.
Hitting the ‘Search’ button without selecting any of the ‘Filter Options’ will return the human orthologous genes (37 proteins, 19 unique in the current version) of mouse genes associated with the phenotype ‘abnormal podocytes’. At this stage, users can either download the results as a spreadsheet by clicking on the link ‘Download the results’ or proceed to view the non-synonymous or synonymous model of each of the protein (see Figure 2 for a description of the output). Selecting the download option presents the user with a list of fields to select from and add to the spreadsheet.
Alternatively, use the ‘Filter options’ to refine the query. For example, from the ‘Filter options’ select ‘Occurring in domain’, ‘Deleterious nsSNP’ and ‘Damaging nsSNP’ and hit ‘Search’. This will return 4 proteins (ARHGDIA, LAMA5, NCK1 and NPHS2), each of which has at least one nsSNP that occurs in a conserved domain and has been predicted as ‘Deleterious/Damaging’ by SIFT/PolyPhen.
The Supplementary Data 2 lists all the mammalian phenotypes along with the associated genes and the number of deleterious and damaging nsSNPs.
Prioritizing candidate nsSNPs
We screened a total of 44 641 (94%) nsSNPs associated with 14 967 protein sequences using SIFT and PolyPhen. Of these, 14 819 (33%) were predicted as ‘deleterious’ by SIFT and 14 622 (33%) as ‘damaging’ by PolyPhen. About 9021 nsSNPs (representing 5436 unique genes) were predicted as both deleterious and damaging indicating a concordance of ∼62% between SIFT and PolyPhen predictions (see Supplementary Data 1 for additional details). Three studies (24–26) thus far have combined both the SIFT and PolyPhen algorithms to screen for deleterious nsSNPs. Xi et al. (26) and Johnson et al. (24) reported a concordance of 62 and 73% between these two programs analyzing the nsSNPs of genes involved in DNA repair and steroid hormone metabolism, respectively. In an earlier analysis of nsSNPs involved in DNA repair, cell cycle regulation, apoptosis and drug metabolism we used both SIFT and PolyPhen and identified 57 potentially deleterious nsSNPs (25). The Supplementary Data 3 lists all the nsSNPs that have been predicted as deleterious and damaging by both SIFT and PolyPhen. The Supplementary Data 4 and 5 list the disease/phenotype-associated genes that have at least one nsSNP predicted as damaging and deleterious by both PolyPhen and SIFT, respectively.
Although useful and widely used, both SIFT and PolyPhen have certain limitations. First, both of these require homologous sequences. Second, both of these algorithms disregard the impacts of a combination of variants (24,27). Third, SIFT and PolyPhen predict the impact of cSNPs only whereas non-coding SNPs (SNPs occurring in promoter or enhancer regions or splicing junctions) can also affect protein levels or protein function (24).
cSNPs resulting in premature stop codons and protein truncation
cSNPs introducing premature termination codons (nonsense SNPs) can alter the stability and function of transcripts and proteins and thus are considered to be biologically important. We retrieved a total of 965 nonsense SNPs (from 830 genes) from dbSNP Build 125 and 416 out of 965 nonsense SNPs affect an amino acid residue that is part of a functional protein domain. This led us to hypothesize that these cSNPs are likely to affect gene/protein function, although their biological relevance needs to be further investigated. However, we have noticed that some of the nonsense SNPs in dbSNP build 125 are either changed or removed from the dbSNP build 126. For instance, in the dbSNP build the number of nonsense SNPs affecting an amino acid residue which is part of a functional domain is 367. These changes will be reflected in our database when it is updated. Supplementary Data 6 lists all the cSNPs (based on dbSNP build 125) resulting in premature stop codons and also includes a comparison with the current dbSNP build 126.
KNOWN MUTATIONS VERSUS nsSNP FUNCTIONAL PREDICTION
To assess the potential for functional consequences of the PolyDoms defined intolerant nsSNPs, we downloaded 1338 SNPs from 611 candidate genes with known disease mutations (ftp://ftp.ncbi.nih.gov/snp/Entrez/snp_omimvar.txt) and subjected them to SIFT and PolyPhen analysis. Of the 1008 nsSNPs analyzed (330 out of 1338 nsSNPs were ignored because some of them were either non-coding SNPs or had erroneous annotations with mismatch of the residues), 568 (56%) nsSNPs showed concordance between SIFT and PolyPhen predictions and were classified both as ‘deleterious’ and ‘damaging’ (Supplementary Data 7). A total of 782 out of 1008 (78%) nsSNPs were either predicted as deleterious or damaging or both. Apart from confirming the utility of these prediction tools in prioritizing the candidate nsSNPs, it also suggests that nsSNPs predicted as damaging and deleterious and already associated with a phenotype/disease (Supplementary Data 4 and 5) represent a pool of candidate loci that should be interrogated further in association studies. We also noticed that only 34 out of the 568 nsSNPs predicted as damaging and deleterious are validated (by frequency) nsSNPs.
RELATED WORK
Although it is beyond the scope of the current article to compare PolyDoms with other resources of similar nature (see Introduction), some of the features that are unique to PolyDoms are related to the management of sets of nsSNPs—the ability to refine, export nsSNP sets as a whole and to create sets of cSNPs through complex queries (such as using pathways or Gene Ontology or mammalian phenotype classes described earlier and in the Supplementary Data 8). The goals of the recently published SNPs3D (14) are similar to ours: to integrate all of the available data relevant for assessing the likely role of particular genes and nsSNPs in a disease and help the researchers in making informed judgments. Additionally, the PolyDoms filter options make the data-mining process and compiling a ‘hit-list’ of nsSNPs relatively easy.
CONCLUSION
We have classified and catalogued the predicted functionality of nsSNPs in human genes to facilitate sequence-based association studies. The current version of PolyDoms however has some limitations. First, the current version of PolyDoms does not contain information on SNP co-occurences, complex haplotype or other relationships among the SNPs. Therefore, one of our future goals is to incorporate the SNP haplotype data (28). This will facilitate retrieving genotype and frequency data, picking tag-SNPs for use in association studies, viewing haplotypes graphically and examining marker-to-marker LD patterns. Second, since PolyDoms is built using multiple sources, keeping it up-to-date and synchronized with external resources, taking into account the different data formats, or the changes in their formats is tedious. However, we will strive to automate this process as much as possible. Third, PolyDoms does not still provide the complete range of analysis tools that can be useful in evaluating and characterizing cSNPs in terms of their potential effects (e.g. relative solvent accessibility of the variant residue). We are in the process of filling this gap using the SABLE server (29). Finally, PolyDoms does not include information about other SNPs (human non-coding SNPs or SNPs from other species). In conclusion, the use of PolyDoms and other resources similar to select functional nsSNPs for epidemiology studies can be an efficient way to explore the role of genetic variation in disease risk or altered response to therapeutic regimens, and to contain cost. However, it should be noted that deleterious effects on protein stability alone may not be sufficient conditions for disease predisposition.
AVAILABILITY
The PolyDoms database can be accessed freely at http://polydoms.cchmc.org.
SUPPLEMENTARY DATA
Supplementary Data are available at http://polydoms.cchmc.org/polydoms/supplementary/
DISCLAIMER
The purpose of this resource is to distribute functional annotations of human cSNP data. These cSNPs and their annotations are meant to be used as guidelines for basic research. Do not use these results to make clinical decisions.
Acknowledgments
The authors would like to thank Drs Deb Nickerson, Robert Livingston and Robert Weiss for super discussions and the Ohio Supercomputer Center for the assistance in using their supercomputing clusters to run whole genome SIFT and PolyPhen analyses. This work was supported by grants NCI UO1 CA84291-07 (Mouse Models of Human Cancer Consortium), NIH R24 DK 064403 (Digestive Diseases Research Development Center—DDRDC), NIEHS ES-00-005 (Comparative Mouse Genome Centers Consortium) and NIEHS P30-ES06096 (Center for Environmental Genetics). Funding to pay the Open Access publication charges for this article was provided by CCHMC, Cincinnati, OH, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chakravarti A. It's raining SNPs, hallelujah? Nature Genet. 1998;19:216–217. doi: 10.1038/885. [DOI] [PubMed] [Google Scholar]
- 2.Collins F.S., Guyer M.S., Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 3.Syvanen A.C., Landegren U., Isaksson A., Gyllensten U., Brookes A. First International SNP Meeting at Skokloster, Sweden, August 1998. Enthusiasm mixed with scepticism about single-nucleotide polymorphism markers for dissecting complex disorders. Eur. J. Hum. Genet. 1999;7:98–101. doi: 10.1038/sj.ejhg.5200291. [DOI] [PubMed] [Google Scholar]
- 4.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savas S., Kim D.Y., Ahmad M.F., Shariff M., Ozcelik H. Identifying functional genetic variants in DNA repair pathway using protein conservation analysis. Cancer Epidemiol. Biomarkers Prev. 2004;13:801–807. [PubMed] [Google Scholar]
- 6.Zhu Y., Spitz M.R., Amos C.I., Lin J., Schabath M.B., Wu X. An evolutionary perspective on single-nucleotide polymorphism screening in molecular cancer epidemiology. Cancer Res. 2004;64:2251–2257. doi: 10.1158/0008-5472.can-03-2800. [DOI] [PubMed] [Google Scholar]
- 7.Mooney S.D., Altman R.B. MutDB: annotating human variation with functionally relevant data. Bioinformatics. 2003;19:1858–1860. doi: 10.1093/bioinformatics/btg241. [DOI] [PubMed] [Google Scholar]
- 8.Stitziel N.O., Binkowski T.A., Tseng Y.Y., Kasif S., Liang J. topoSNP: a topographic database of non-synonymous single nucleotide polymorphisms with and without known disease association. Nucleic Acids Res. 2004;32:D520–D522. doi: 10.1093/nar/gkh104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng P.C., Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reumers J., Maurer-Stroh S., Schymkowitz J., Rousseau F. SNPeffect v2.0: a new step in investigating the molecular phenotypic effects of human non synonymous SNPs. Bioinformatics. 2006;22:2183–2185. doi: 10.1093/bioinformatics/btl348. [DOI] [PubMed] [Google Scholar]
- 11.Karchin R., Diekhans M., Kelly L., Thomas D.J., Pieper U., Eswar N., Haussler D., Sali A. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 12.Ferrer-Costa C., Gelpi J.L., Zamakola L., Parraga I., de la Cruz X., Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 13.Stone E.A., Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15:978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yue P., Melamud E., Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mooney S. Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Brief Bioinformatics. 2005;6:44–56. doi: 10.1093/bib/6.1.44. [DOI] [PubMed] [Google Scholar]
- 16.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horii A., Nakatsuru S., Miyoshi Y., Ichii S., Nagase H., Kato Y., Yanagisawa A., Nakamura Y. The APC gene, responsible for familial adenomatous polyposis, is mutated in human gastric cancer. Cancer Res. 1992;52:3231–3233. [PubMed] [Google Scholar]
- 18.Henikoff S., Henikoff J.G. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 1991;19:6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z., Moult J. SNPs, protein structure, and disease. Hum. Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- 20.Sunyaev S., Ramensky V., Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 21.Hamosh A., Scott A.F., Amberger J., Bocchini C., Valle D., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker K.G., Barnes K.C., Bright T.J., Wang S.A. The genetic association database. Nature Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 23.Smith C.L., Goldsmith C.A., Eppig J.T. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson M.M., Houck J., Chen C. Screening for deleterious nonsynonymous single-nucleotide polymorphisms in genes involved in steroid hormone metabolism and response. Cancer Epidemiol. Biomarkers Prev. 2005;14:1326–1329. doi: 10.1158/1055-9965.EPI-04-0815. [DOI] [PubMed] [Google Scholar]
- 25.Livingston R.J., von Niederhausern A., Jegga A.G., Crawford D.C., Carlson C.S., Rieder M.J., Gowrisankar S., Aronow B.J., Weiss R.B., Nickerson D.A. Pattern of sequence variation across 213 environmental response genes. Genome Res. 2004;14:1821–1831. doi: 10.1101/gr.2730004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xi T., Jones I.M., Mohrenweiser H.W. Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics. 2004;83:970–979. doi: 10.1016/j.ygeno.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 27.Rebbeck T.R., Spitz M., Wu X. Assessing the function of genetic variants in candidate gene association studies. Nature Rev. Genet. 2004;5:589–597. doi: 10.1038/nrg1403. [DOI] [PubMed] [Google Scholar]
- 28.Thorisson G.A., Smith A.V., Krishnan L., Stein L.D. The International HapMap Project Web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adamczak R., Porollo A., Meller J. Accurate prediction of solvent accessibility using neural networks-based regression. Proteins. 2004;56:753–767. doi: 10.1002/prot.20176. [DOI] [PubMed] [Google Scholar]
- 30.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchler-Bauer A., Anderson J.B., Cherukuri P.F., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M. The KEGG database. Novartis Found. Symp. 2002;247:91–101. discussion 101–103, 119–128, 244–152. [PubMed] [Google Scholar]
- 34.Karp P.D., Ouzounis C.A., Moore-Kochlacs C., Goldovsky L., Kaipa P., Ahren D., Tsoka S., Darzentas N., Kunin V., Lopez-Bigas N. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann R., Valencia A. A gene network for navigating the literature. Nature Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- 37.Hsu F., Pringle T.H., Kuhn R.M., Karolchik D., Diekhans M., Haussler D., Kent W.J. The UCSC Proteome Browser. Nucleic Acids Res. 2005;33:D454–D458. doi: 10.1093/nar/gki100. [DOI] [PMC free article] [PubMed] [Google Scholar]